Unit 2 - Protein Sorting
1/53
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
54 Terms
Nucleus
storage of DNA; RNA transcription happens here transcription factor binding and transcription and splicing); DNA replication; structure - nuclear envelope, perniculear space, and nuclear pore
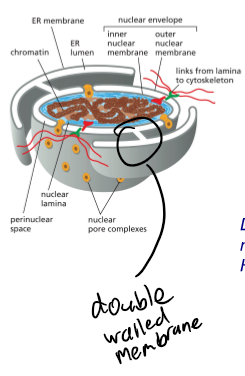
Nuclear Envelope
double membrane enclosing the nucleus; each one is a lipid bilayer
Perinuclear space
contigous with ER lumen; white space between the lipid bilayers
Nuclear Pore
opening across nuclear envelope; can move 1,000 marcomolcules/sec and in both directions at the same time; small molecules will just diffuse across (<5,000 Da - ATP, ions, Ca, nucleotides); large molecules require karyopherins (>5,000 Daltons)
Nuclear Pore Complex (NPC)
large complex (30 diff. proteins); 8-fold symmetry; huge (internal dia. of 40 nm); fully folded and processed proteins move across this (w/ posttranslational modifications and protein complexes → move them as a singular unit)
Nuclear Transport
phenylalanine-glycine (FG) repeats form a mesh → keropherins interact w/ FG repeats and move across along with any proteins bound to it (normal proteins can’t cross nuclear envelope)
Karyopherins
import and export receptors; bind cargo proteins; shuttle between nucleus and cytoplasm (diffused across the nuclear pore); superfamily consisting of import and export receptors (each carries out only 1 type though so either in or out)
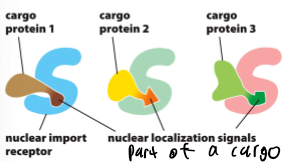
Nuclear Localization Signals
NLS; part of a cargo protein that interacts with the nuclear import receptor
Mitochondria/Chloroplasts
Krebs cycle; oxidative phosphorylation
Endoplasmic Reticulum
ER; protein synthesis; connected to nucleus; Ca storage; site of synthesis for all proteins (secreted, inside ER, golgi, endosomes, or lyosomes, reside in membrane of above, and once in ER, move to other sites by vesicle; site of covalent protein modifications (glycosylation - secreted proteins always have this, lipid mods., and disulfide-bond formation); rough ER (RER) - has ribosomes
Golgi Complex
protein maturation and sorting; acts as cell’s post office; pancake looking thing
Endosome and Lysosome
protein sorting and processing and degradation of macromolecules; material degradation and imports
Cytoplasm
‘non-nuclear’ liquid; contains organelles
Cytosol
‘non-nuclear’ liquid w/o any organelles
Organelle
membrane-enclosed compartment inside euk cell; can create physically isolated environments; have control of what goes in and out
What are the 3 ?’s to study gene function?
find it - what and where is it (identify it)
lose it - what happens when it is gone? Is it necessary for normal function?
move it - what happens when it is active at the wrong time and place? (what happens when adding it to a part that doesn’t have it?
Signal Sequence
conserved aa sequence that controls subcellular localization of final protein; short stretches of aa; direct protein to a particular location; short stretches of amino acids that direct proteins to a specific subceullar location; ex, nuclear import and export, and secretion from the cell; some proteins may have multiple (including w/ same function)
Lose it
KO import sequence (will protein still go to the nucleus w/o it?); signal is necessary for import of a nuclear protein (test if a specific sequence is necessary for import)
Move it
take protein not normally in the nucleus and add the import sequence to it and see if it enters the nucleus; signal is sufficient for nuclear import of an otherwise cytosolic protein (add NLS and see is it changed the localization of target protein)
Nuclear Import
have to do both lose it and move it to see if sequence is neccessary and sufficient of importing the protein into the nucleus
NLS
nuclear localization signal
Nuclear Export
lose it is necessary for export of a protein that is normally exported and move it is sufficient for export of a normally nuclear protein
NES
nuclear export signal; phosphorylation can turn both this and NIS on and off so can control export vs import being in precidence
Ran GTPase
small G-protein; has a distribution across nuclear envelope/pore; GEF is limited to inside the nucleus and GAP is limited to the cytosol
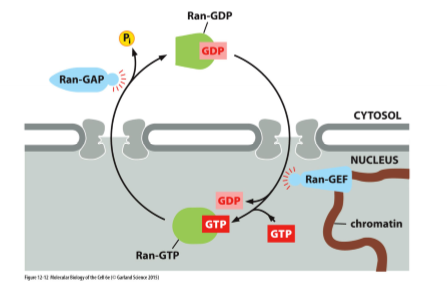
G-Proteins (GTPases)
non-covalent bonding of GDP/GTP (not a post-translational modification); enzyme controlled (GTP hydrolysis by GAP and GTP exchange by GEF); gradient if Ran-GTP/Ran-GDP across envelope
GTP Hydrolysis
by GTPases activating protein (GAP); enzymatic catalysis by G-protein activation by GAP; tells G-protein to activate hydrolysis
GTP Exchange
guanine-nucleotide exchange factor (GEF); remove GDP, fill with cytosolic GTP; kick out GDP and then GTP binds because there is more of that in the cell
Ran Conformational Change
happens when Ran-GTP → Ran-GDP; Ran-GAP protein in cytosol while Ran-GEF in nucleus; creates binary system (Ran-GTP in nucleus and Ran-GDP in cytosol); Ran shuttles itself between cytosol and nucleus; red change to blue in pic
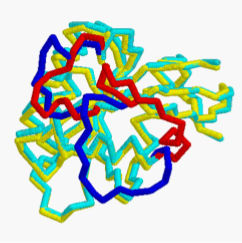
Ran Controls Cargo Binding and Release
Ran-GTP conformation different from Ran-GDP; import receptor binds either Ran-GTP OR cargo (mutually exclusive)
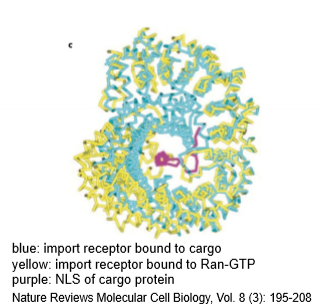
Import Receptor Structure
2 states - bound to cargo NLS or to Ran-GTP; mutually exclusive states because the conformational change for Ran-GTP occupies same space as the cargo protein; Ran-GTP binding disrupts NLS on a cargo protein binding; forms central pocket where NLS binds (binding pocket)
RECAP
Ran-GTP in nucleus; Ran-GDP in cytoplasm; karyopherins either import or export receptors and shuttle, w/ or w/o cargo; import receptors can bind either cargo NLS or Ran-GTP, but not both (mutually exclusive)
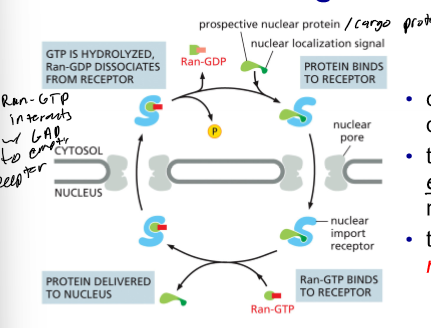
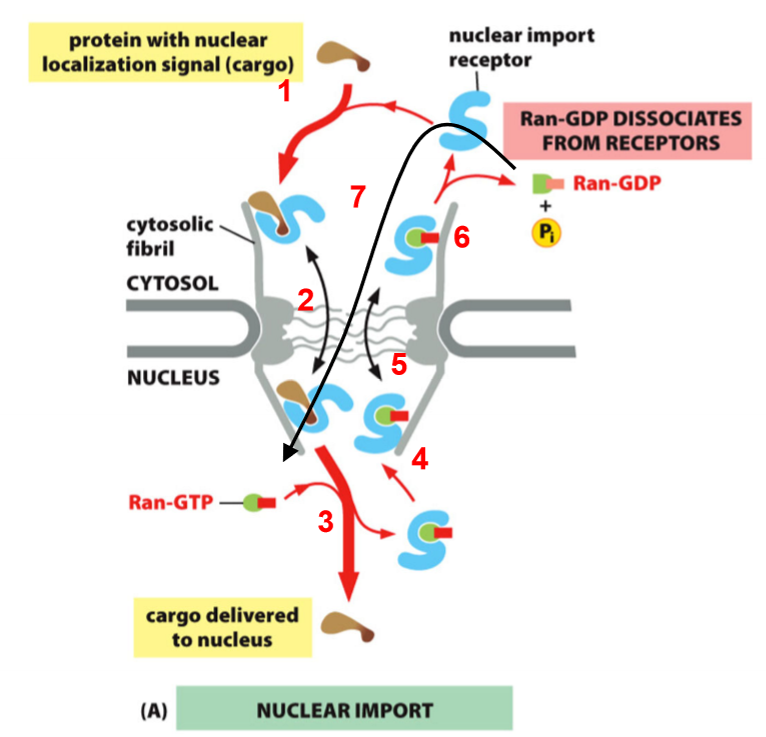
Ran Controls Nuclear Import
import receptors bind cargo in cytosol
some move into nucleus
Ran-GTP dissociates complex (import receptor and cargo come apart)
Ran-GTP binds to import receptor
Import receptor w/ Ran-GTP goes into cytosol
Hydrolysis to Ran-GDP (complex comes apart and recycles import receptor)
Ran goes back into nucleus on its own
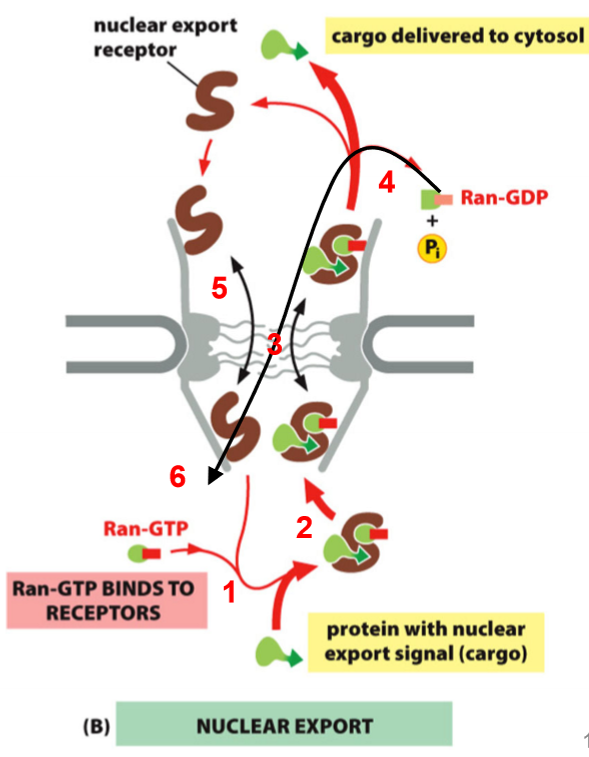
Ran Controls Nuclear Export
in nucleus, Ran-GTP binds to export receptor
complex then binds cargo
some moves to cytosol (out of the nucleus)
Hydrolysis to Ran-GDP dissociates complex
Export receptor and Ran-GDP return to nucleus
Ran-GEF returns Ran-GDP to Ran-GTP
*Export receptor MUST bind to Ran-GTP before cargo can bind
Regulated Nuclear Import
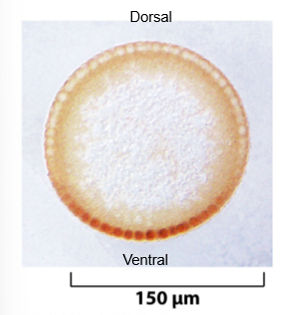
dorsal protein in fruit flies example: transcription factors must be in the nucleus to be active, determines ventral (belly) side of embroyo, generally cytosolic in embryo but moves into nucleus on ventral side, initiates transcription of genes necessary for ventral development (when in the nucleus); dorsal - protein that controls cell identity (active when in nucleus so only on ventral side of the picture)
mammalian T cells example: adaptive immune response use B and T cells, B cells make secreted antibodies and T cells activate cellular response (generally at rest unless activated by binding antogen); have both a nuclear import and export factor
Mammalian T Cells and NF-AT (Regulated Nuclear Import)
NF-AT; regulation - phosphorylation controls which nuclear localization signal is active; if NF-AT cytosolic = TF inactive and if it’s nuclear = TF active
NF-AT
Nuclear Factor in Activated T Cells; controls response of T cells to infections; transcription factor (TF) regulates expression of over 4,000 genes and is important to be turned on when there is an infection and off when there is not one; has both import and export sequences
Summary Nuclear Transport
NPCs allow import/export of biomolecules in/out of the nucleus; small molecules diffuse but large ones have to be actively transported; karyopherins bind nuclear import/export sequences and they shuttle; Ran-GTPase controls association/dissociation between karyopherins and cargo (localization - Ran-GTP is nuclear and Ran-GDP is cytoplasmic); directed transport by equilibria and localized dissociation; transported proteins are fully translated and folded
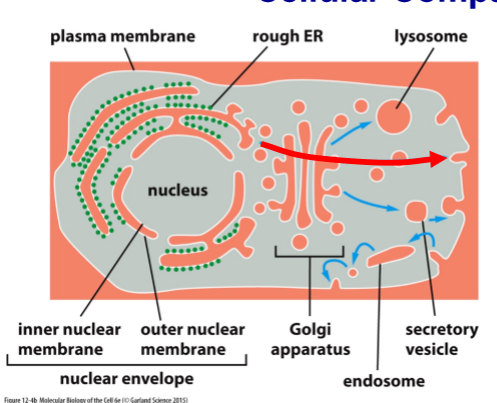
Relationship between Cellular Compartments
3 liquid spaces - cytosol (gray), nucleoplasm (gray), and everything else (orange (connected and have free exchange w/ the extracellular space) - perinuclear space, ER, golgi, endosomes and lysosomes, and extracellular space); transport between orange spaces by vesicles (move liquid from one space to another); exchange of materials fro the membranes too
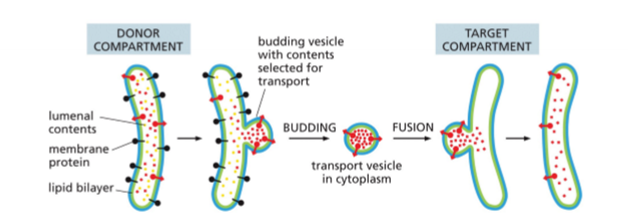
Moving Material from ER to other Organelles
material in lumen of ER will be in lumen of any vesicle and any organelle and in the extracellular space; membrane material moves between ER and plasmamembrane
RER vs SER
RER - ribosomes; synthesizes secreted proteins, ones in organelles, and transmembrane ones; usually major portion of ER
SER - no ribosomes; synthesis/metabolism of lipids, drug detox, and steriod/sugar metabolism; minor portion of ER; budding of transport vechicles; sarcoplasmic reticulum
Sarcoplasmic Reticulum
special SER; Ca2+ storage site in muscle
ER Signal Sequence Detection
SRP binds emerging signal sequence which pauses translation; steps: SRP binds emerging SS → SRP pauses translation → complex binds SRP receptor and inserts SS into translocator (Sec61) → SRP leaves ribosomal complex → translation resumes and pushes peptide through the membrane; folding and stuff happens in ER (after getting pushed through); SS doesn’t go into ER (gets degraded)
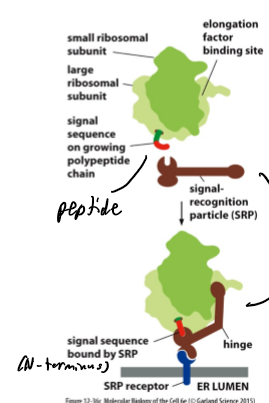
Signal Sequence
SS for short; 15-25 hydrophobic amino acids; KNOW HOW TO FIND IT, LOSE IT, AND MOVE IT
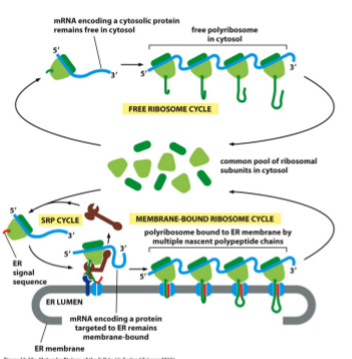
Ribosomal Clustering
cytoplasmic and ER-bound mRNAs bound by multiple ribosomes at the same time; once RNA tethered to ER, will remain attached even when the first ribosome finishes translation
Sec61
translocator; allows peptides to be pushed across membrane into lumen and into the membrane itself; will be closed with a protein plug when not in use; can open just straight through membrane and can open up laterally so that the ss can slide into the membrane
Co-Translational Transport
into ER lumen; Sec61 opens into and across membrane; chaperone proteins help; N-terminal SS chopped off and degraded (generate a new one though); soluble protein now in lumen of ER
Transmembrane Proteins Making
N-terminal SS and transmembrane domain are structually the same just the N-terminal is on the very end and the TD or TM is internal (localization is different); peptide still pushed through Sec61 and TM slides into membrane at next TD; remaining SS degraded
Multipass Transmembrane Protein Making
same steps as a transmembrane protein but just keep repeating; pushing peptide into ER → sliding into membrane → pushing peptide into cytosol → sliding into membrane; has multiple hydrophobic aa stretches and when next one comes along, it pushes the one in the lateral gate into membrane of ER
2 Modes of Import into ER
co (emerging peptide pushed through Sec61 as it comes out of ribosome) and translational import (chaperone proteins bind the peptide backbone and prevent folding, keep capable import, no N-terminus SS, recognizes internal TD)
Determining Orientation of First TD for Post-Translational Import
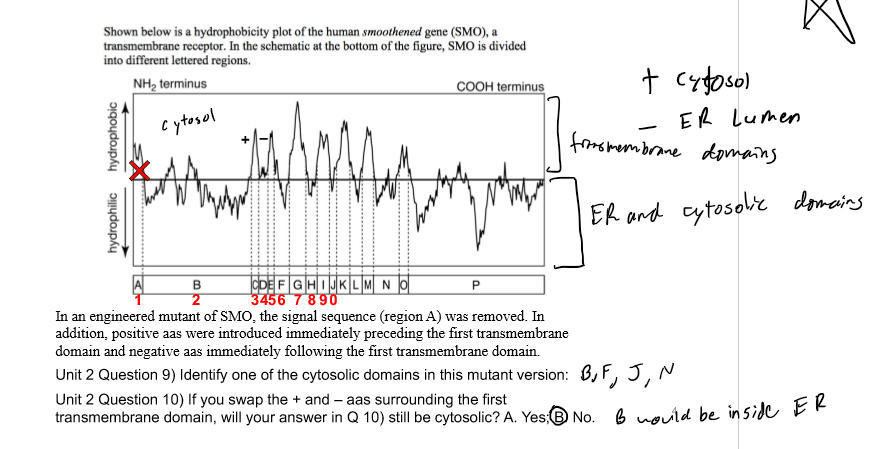
first TD surrounded by charged (T/-) aas which control how a domain will insert; negative prefers inside ER lumen; + = cytosol and - = ER; if have an N-terminal SS will push rest into ER lumen and translate until get to next TD (can have both N and C in the lumen)
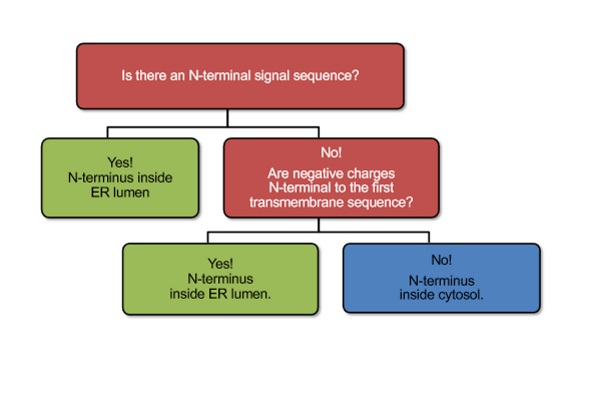
Summary
most proteins trasnported into ER are translocated co-translationally; stretches of 15-25 hydrophobic aas function as TD; N-ternimal TD domain is a SS and recognized by SRP complex (co-translational import); SRP causes ribosome to pause and SS to insert into Sec61; SS removed by signal peptidase; additional TDs make single and multipass transmembrane proteins; once direction of the first TD is determined, direction of all following TDs is fixed; if no N-terminal SS is present, charge distribution around the first TD domain sets orientation; polypeptides fold inside ER
Orientation Matters
cell needs to know because it determines ion flow (into/out of ER/SR); gradients can not be established if all pumps in same direction; first transfer sequence (- ER and + cytosol) determines orientation of protein but only if no N-terminal SS
Hydrophobicity Plot 1 Pic
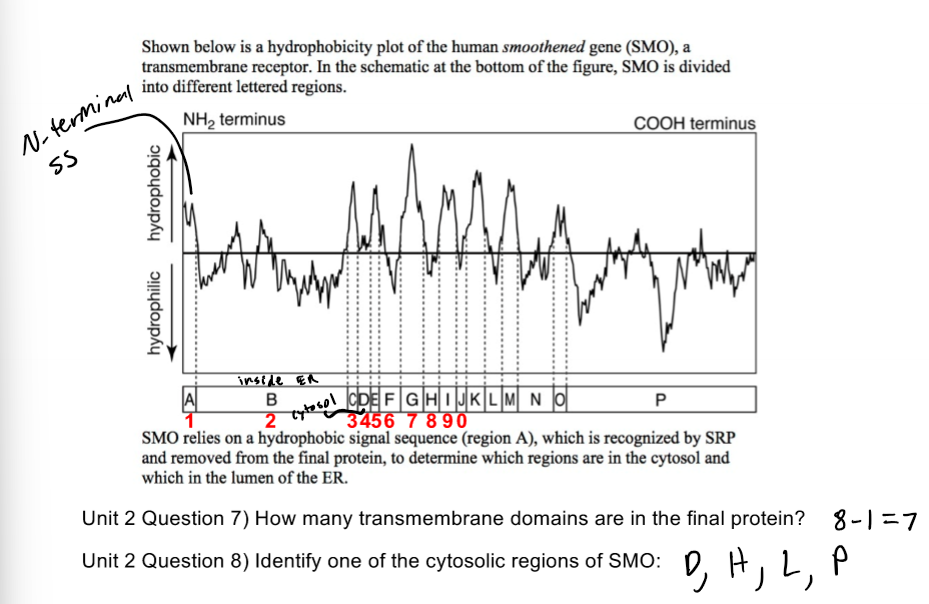
Hydrophobicity Plot 2 Pic