pt 2 genome and genomics review
1/29
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
30 Terms
transposase
what do autonomous and nonautonomous elements both require for transposition?
autonomous
which elements encode transposase for their own movement?
they do not encode transposase, but can “borrow” transposase encoded by autonomous elements from their family (using a transacting enzyme)
how do nonautonomous elements get transposase?
yes, by mutation
can autonomous elements can turn into nonautonomous elements?
Class 1: Retrotransposons – transpose via an RNA intermediate
Class 2: DNA Transposons – transpose via a DNA intermediate
what are the two types of transposable elements in the human genome?
LTR and non-LTR
what are the two types of retrotransposons?
introns: they are less likely to cause a deleterious mutation
do TEs primarily insert into introns or exons?
differences in genome size
TEs in plants are responsible for . . .
no
Is the distribution of TEs in eukaryotic genomes random?
it considers an organism’s full complement of hereditary material, rather than one gene or one gene product at a time
how does genomics differ from classical genetics?
Forward genetics
what does this describe: Study the genetic basis of a phenotype or trait;
Genetic mapping
Map based cloning
RNA seq
reverse genetics
what does this describe: Study what phenotypes arise as a result of particular genes;
RNAi
CRISPR/Cas genome editing
Transposon mutagenesis
bioinformatics
a subdiscipline of biology and computer science concerned
with the analysis of the information content of entire genomes.
comparative genomics
Considers the genomes of closely and distantly related species for evolutionary insight
functional genomics
The use of expanding variety of methods including reverse genetics, to understand gene and protein function in biological processes
paralog
Genes that are related by gene duplication within a single genome
ex) Hemoglobin and Myoglobin — both came from an ancestral oxygen-binding gene that duplicated and evolved into different roles.
ortholog
Genes in different species that evolved from a common ancestorial gene by speciation
homolog
Genes that share a common ancestor. — can be paralogs or ortholog
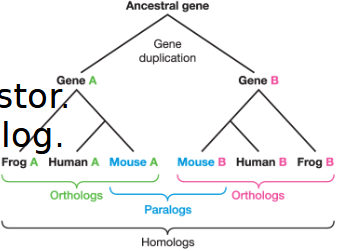
yes
can you envision the homolog tree?
functional genomics
Global approach to the study of the function, expression, and interaction of gene products
transcriptomics
the study of RNA molecules present in a sample
proteomics
the study of proteins present in a sample
interactomics
The complete set of physical interactions between proteins and DNA segments, between proteins and RNA segments, and between proteins
ChIP-seq
what has been devised to identify all the binding sites of a protein in a sequenced genome?
disrupt its function and to understand phenotypes in native conditions (Reverse Genetics)
Study what phenotypes arise as a result of particular/specific genes
The gold standard for establishing the function of a gene or genetic element is to . . .
its genetic deviation (g) and its environmental deviation (e)
The phenotypic deviation (x) of an individual is composed of two parts:
the genetic (VG) and the environmental (VE) variances.
(This decomposition assumes that the genotypes and environments are uncorrelated)
The phenotypic variation in a population for a trait (Vx /VP) can be decomposed into . . .
the genetic (VG) to the phenotypic (VP) variance.
Broad-sense heritability (H2) is the ratio of . . .
the extent to which differences among individuals within a population are due to genetic versus environmental factors
(only applying to the population and environment in which they
were made)
Broad-sense heritability (H2) measures . . .
environmental AND genetic factors
Complex inherited traits are affected by