Lecture 3/4: Assays, DNA repair, and DNA recombination
1/85
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
86 Terms
Helicase assay
start with a circular ssDNA with a labeled primer, helicase, ATP (for helicase to work) and buffer with Mg. active helicase will displace the primer. Run the sample on non-denaturing gel. If the helicase does not work, the primer will not move far on the gel as it is attached to a large fragment of DNA. If it did work, it would move farther on the gel.
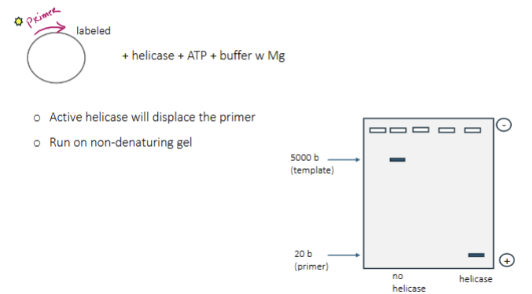
What would happen if you run a helicase assay on a denaturing gel?
the primer will be displaced whether or not the helicase is working, making both bands go far on the gel. This would not tell us anything about the nature of a helicase.
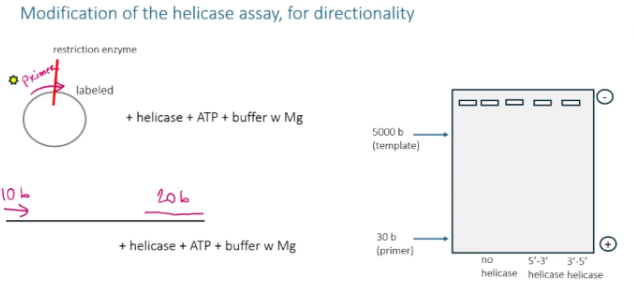
Helicases have polarity. What does this mean?
some move 5’-3’ and others move 3’-5’
How can you use a helicase assay to find the direction of movement of a helicase?
set up a non-circular DNA template with a primer on either side of where the helicase attaches (one in the 5’ direction, one in the 3’ direction) . Make sure each primer has a difference in size. Using the size difference on an agarose gel, you can see whether or not the helicase knocked off the primer in the 5’ direction or in the 3’ direction.
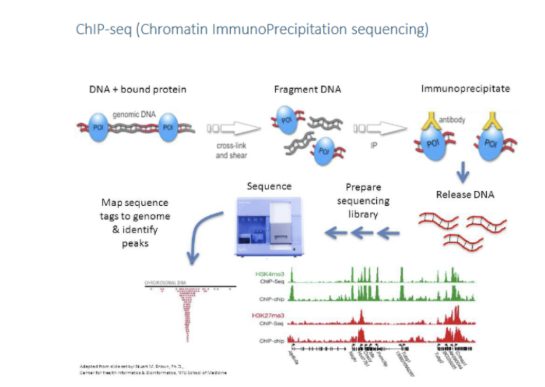
2 methods to check if proteins interact with DNA
chip-seq and EMSA
Chip-seq
chromatin immunoprecipitation sequencing. Identifies the position on DNA where a certain protein binds.
Benefits of ChIP-seq
may be done for full genome or for any protein that binds to DNA
Draw out the steps of Chip-seq
EMSA assay
electrophoretic mobility shift assay. On a nondenaturing gel, proteins attached to DNA do not detach. Using this, you can see which proteins are attached to a DNA sequence based on their relative size in a gel
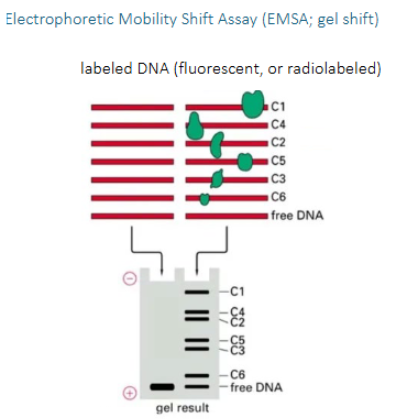
Advantages and disadvantages of EMSA
advantages are that it's simple, fast, and quantitative. The disadvantage is that it cannot tell the sequence, you must know the sequence you are probing. Cannot be done genome wide
Diminished capacity for DNA repair is linked with —
disease
Main repair pathway for replication errors
strand-directed mismatch repair.
Direct base repair repairs what DNA damage? What causes that type of damage?
O6-methylguanine and pyrimidine dimers. UV light and spontaneous reactions
Base-excision repair (BER) repairs what DNA damage? What causes that type of damage?
Uracils, abasic sites, single strand breaks, and 8-oxoguanine. X-rays, oxygen radicals, alkylating agents and spontaneous reactions
Nucleotide-excision repair (NER) repairs what DNA damage? What causes that type of damage?
bulky adduct and pyrimidine dimers. UV light and polycyclic aromatic hydrocarbons
Recombinational repair (HJ, EJ) repairs what DNA damage? What causes that type of damage?
interstrand cross-link and double-strand breaks. X-rays and anti-tumor agents
Mismatch repair (MMR) repairs what DNA damage? What causes that type of damage?
A-G mismatch, T-C mismatch, insertion, deletion. Replication errors
Most common reason for incorporation of the incorrect nucleotide
tautomerization
Two most frequent spontaneous chemical reactions that cause DNA damage
depurination and deamination of cytosine
In mismatch repair, what generally happens?
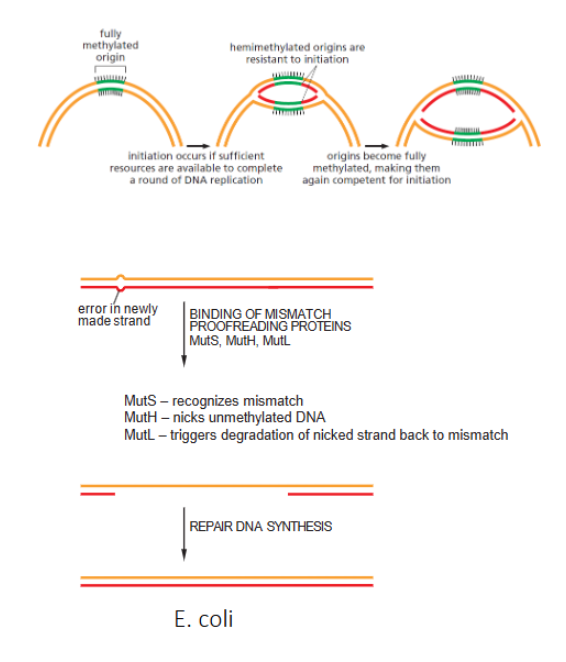
MutS, MutH and MutL in E.coli
part mismatch repair. mutS recognizes mismatch, mutH nicks unmethylated DNA and muTL triggers degradation of nicked strand back to mismatch
How does E.coli recognize which strand is old and which is new in repair mechanisms
old strand is methylated while new one isnt
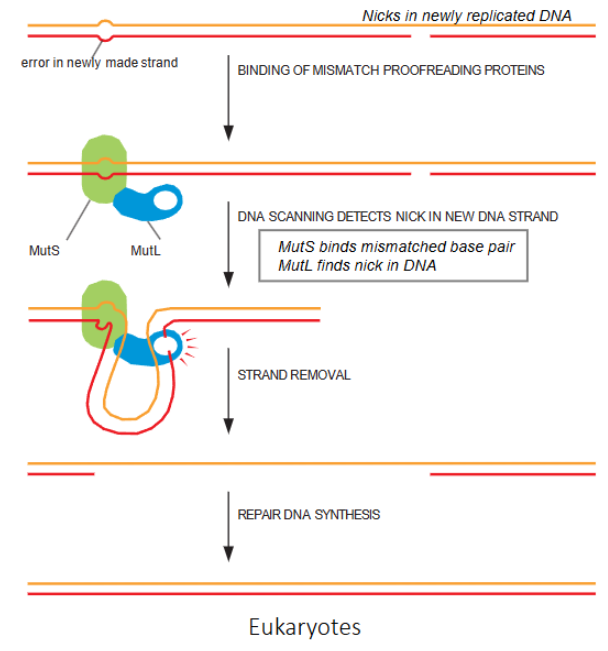
How do eukaryotes recognize which strand is old and which is new in mismatch repair
it finds the strand with a nick in and that is the new strand
MutS and MutL in eukaryotes
part of mismatch repair. mutS binds mismatched base pair while MutL finds nick in DNA
strand -directed mismatch repair in E.coli
strand -directed mismatch repair in Eukaryotes
Ribonucleotide excision pathway
RNase H2 cuts next any rNMP then DNA polymerase starts DNA synthesis from free 3’ OH and displaces the strand with rNMP. Flap nuclease cuts off the displaced part and ligase seals the nick
Depurination
spontaneous reaction which can release guanine or adenine from DNA. results in abasic site.
Deamination
replacement of an amino group with a carbonyl group on a DNA base. Most common deamination is cytosine to uracil which is easily recognized b/c uracil is not a DNA base.
When cytosine is deaminated, what is the resulting base? when 5- methyl cytosine is deaminated?
uracil; thymine
When 5-methyl C is deaminated what is made? What may this lead to?
T; mutation if not repaired before replication
Consequence of deamination of C if not repaired before replication
it creates one unchanged DNA strand and one mutated new strand that has G changed to A.
If DNA repair mechanisms encounter a G-T pair it automatically assumes that the — in the pair was created via —-- of —- and replaces it with —-
assumes that the thymine in the pair was created via deamination of 5-methyl cytosine and replaces it with cytosine
Consequence of depurination if not repaired before replication
it creates one unchanged new strand and one new strand with a deletion or a 75% chance of mutation (as a random base is added to replace the missing purine)
O6-methylguanine
G base with an unexpected methyl group. Pairs with T causing G
Alkyl transferase
“suicide enzyme” that works in direct base repair. interact with bases that have damage in the form of an unexpected methyl group (like O6-alkyl-guanine). Transfers the methyl group to the cysteine in their active site. Not truly an enzyme by definition since they can only do their job once before being discarded.
MGMT
alkyl transferase that is overexpressed in some cancers
Why are alkyl transfereases considered “Suicide enzymes”
they function only once ( they are not reused)
What are unnatural DNA bases recognized by? Give examples of these unnatural bases. What DNA repair mechanism will repair these bases?
DNA glycosylases; hypoxanthine, xanthine and uracil; Base excision repair (BER).
Base excision repair (BER)
DNA glycosylases recognize unusual bases, damaged bases, and cut out damaged bases. Specific for particular damaged bases. This creates a abasic site. The glycosylases then recruit the next enzymes needed for repair. Endonucleases and phosphodiesterase removes sugar phosphate of abasic site (and more bp around the site). DNA polymerase adds DNA nucleotide where the cut occurs and DNA ligase seals the nick.
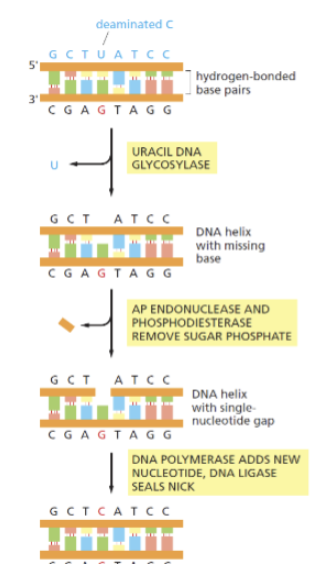
DNA glycosylases
scans genome by flipping bases out one by one and recognizes whether or not it is damaged (it recognizes this by strongly bonding to its specific damaged base type). If damaged they will cut out the base and trigger BER. Specific for particular damaged bases (ex. One would scan for uracil while another would scan for hypoxanthine).
Monofunctional glycosylases
hydrolyze glycosidic bond when they detect unusual or damaged base
Bifunctional glycosylases
hydrolyze glycosidic bond when they detect unusual or damaged base AND they can also hydrolyze the phosphodiester bond in the backbone
Nucleotide excision repair (NER)
repairs bulky lesions such as large adducts, crosslinked strands and pyrimidine dimers (thymine dimers are most common). Recognizes change in helix not particular type of damage. DNA is scanned constantly for distortion of helix. When found the DNA strand is nicked on both sides of the damage, helicase peels off the excised DNA, and DNA polymerase and ligase fill in the gap.
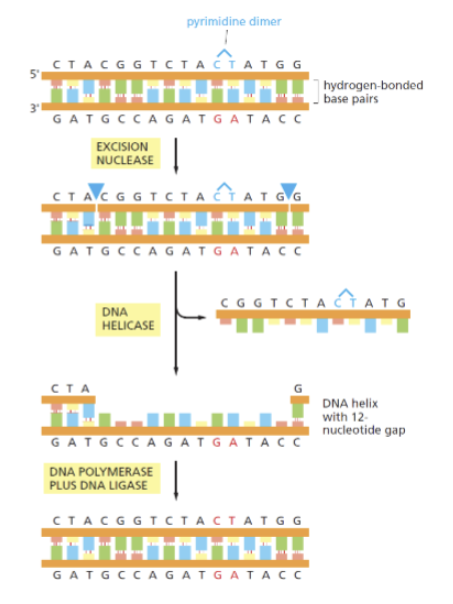
Pyrimidine dimers (show the two types that can form)
usually caused by UV light on DNA. Thymine dimers are the most common type. Repaired by NER.
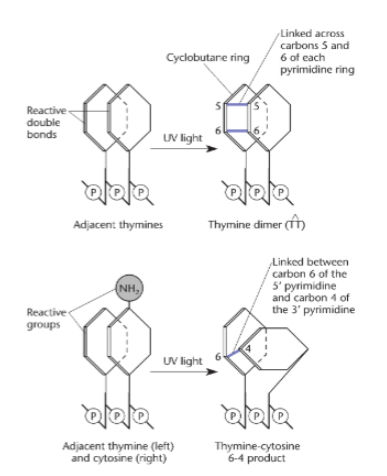
NER most commonly repairs what damage?
thymine dimers
Thymine dimers are most common in what type of cell? why?
skin cells; it's the type of cell most exposed to UV
Proteins that scan helix as part of E.coli NER
Uvr,A,B,C, and D
Some organisms (not humans) may repair pyrimidine dimers by —--. What enzyme does this?
direct repair; photolyase
Global genomic NER (GG-NER or GGR)
NER that removes a wide variety of bulky DNA damages from across the entire genome, including silent and untranscribed regions
Transcription coupled NER (TC-NER or TCR)
NER that removes bulky damage on the template strand of actively transcribed genes. RNA Pol stalls at DNA lesions, recruits repair factors and directs excision to lesion sites.
Photolyase
reverses pyrimidine dimers via direct repair. Activated by visible light. Not active in placental mammals
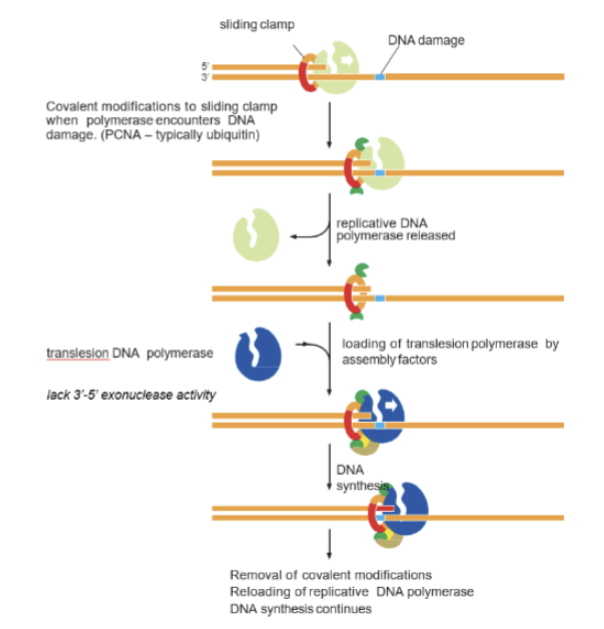
Translesion polymerases
if damage is not repaired before DNA replication, this is the last resort. Replication fork stalls when it encounters DNA damage and these are recruited to go through the lesion. Low fidelity, no 3’-5’ exonuclease (no proofreading). Less discriminatory about what they incorporate. Low processivity. Can introduce mutations.
Show how translesion polymerases take over DNA replication when DNA polymerase encounters damage
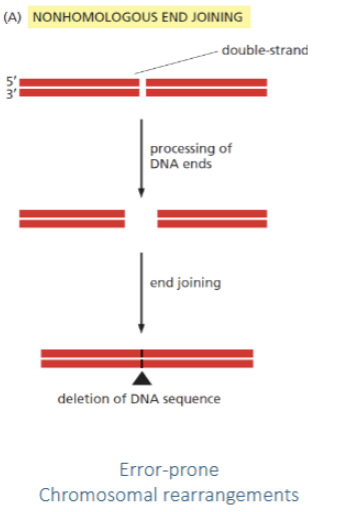
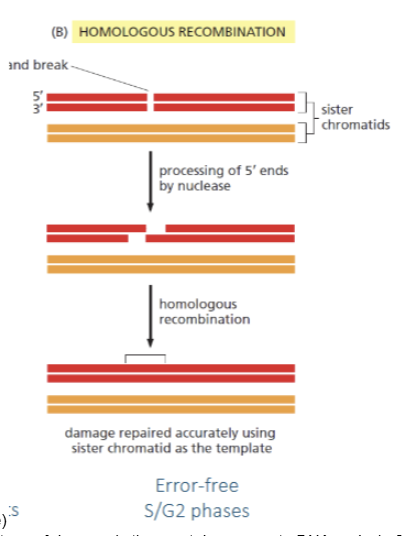
NHEJ vs HR
Both are repair mechanisms for double strand breaks. NHEJ is more error prone while HR is error-free. NHEJ occur before replication while HR occurs after replication (in S/G2) as it uses the sister chromatid as template for repair.
NHEJ
non-homologous end joining. Double strand break repair in which DNA strands are reattached with deletion where the break occurred. Error-prone and may result in chromosomal rearrangements.
HR
homologous recombination. Double stand break repair which uses sister chromatid as template to repair the break. Error free and may occur after replication (in S/G2 phase)
What type of damage is the most dangerous to DNA and why?
double strand breaks as open ends on the DNA allows the DNA to be destroyed by exonucleases
t/f double strand breaks can easily kill a cell if not repaired
true
Most common cause of double strand breaks in DNA?
radiation
Besides NHEJ and HR, what other double-strand break repair mechanisms are there? Which one are they more similar to, NHEJ or HR?
alternative end joining (more similar to NHEJ) and single-strand annealing (more similar to HR)
First step of any double strand break repair mechanism
end resectioning so that there is a clean break where ligase may work
Ku70-ku80 protein
proteins bound together that recognize double strand breaks before replication and trigger NHEJ pathway. Holds chromosome strands together so that they do not float away.
What is the pathway for double strand break repair if end resectioning is around less than 20 nt?
NHEJ
Steps of NHEJ
resection occurs until there is a small region of homology (2 or 3 nucleotides) that may be filled in by a polymerase then ligated via ligase. Some nucleotides are lost in the resectioning which introduces deletions. There may be additions as well if the small region of homology unbinds while DNA polymerase is extending (this would cause the process of resectioning and extending to start over again)
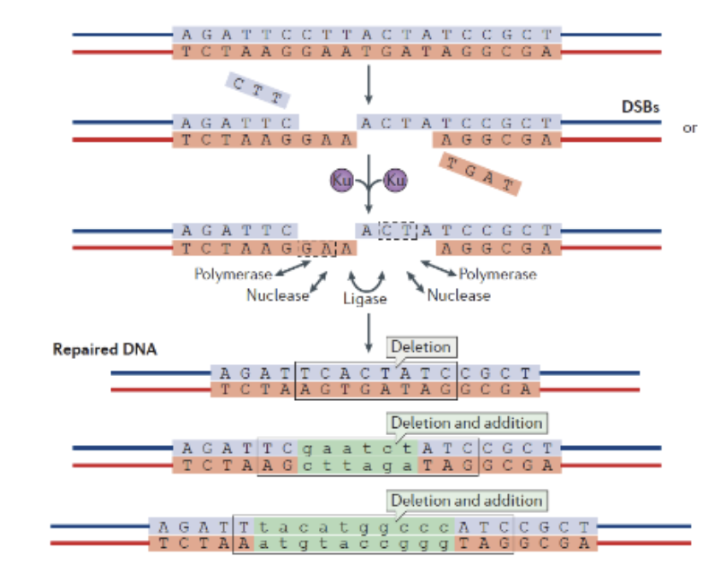
Why may additions or deletions caused by NHEJ not matter at all?
most DNA is noncoding, so it matters more to repair the double strand break
D-loop
DNA structure formed when a single strand of DNA invades a homologous double-stranded DNA molecule, displacing one of the original strands to form a hybrid duplex. This D-loop allows for DNA synthesis to repair the broken DNA
How does HR occur in E.coli
RecBCF complex has 2 helicases and an endonucleus. Helicases have opposite polarities and different speeds. RecA coats a single strand and stretches it to expose 3-4 bases that are able to form a base pair which facilitates strand invasion of the sister chromatid and formation of D-loop. Strand is the extended by DNA pol1
Show the process of HR
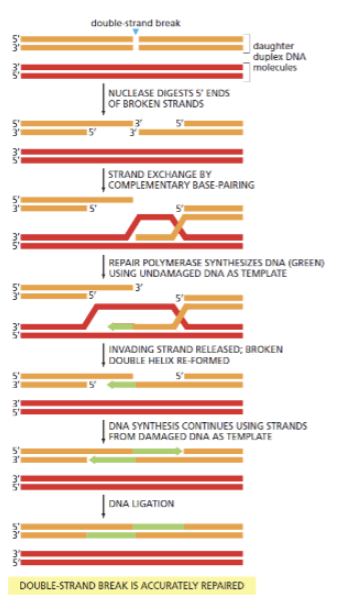
Show how HR may repair a broken replication fork
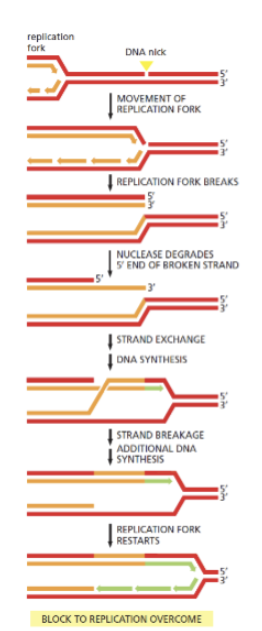
Collision of replication fork with —- may result in a double stranded DNA break. This can be repaired by —- using —- as a template
a single-strand break (nick) ; HR; using the undamaged DNA duplex as a template
What process is HR crucial for (apart from repair)?
crossing over in meiosis
First step of HR during crossing over of meiosis
intentional double-strand break
Difference between HR for repair and HR for crossing over
in repairs, general sequences are identical while in crossing over the recombination will be between chromosomes that are not necessarily the same
Show the process of how HR occurs in meiosis (show the two alternative pathways)
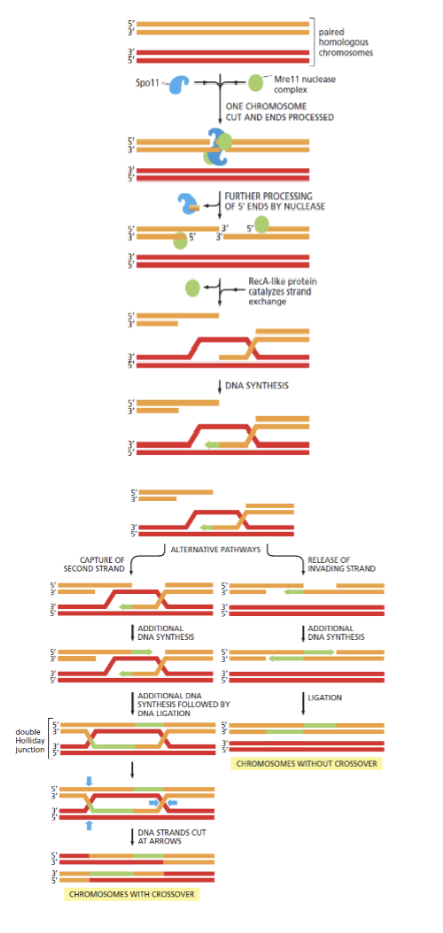
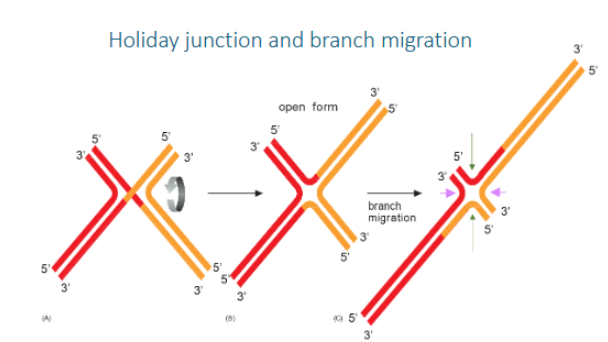
Holiday junction
a cross-shaped DNA structure formed during homologous recombination, where four DNA strands from two homologous DNA duplexes are exchanged to create a cross-linked intermediate
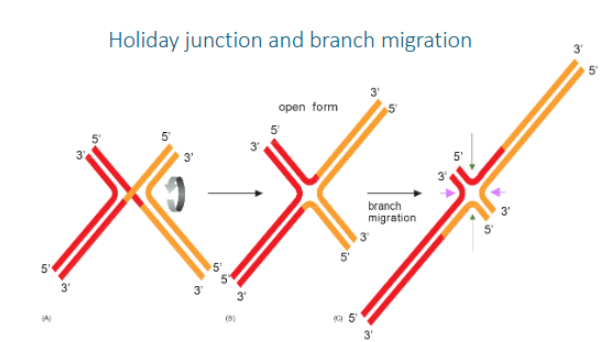
Branch migration
in a Holliday junction is the process where a DNA strand is progressively exchanged for a homologous strand, effectively moving the crossover point along the DNA
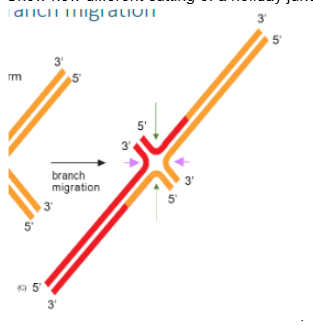
Show how different cutting of a holiday junction may or may not lead to crossover
if cut at the pink arrows gene conversion and CO occurs but when cut at green arrows it is non-CO with just a heteroduplex patch (gene conversion without crossover)
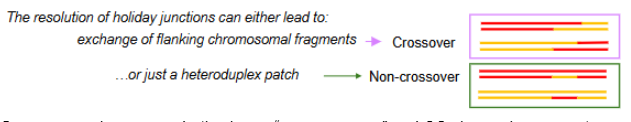
Gene conversion
occurs both when a “non-crossover” and CO change happens at a holliday junction. Seen when the holliday junction is cut and a heteroduplex patch is made between the two dna strands of each chromosome during HR.
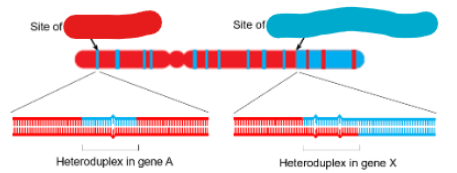
label the image
red- site of gene conversion and blue- site of crossover and gene conversion
After crossing over occurs there may be some —-- caused by differences in alleles. These are resolved via what repair mechanism?
DNA mismatches; Mismatch repair
RuvABC complex mediates —-- and —-- in E.coli
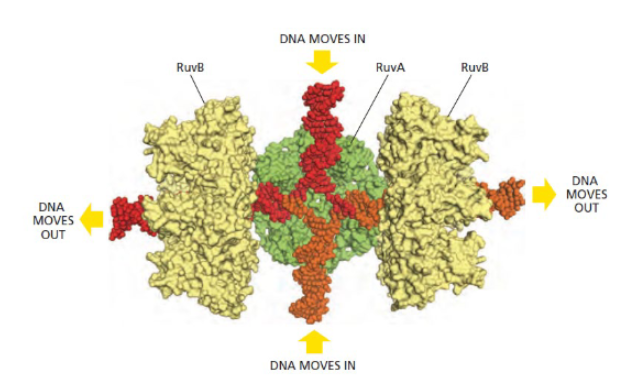
branch migration and resolution of Holiday junctions
RuvABC complex of E.coli
proteins which stabilizes the holliday junction and allows for the the junction to move (branch migration)
When heteroduplexes formed from crossover have genes with different alleles (different sequence) on two DNA strands, what happens?
mismatch repair excises a random portion (either from one strand or the other) and DNA synthesis fills the gap while ligase seals nicks
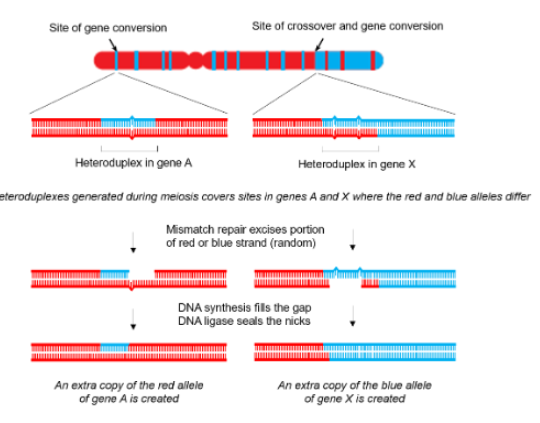
How does mismatch repair choose which strand portion to excise when there are mismatches due to allele difference after crossovers?
it chooses randomly
what repair mechanisms may repair pyrimidine dimers?
NER, direct repair, and translation polymerases