mitochondria and chloroplasts
1/28
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
29 Terms
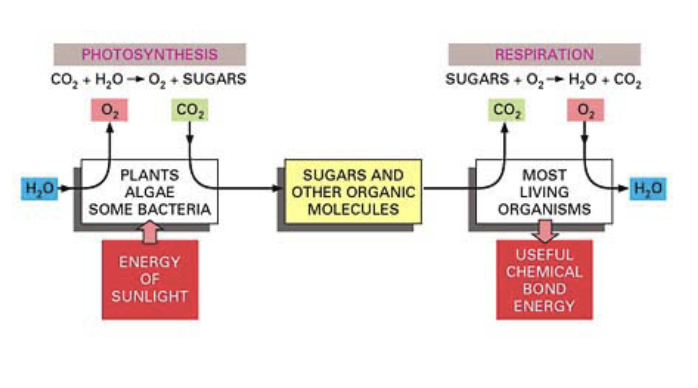
both energy producing organelles
Chloroplasts
Fixation of co2 into sugars and other organic molecules
Requires energy (from sunlight)
Splitting of water and release of water
Mitochondria
Release co2
Consume oxygen
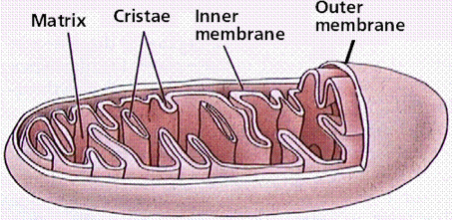
mitochondria structure
2 different membranes
Outer = porous
Intermembrane proteins
Nutrients and small organic molecules can move freely across
Inner = highly folded
Folds are called cristae
Increases surface area
Matrix
Where proteins are present (e.g. enzymes for citric acid cycle)
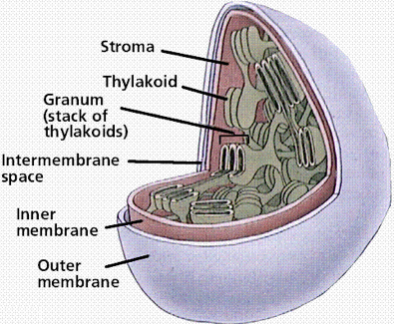
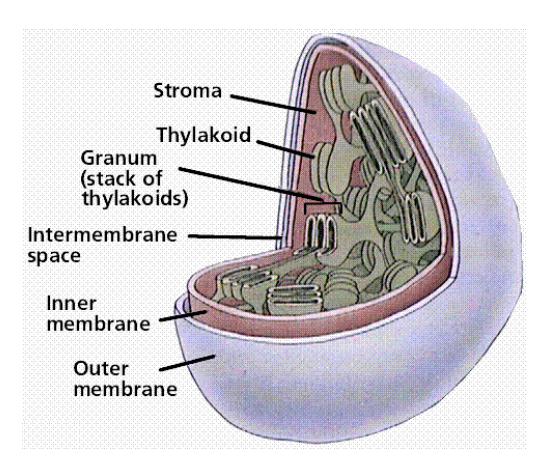
chloroplast structure
3 membranes
Inner
Outer
Thylakoid
Into stacks of grana
Large areas of membrane
Where ATP synthase sits
Where most photosynthetic machinery is
e.g. photosystems
mitochondria ATP synthesis
Anabolic reactions of cells responsible for growth and repair processes
Catabolic reactions release energy needed to drive anabolic reactions
Must be an efficient linking or coupling of energy yielding to energy requiring processes
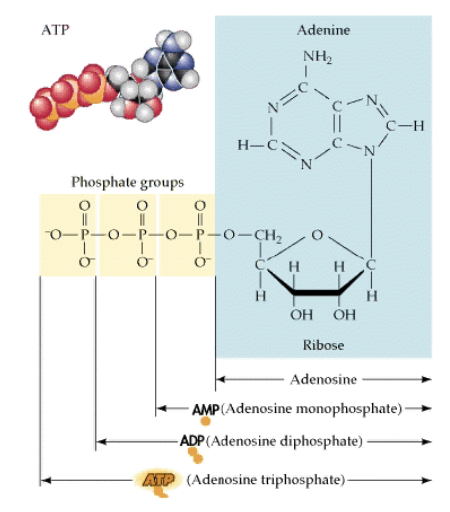
ATP is most commonly used as this energy intermediate
Energy currency of the cell
Transfers the energy captured during cellular respiration to the cellular sites that use energy
Cleavage of phosphate bonds provides energy
Cells obtain most of their energy from membrane bound mechanisms
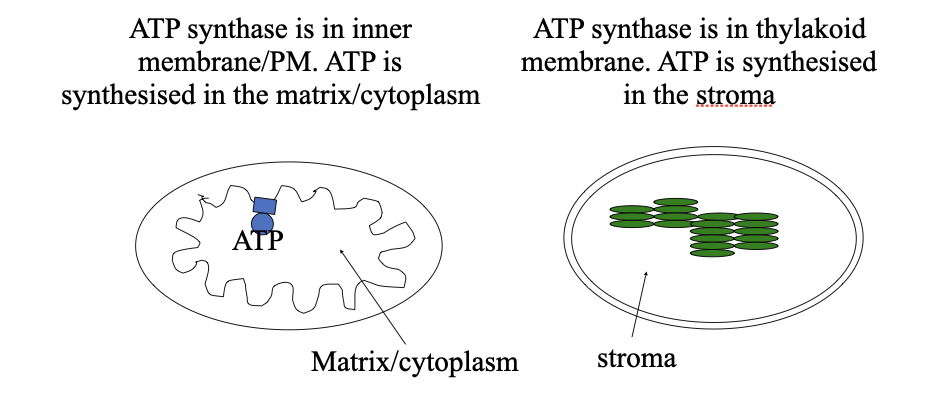
ATP synthase is found in the mitochondrial inner membrane, the chloroplast thylakoid membrane and the inner membrane of eubacteria
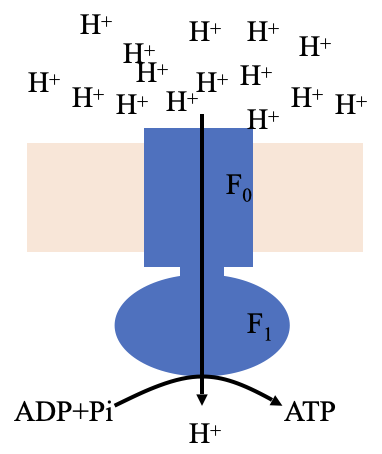
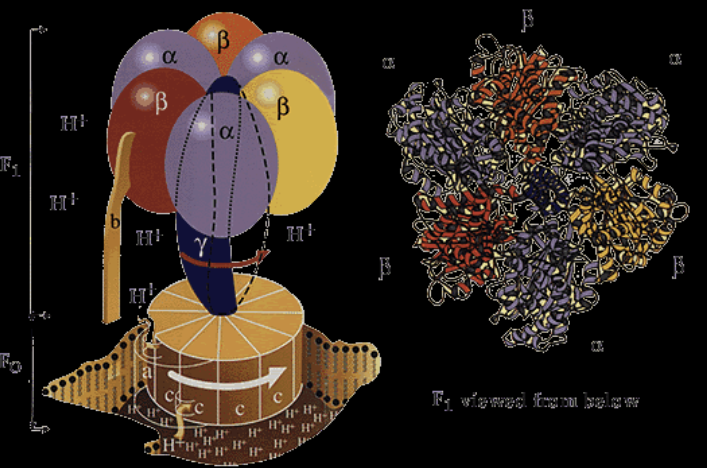
Large multisubunit F-type ATPase made up of an F0 which in integral in the membrane and F1 which is peripheral
structure and function of ATP synthase
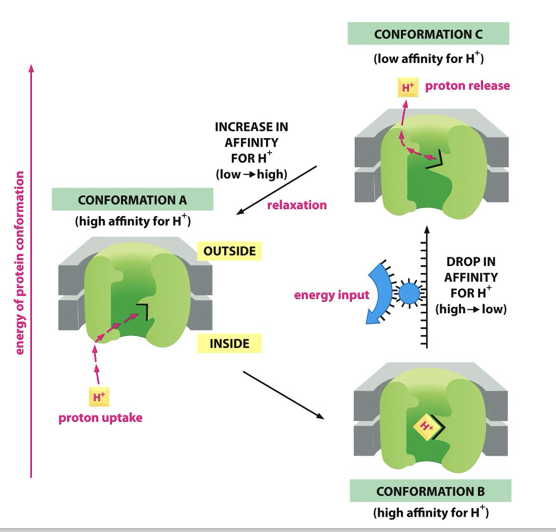
Large head is attached through a stalk to the transmembrane carrier for protons
As protons pass through the carrier it is thought that the stalk spins inducing the head to produce ATP
As protons move through ATP synthase, the stalk rotates
Causes conformational change of shape (distorts F1) as gamma stalk is asymmetrical
Provides energy for production of ATP from ADP and Pi
From slides
This rotation drives the conformational transitions of the catalytic subunits which, in turn, alters the nucleotide binding site affinities. As a consequence, conformational energy flows from the catalytic subunit into the bound ADP and Pi to promote their dehydration into ATP.
more on ATP synthase
The proton gradient is a form of stored energy
Determines pH
Across mitochondria
Intermembrane space = pH 7
Matrix space = pH 8
Can produce around 100 ATP per second
Around 3 protons are needed to synthesise 1 molecule of ATP
Can be reversed
Use the hydrolysis of ATP to pump protons across membrane in the opposite direction
Location in mitochondria and chloroplasts
how is the proton gradient across the mitochondrial membrane generated
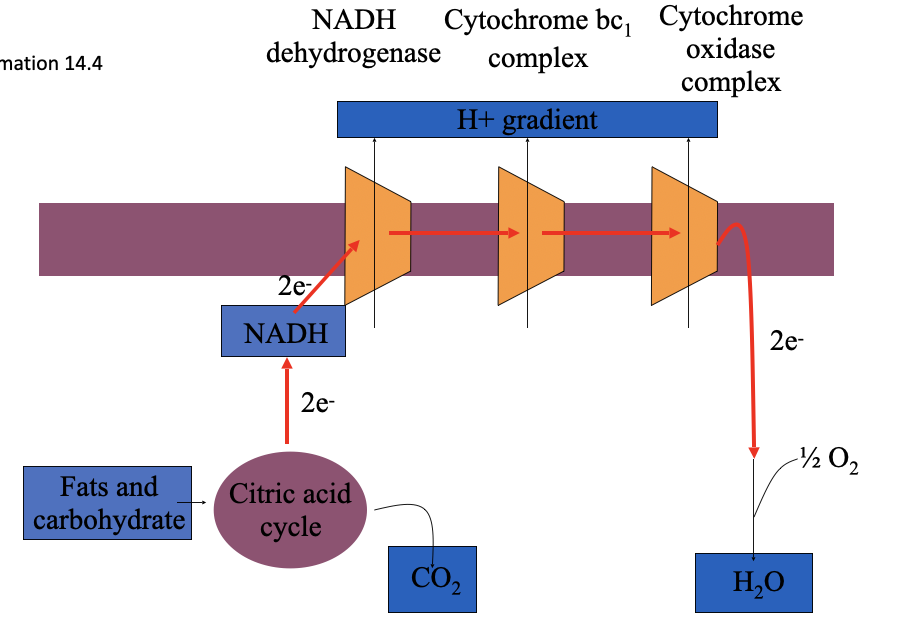
High energy electrons are passed along an electron transport chain
An electron can bind and release a proton at each step in the chain
When an electron is lost, the affinity for the proton is reduced and it is released
This is oxidation
These electron transfers release large amounts of energy which is used to pump H+ across the membrane
Creates an electrochemical proton gradient
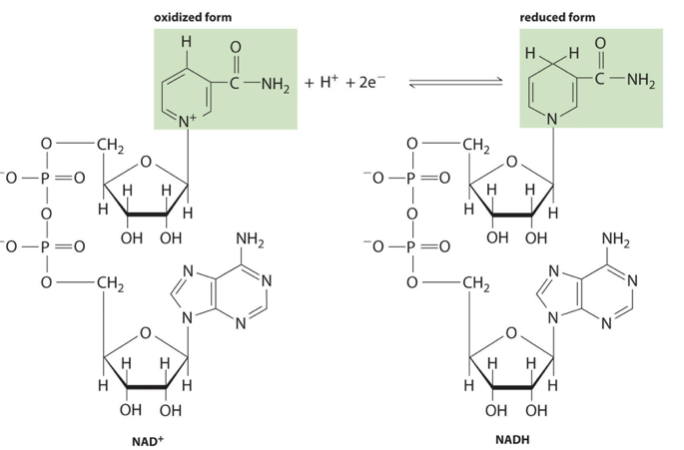
NAD in mitochondrial membrane
NAD + e- + H+ -> NADH
Reduction
Loses electron at NADH dehydrogenase
oxidation
Goes back to krebs
Electron is transferred along electron transport chain
Electron goes to final electron acceptor = water
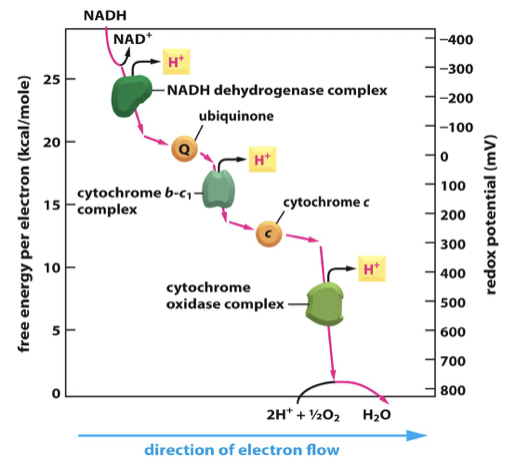
mobile electron carriers
Ubiquinone carries electrons from the NADH dehydrogenase to the cytochrome b-c1 complex
Cytochrome c carries electrons from the cytochrome b-c1 complex to the cytochrome oxidase complex
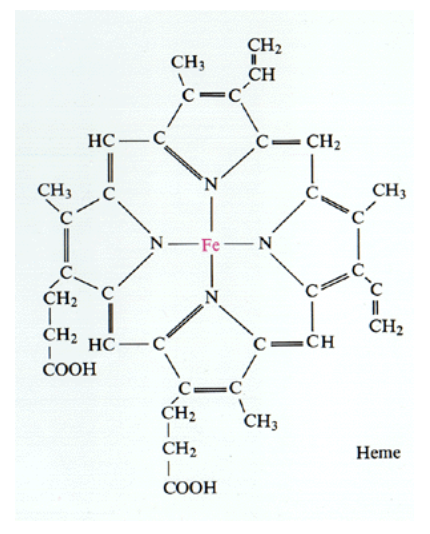
Has haem groups
chemiosmotic coupling
The linkage of electron transport, proton pumping and ATP synthesis
In mitochondria this is known as oxidative phosphorylation
Consumption of oxygen
how do electrons move along the electron transport chain?
By a series of oxidation, reduction reactions
As one reactant is oxidised (loses electrons), another is reduced (gains electrons)
Reducing agents ranked according to electron transfer potential
NADH has high electron transfer potential (-ve value)
H2O has low electron transfer potential (+ve value)
Standard redox potential E’0 (measured in Volts)
ΔG0’ = -nFΔE’0
Redox potential (electron affinity) increases along the mitochondrial electron transport chain
Important as energy decreases down chain
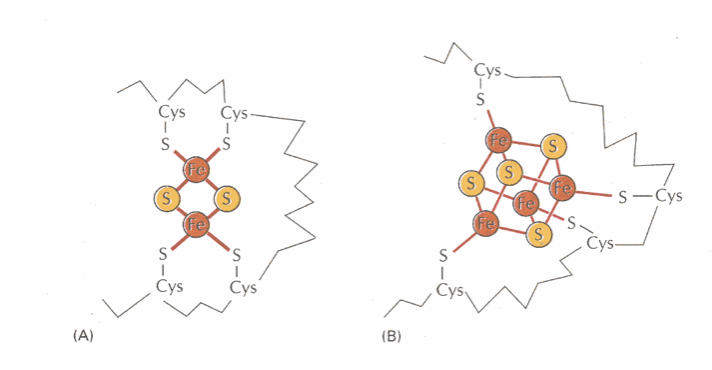
the nature of electron carrying groups
Prosthetic groups are associated with these
Metal associated with them have different redox potentials
Have different electron affinities
NADH dehydrogenase
Flavin nucleotides
Fe-S
Cytochrome b-c1 complex
Haem
Fe-S
Cytochrome c
Haem
Cytochrome oxidase complex
Haem
CuA
CuB
iron sulfur (Fe-S)
haem
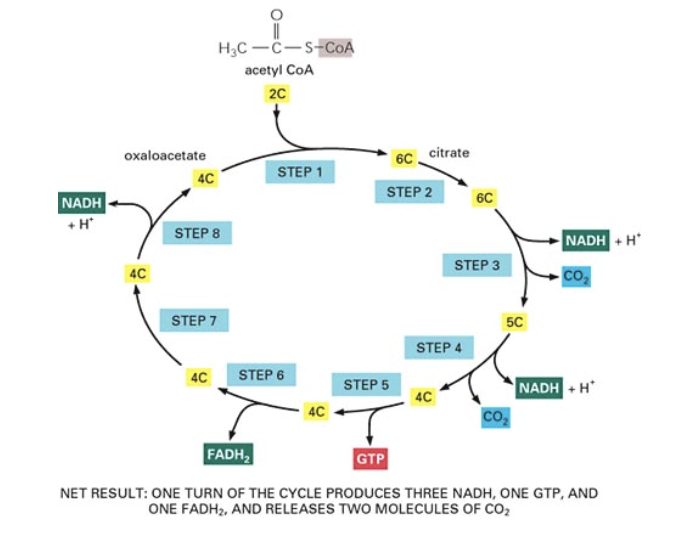
the citric acid cycle
mitochondria structure
Matrix
Enzymes of citric acid cycle
Mitochondrial DNA
etc
Inner membrane
Electron transfer proteins
ATP synthase
Transport proteins
Outer membrane
Has large pores
Lipid synthesis
Conversion of lipid substrates into forms that can be metabolised in the matrix
Intermembrane space
Several enzymes that use ATP passing out of the matrix phosphorylate other nucleotides
Cristae
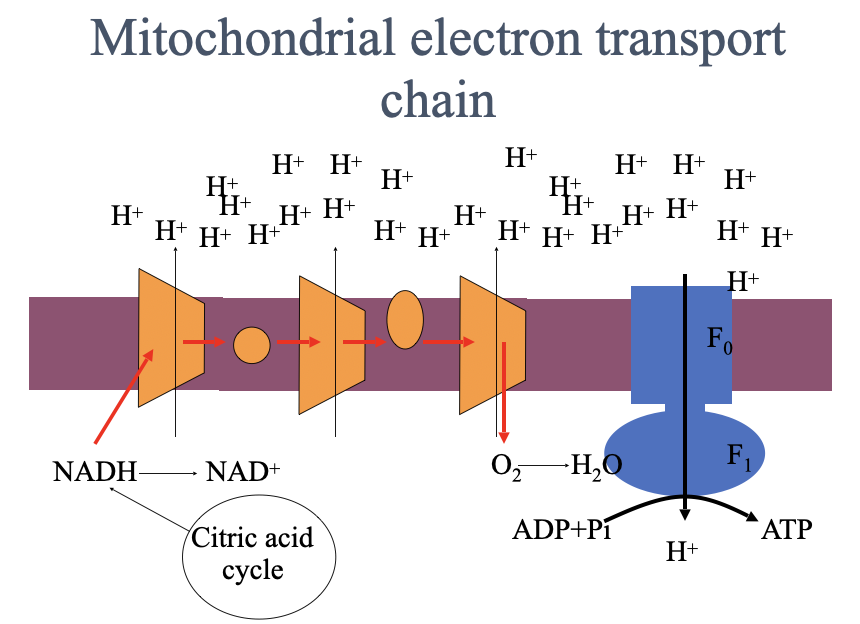
agents that interfere with oxidative phosphorylation
Cyanide and carbon monoxide inhibit cytochrome oxidase
Block the passage of electrons to oxygen
ATP synthesis grinds to a halt
uncoupled mitochondria generate heat
In most newborn mammals including humans a type of adipose tissue (brown fat) uses fuel oxidation to produce heat and not ATP
This is achieved by a protein (thermogenin) which provides a path for protons to return to the matrix without passing through the F0F1 complex
The energy is dissipated as heat
Protons flow through mitochondrial membrane but not through ATP synthase
chloroplasts use energy from sunlight to fix carbon
features of photophosphorylation
Unlike NADH, water is a poor donor of electrons
Requires energy input in the form of light to create a good electron donor
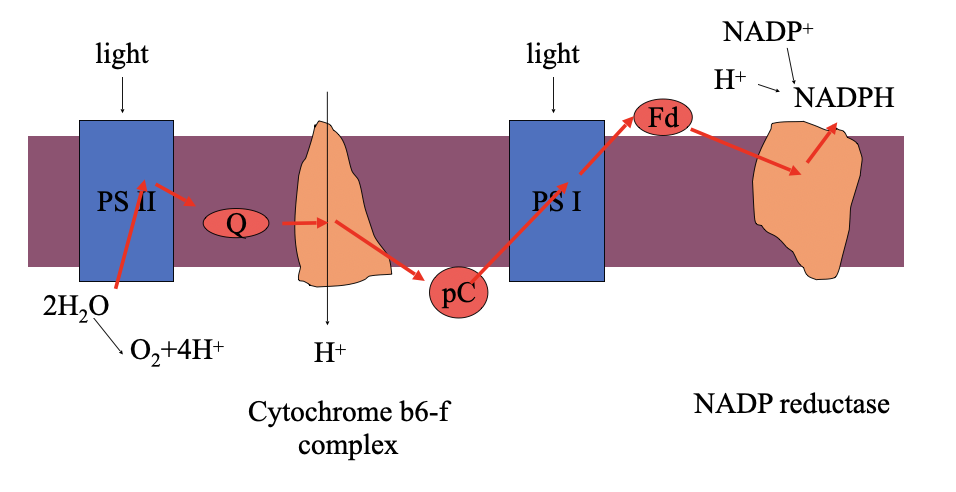
how is the proton gradient across the thylakoid membrane generated
Again electron transfer is coupled to proton pumping
Also protons released upon water oxidation contribute to the electrochemical proton gradient
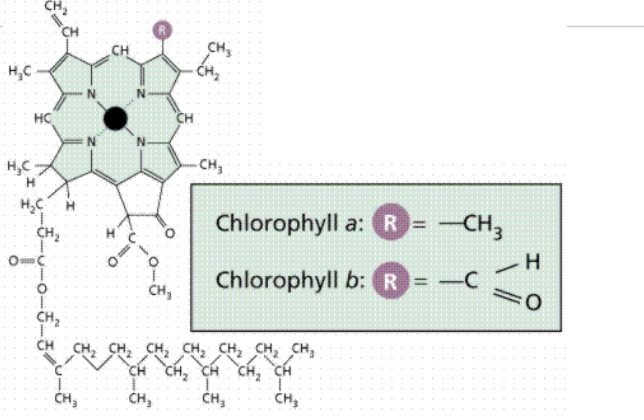
Sunlight is absorbed by chlorophyll molecules and electrons interact with photons of light raising them to a higher energy level
The energy from hundreds of chlorophyll molecules (in the antenna complex) is channelled into a special pair of chlorophyll molecules in the reaction centre
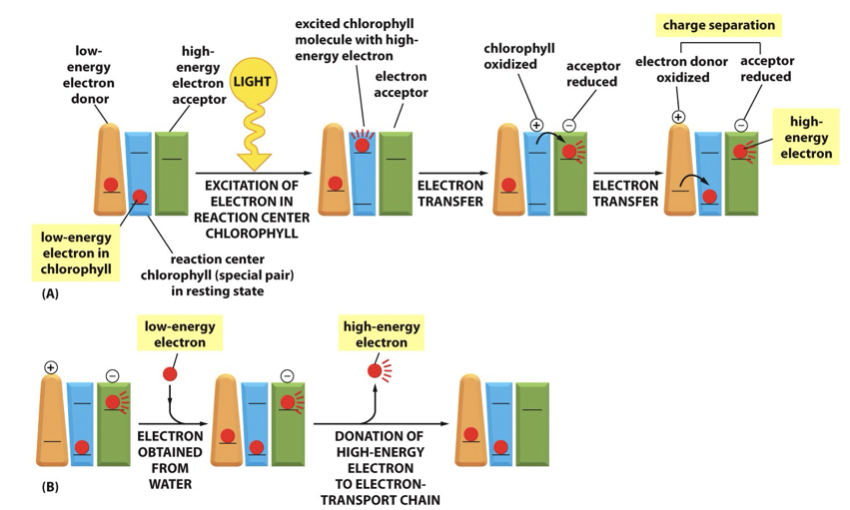
photosystems
Reaction center chlorophyll
PS II: Uses P680 (absorbs light best at 680 nm)
PS I: Uses P700 (absorbs light best at 700 nm)
Order in the light reactions
PS II: Acts first in the light-dependent reactions
PS I: Acts second
Primary function
PS II: Splits water (photolysis) to release oxygen, protons (H⁺), and electrons
PS I: Produces NADPH by transferring electrons to NADP⁺
Electron source
PS II: Electrons come from water (H₂O)
PS I: Electrons come from PS II via the electron transport chain
Contribution to ATP formation
PS II: Helps create a proton gradient used for ATP synthesis
PS I: Does not directly contribute to the proton gradient (in non-cyclic photophosphorylation
the two photosystems work in series
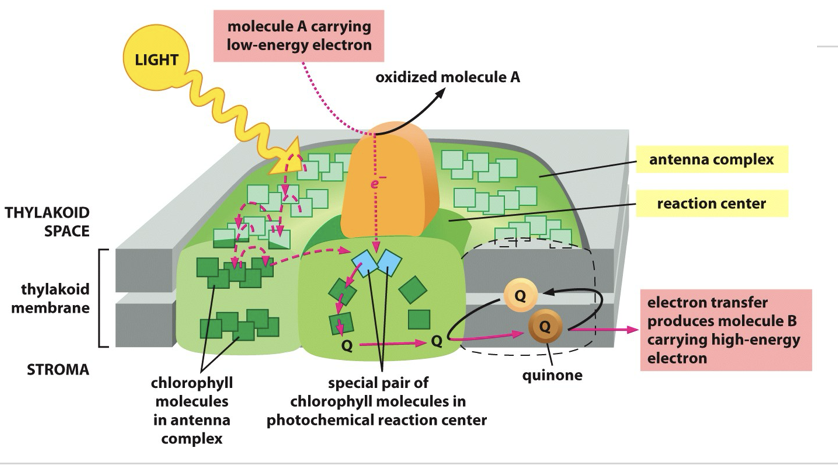
mobile electron carriers
Plastoquinone (closely resembles ubiquinone of mitochondria)
Plastocyanin (a small copper containing protein)
Ferredoxin (a small protein containing an iron-sulphur centre)
redox potentials
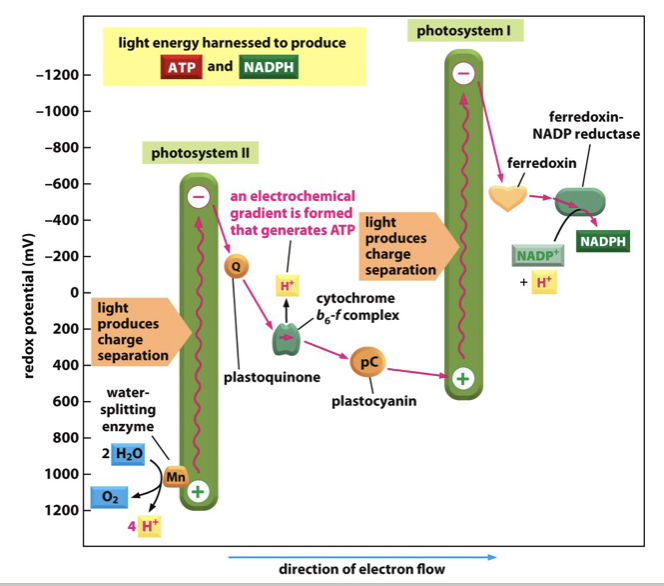
light energy is converted to chemical energy
ATP is generated by the proton gradient across the thylakoid membrane in the same way as in mitochondria
H+ generated by the splitting of water also contributes to the proton gradient
The high energy electrons are ultimately passed on to form the high energy compound NADPH
ATP and NADPH are used for carbon fixation in the calvin cycle
For every 3 carbon sugar produced 9 molecules of ATP and 6 of NADPH are required
rubisco
Ribulose bis phosphate carboxylase
Catalyses the initial reaction in carbon fixation
It is a sluggish enzyme, processing only about 3 molecules of substrate per second
Can make up to 50% of total chloroplast protein
Claimed to be the most abundant enzyme on earth
Rubisco fixes carbon
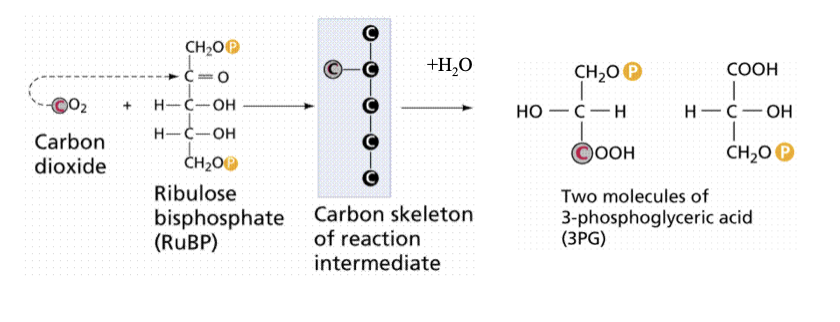
carbon fixation
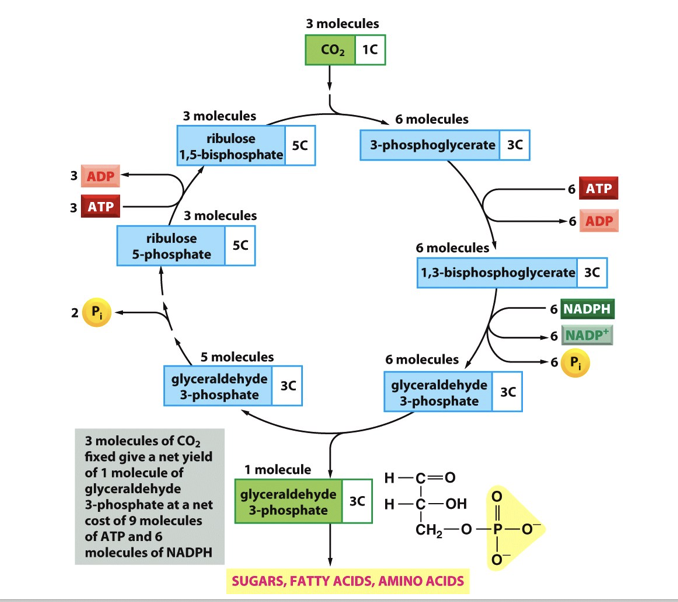
chloroplasts
Thylakoid
Photosystems 1 and 2
ATP synthase
NADP reductase
Stroma
ATP synthesised
NADPH synthesised
Carbon fixation
DNA
similarities between processes in mitochondria and chloroplasts
Both use proton gradients across membranes to produce ATP using ATP synthase
Electron transport along an electron transport chain drives proton pump
Similarities between some of the components of electron transport chain (cytochrome bc and b6f show sequence similarity and ubiquinone and plastoquinone resemble one another)
differences between chloroplasts and mitochondria
Chloroplasts
Low energy electrons come from water but are excited to higher energy by light
Ultimate electron acceptor is NADP+
Chemical bond energy and reducing power utilised in carbon fixation
Mitochondria
High energy electrons come from NADH
Ultimate electron acceptor is oxygen
Chemical bond energy used in cellular processes