D1.2: Protein Synthesis
1/73
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
74 Terms
Transcription
synthesis of an RNA molecule from a DNA template using RNA Polymerase II
Purpose of Using mRNA (2)
Protection and Speed
Protection
DNA is kept safely in nucleus
Speed
one gene can be converted to many mRNAs, one mRNA can be converted to many polypeptides
Therefore speeds up process
RNA Polymerase II
transcribes mRNA
RNA Polymerase II Process
1. binds to promoter (DNA sequence) to begin transcription
2. unwinds/separates DNA
3. Moves along template DNA, produces single mRNA strand, complementary to DNA by joining ribonucleoside triphosphates (rNTPs) covalently, growing in the 5' --> 3' direction
4. Detaches at terminator and allows DNA to reform double helix, releases mRNA which eventually leaves via nuclear pores
Complementary Base Pairing
Each RNA nucleotide added is complementary to the corresponding base on template DNA (A-U, C-G)
*cannot form h-bonds w/ other bases*
Sense vs Antisense Strand
Either of two DNA strands may contain the gene of interest, so the designation varies
Sense Strand
The coding strand (NONTEMPLATE), contains the gene to be transcribed into mRNA and identical to the mRNA but w/ T instead of U
Antisense Strand
The TEMPLATE strand (noncoding), contains the complementary base sequences to the mRNA
This is the actual strand that RNA Polymerase II moves along, and is the location of the promoter and the terminator
Stability of DNA
Stability is essential bc DNA is transcribed many times during a cell's life, therefore transcription should not impact DNA
Once transcription completed, 2 DNA strands close again by reforming hydrogen bonds to complementary bases
DNA only vulnerable to chemical changes (mutation) when RNA Polymerase II moves along DNA
**if mutations were common, mRNA would contain increasing amounts of errors*
Gene Expression
Process in which information in genes has observable effects on organisms
Function of most genes is to produce AAs in a polypeptide, which result in proteins that determine observable traits
Transcription is the first stage of gene expression where genes can be switched on or off
Variability of Expressed Genes
Some genes are always transcribed bc their proteins are required (housekeeping genes)
Some genes are never expressed in the life of a cell
ex. insulin gene in a skin cell
Housekeeping Genes
a gene that is transcribed continually because its product is needed at all times and in all cells
Transcriptome
all the RNA molecules transcribed from a genome
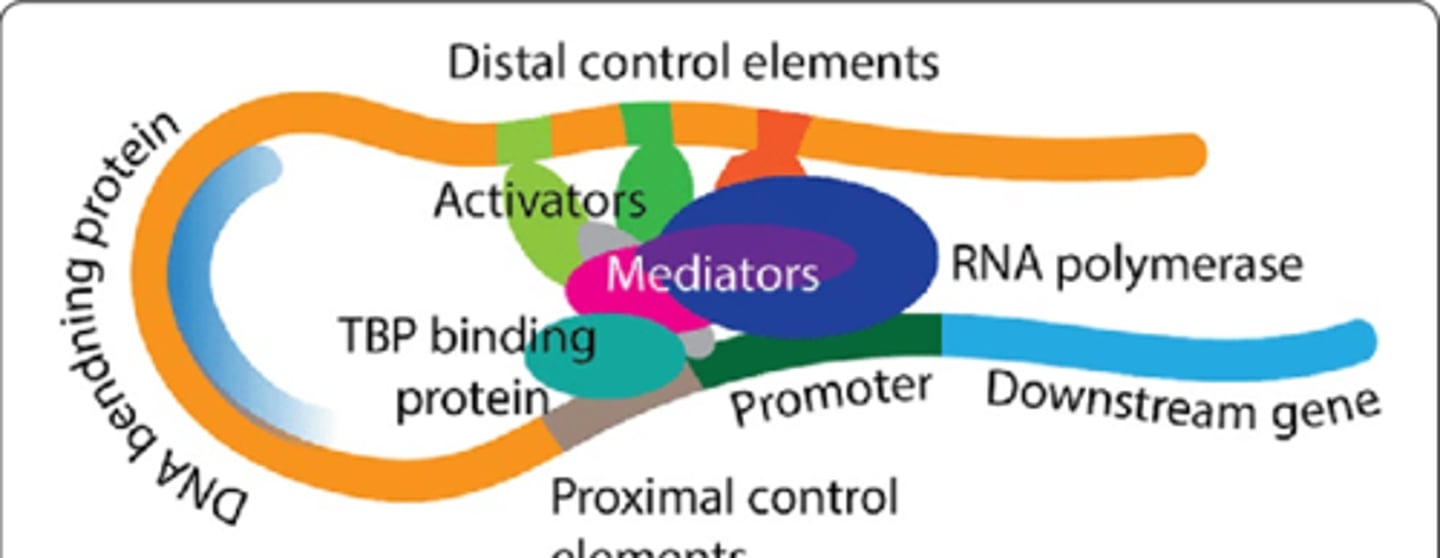
Transcription Initiation
RNA polymerase binds to a promoter
Promoter
DNA sequence that RNA Polymerase II binds to, to begin transcription
Found upstream of gene of interest on template strand, regulated via transcription factors and regulatory proteins
Transcription Factors
Group of proteins that guide RNA Polymerase II to the promoter, making it easier to bind
Can change gene expression
RNA Polymerase II usually cannot begin transcription without them
Regulatory Proteins
Bind to DNA upstream of promoter and affects transcription factors
Either activators or repressors
Activators
Proteins that encourage transcription factors and RNA Polymerase II to bind to DNA, increases transcription
Repressors
Proteins that prevent transcription factors and RNA Polymerase II from binding, decreases transcription
Control Elements
DNA sequences that regulatory proteins bind to (most genes have multiple to control gene expression)
Either Proximal elements or Distal elements
Proximal Elements
Located close to promoter, transcription factors typically bind here
Distal Elements
Located farther upstream from promoter, activators and repressors typically bind here
Transcription Factors Image
Types of Non-Coding DNA (4)
1. Base sequences that do not code for polypeptides but have other functions like RNA genes that are not translated into proteins (make rRNA/tRNA/snRNA)
2. Introns
3. Regulators of Gene Expression
4. Telomeres
Regulators of Gene Expression
Turns genes on/off by regulating transcription (promotors, proximal elements, distal elements)
Telomeres
Repetitive DNA at the end of linear chromosomes, postpones deterioration that occurs during replication
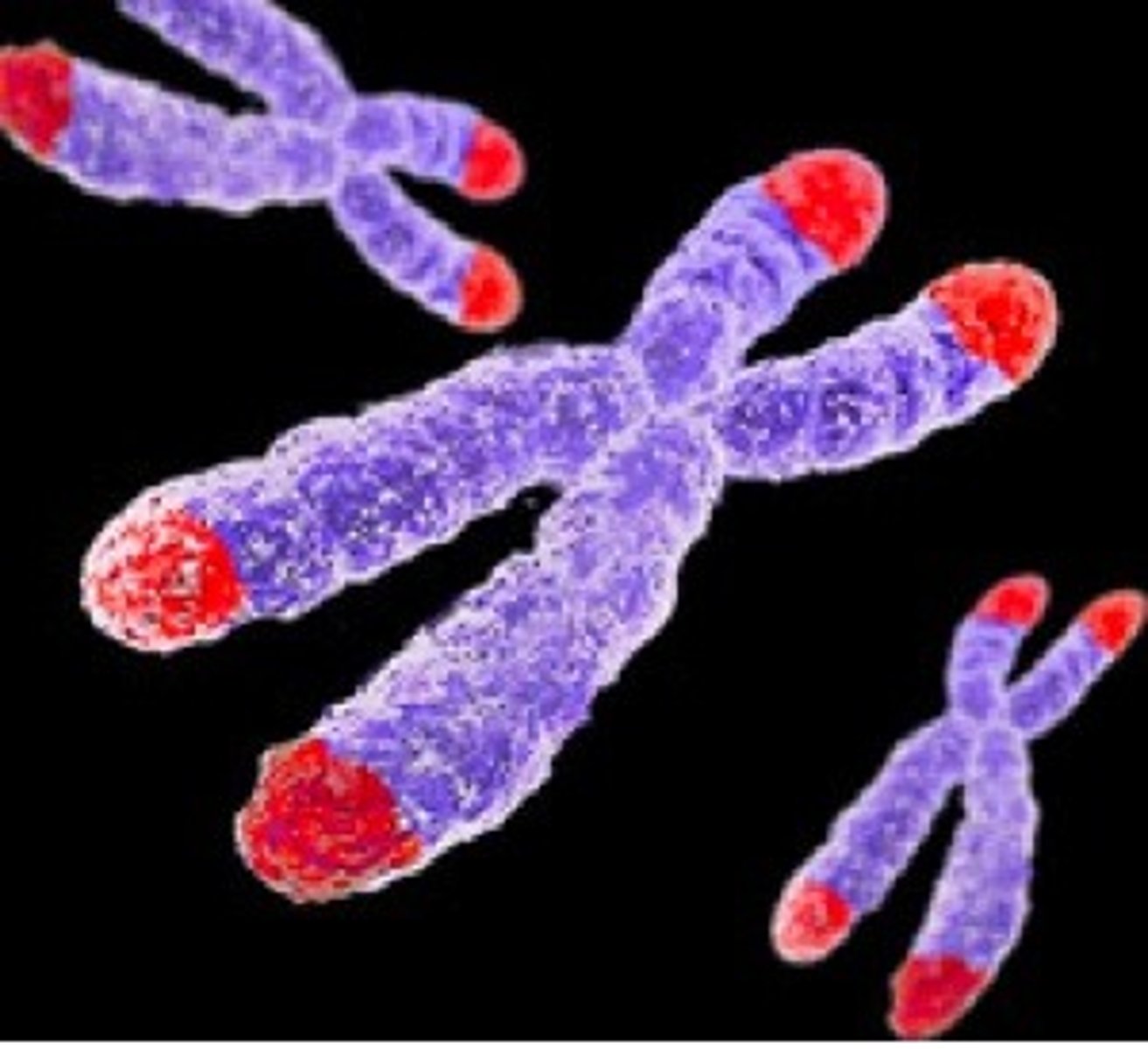
Why are telomeres necessary?
DNA replication cannot extend all the way to the end of the chromosome and as a result, we lose sequences of information with each round of replication (can't replace RNA primer at end of DNA strand being replicated)
Advantages of Separate Nucleus
Transcription and Translation occur independently (eukaryotes)
Translation happens after mRNA leaves the nucleus
This allows for post-transcriptional modification, which isn't possible in prokaryotes bc translation occurs before transcriptions finishes
Post-Transcriptional Modification
Changes in mRNA after its production, before translation, allows new mRNA to convert into mature mRNA before exiting nucleus
5' Cap
Adds a guanine w/ an extra methyl group to 5' end
Polyadenylation
Adds a poly-A tail (100-200 Adenines) to 3' end
Function of 5' Cap/Polyadenylation
Stabilizes transcript and helps it move out from nucleus
Protects against digestion from exonucleases
Exonucleases
Cut up DNA (typically viral DNA)
Newly transcribed RNA contains alternating...
Exons and Introns
Exons
Coding sequences expressed into amino acids (about 8.8 per gene), fused together to produce final transcript
Exons Exit nucleus
Introns
intervening, unexpressed sequences (about 7.8 per gene), removed from mRNA and digested into single nucleotides
Introns remain In the nucleus
Alternative Splicing
Allows for one gene to be coded into many different polypeptides, producing polypeptides w/ differing functions w/out need for second, similar gene
Most frequent change is exon skipping
Exon Skipping
when an exon is spliced out of a pre-mRNA
ex. Exon 1, Exon 2, Exon 3 in one mRNA and Exon 1, Exon 3, Exon 4 in another mRNA
Central Dogma
DNA -> RNA -> Protein
Translation
mRNA--> amino acid sequence of proteins
mRNA holds codes necessary for polypeptides, copied from a gene via transcription and translated by ribosomes in cytoplasm
Direction of Transcription/Translation
Enzymes involved in transcription and translation move in 5'-->3' direction
Transcription Direction
RNA Polymerase II adds phosphate group of RNA nucleotides to ribose sugar of growing mRNA strand
Translation Direction
Ribosomes move from 5'-->3' of mRNA
mRNA
consists of codons that specify for amino acids in a polypeptide AND a ribosome binding site
includes START and STOP codons, where translation begins and ends
Can be used multiple time unless damaged or no longer needed
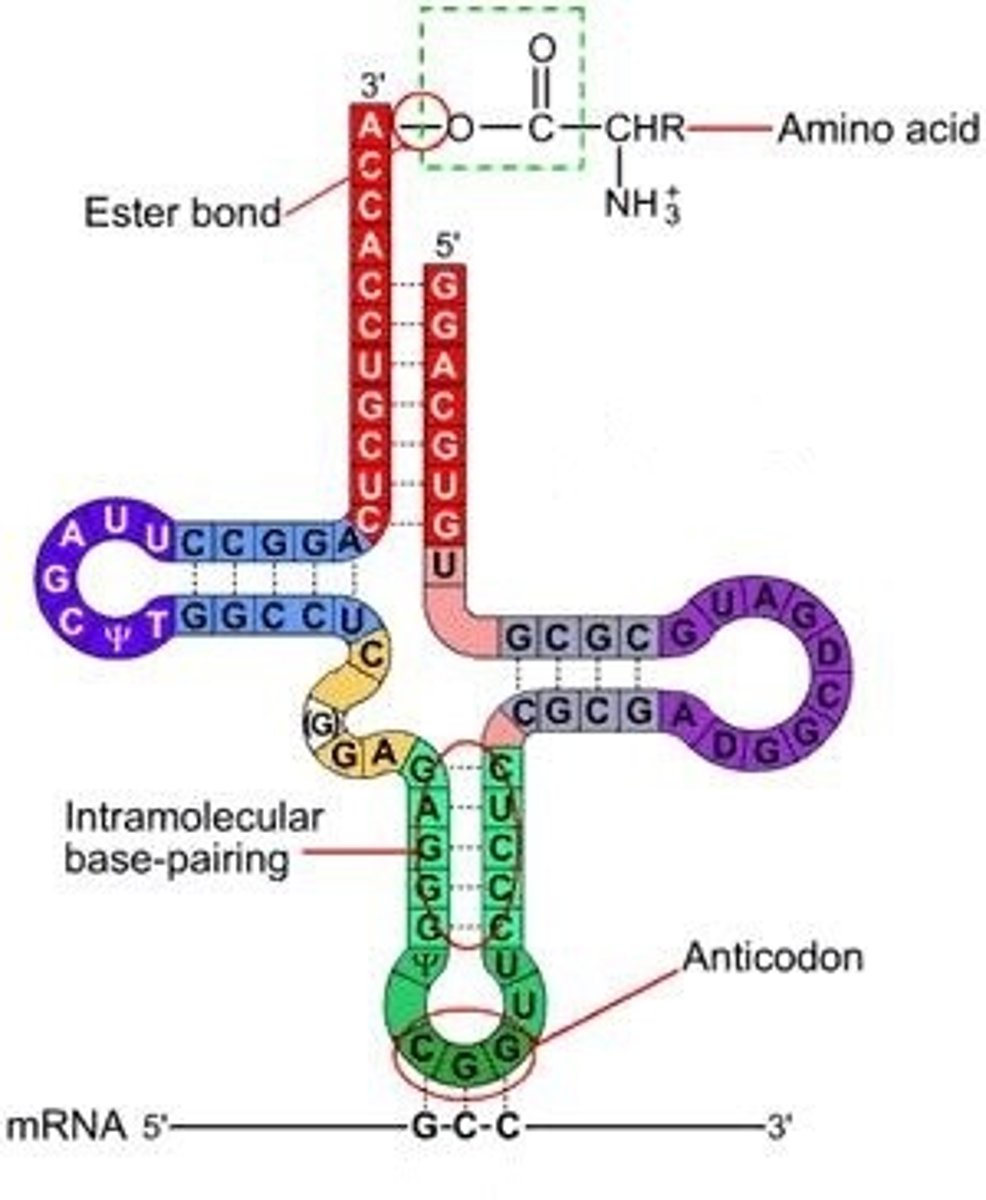
tRNA
Matches codon in mRNA w/ proper amino acid, has anticodon at one end and amino acid attachment site at the other (3')
Each tRNA has a distinctive shape that can be recognized by aminoacyl tRNA synthetase (activating enzyme), that attaches the correct AA to tRNA
Ribosomes
Composed of protein and rRNA (ribozyme)
2 subunits: large and small
Size: 70S in prokaryotes and 80S in eukaryotes
Large Ribosome Subunit
Has A (aminoacyl, 3' end), P (peptidyl), and E (exit, 5' end) sites
Small Ribosome Subunit
mRNA binding site
Adaptation of ribosomes
Large (30 nm) structure of rRNA and proteins
Either free or bound
Free Ribosomes
Make polypeptides in cytoplasm that are either utilized there or enter the nucleus (histones in octamer)
Produce proteins means to be used in cell, including for housekeeping (glycolysis)
Bound Ribosomes
Make polypeptides that must be transported somewhere else in cell or secreted
Placed into lumen (inside space) of rER, transported in vesicles that bud off
Genetic Code
Set of rules by which mRNA is converted to AA sequences in polypeptides
mRNA contains CODONS
Codons
*non-overlapping, triplet bases that code for one AA*
64 possibilities (4^3), codes for 20 AAs
Start Codon
AUG, codes for methionine and begins translation
Stop Codon
UAA, UGA, UAG, codes for release factor which releases polypeptide chain from ribosome by adding H2O for hydrolysis
Genetic Codes are Degenerate
Means that different codons can code for same AAs (ex. GUU and GUC both code for valine)
But, genetic codes are NOT ambiguous (one codon can't mean two different AAs)
Genetic Code must be read in correct reading frame
Reading Frame
On an mRNA, the triplet grouping of ribonucleotides used by the translation machinery during polypeptide synthesis.
Genetic Code is Universal
Same codon can be translated into same AA in all living organisms and viruses
Therefore must have originated EARLY in life to be present in all modern organisms
ex. Human insulin can be made by bacteria
Translation Initiation
Assembly of Components:
1. Aminoacyl tRNA synthetase attaches methionine to initiator tRNA w/ anticodon UAC (activates tRNA)
2. Initiator tRNA binds to small ribosome subunit forming a Ternary Complex, along w/ AA
3. Ternary Complex binds to 5' end of mRNA
4. Scans along until it finds start codon (AUG), forms hydrogen bonds between codon and anticodon
5. Large subunit sets w/ tRNA at P site forming initiation complex
Ternary Complex
Complex formed by binding of activated initiator tRNA to small ribosome subunit
Initiation Complex
When the large ribosome subunit sets w/ tRNA at P site at the beginning of translation
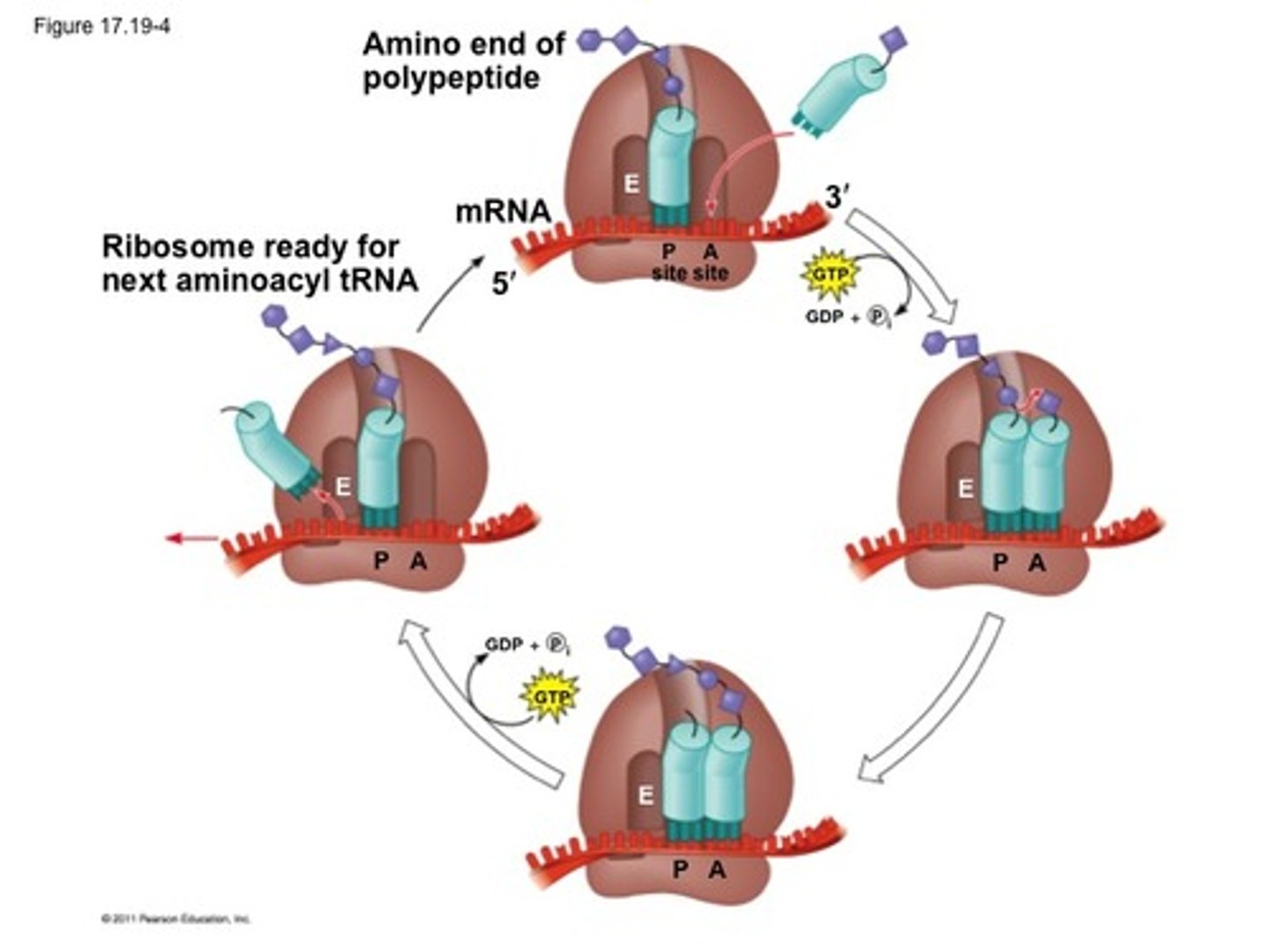
Translation Elongation
**One cycle adds ONE AA to growing polypeptide chain
1. tRNA w/ complementary anticodon binds to A site (on right, near 3'), pairing w/ mRNA via hydrogen bond
2. AA on tRNA in P site forms a peptide bond TO AA in A site (attaches polypeptide chain to amino acid on A site)
3. tRNA in P site becomes in active, all AA on tRNA in A site
4. Ribosome moves down mRNA, codon by codon (5'-->3')
5. Inactivated tRNA moves to E site and exits, tRNA w/ polypeptide in A site moves to P site
6. Different tRNA entered A site and process repeats
Translation Termination
Disassembly of Components
Elongation continues until stop codon
Release factor enters A site, adds water instead of amino acid, releasing polypeptide and disassembling ribosome
Modification of Polypeptides (5)
1. Remove methionine
2. Change AA's r-chains, including phosphorylation or addition of carbohydrates (to help w/ tertiary bonding)
3. Fold polypeptide to stabilize its tertiary structure, using disulfide bonds
4. Remove part of polypeptide to convert propeptide into mature peptides
5. Combine two or more polypeptides to form quaternary structure, can also add non-polypeptide groups (prosthetic) to quaternary structure to make conjugated proteins
Propeptide
A polypeptide created by a ribosome that has not yet been modified
Pre-proinsulin
Inactive insulin precursor (110 AAs) just formed by ribosome, still contains signal peptide
Proinsulin
Precursor of insulin, formed after protease removes 24 AAs (signal peptide) from n-terminal
Pre-proinsulin to Insulin Conversion
1. Insulin gene produces pre-proinsulin
2. protease removes some AAs (called signal peptide) from n-terminal to form proinsulin
3. Proinsulin folded w/ 3 disulfide bonds to stabilize tertiary structure
4. Other proteases break peptide bonds at 2 points, produces 2 separate chains (1 removed)
5. A-chain and B-chain (2 formed chains) are held together by disulfide bonds made earlier
6. 2 AAs removed from B-chain to yield mature insulin
Recycling Amino Acids (Reasons, 3)
Sustaining a functional proteome requires constant protein breakdown and synthesis bc most proteins have a relatively short life.
Cell's activity changes and protein is no longer necessary (ex moving from 1 stage in cell cycle to the next)
Protein structure is easily affected by free radicals or reactive chemicals, proteins become misfolded or denatured and not functional
Proteasomes
Break down proteins that no longer function
Identifies proteins for destruction by tagging them w/ ubiquitin (small proteins, acts as signals for proteasomes)
Proteasomes unfolds and feeds protein into central chamber, active sites of multiple proteases within proteasome break down proteins into oligopeptides, which move out of proteasome
Oligopeptides further digested into AAs in cytoplasm
Ubiquitin
A protein that attaches itself to faulty or misfolded proteins and thus targets them for destruction by proteasomes
Oligopeptides
Short polypeptides