L4: Regulation of mitochondrial and chloroplast biogenesis
1/55
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
56 Terms
How do both mitochondria and plastids develop
By division of pre-existing organelles
i.e→ there is not de- novo synthesis
But theri functional state is affected by their
Developmental stage
Environmental conditions
Example of Environmental conditions affecting this
Oxygen
Light
Affect on mitochondrial with the absence of oxygen
mitochondria electron tranfer cannot occur as O2 is the final electron acceptor
Catalysed by cytochrome oxidase (complex IV)
COX
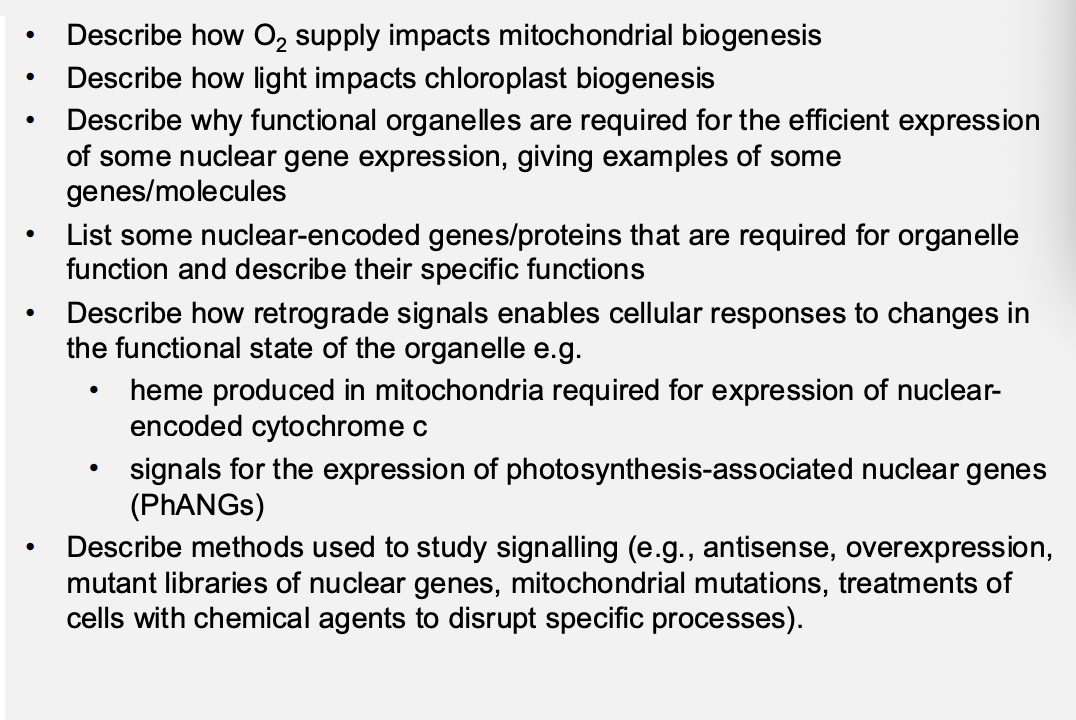
Yeast show how mitochondria development can be alted by O2 levels
Anaerobic conditions (or on glucose)→ use fermentaion and mitochondria are small with few internal membranes and no respiratory complexes→ PROMITOCHONDRIA
Aerobic conditions→ Mitochondria are functional→ fully develop when O2 is supplied and/or all glucose consumed
Why does O2 have this affect on mitochondria?
Because O2 is reuired by haem biosynthesis
and haem is a cofactor for cytochromes
therefore→ when there is oxygen→ mitochondria can develop fully
note: Haem is produced in the mitochondria
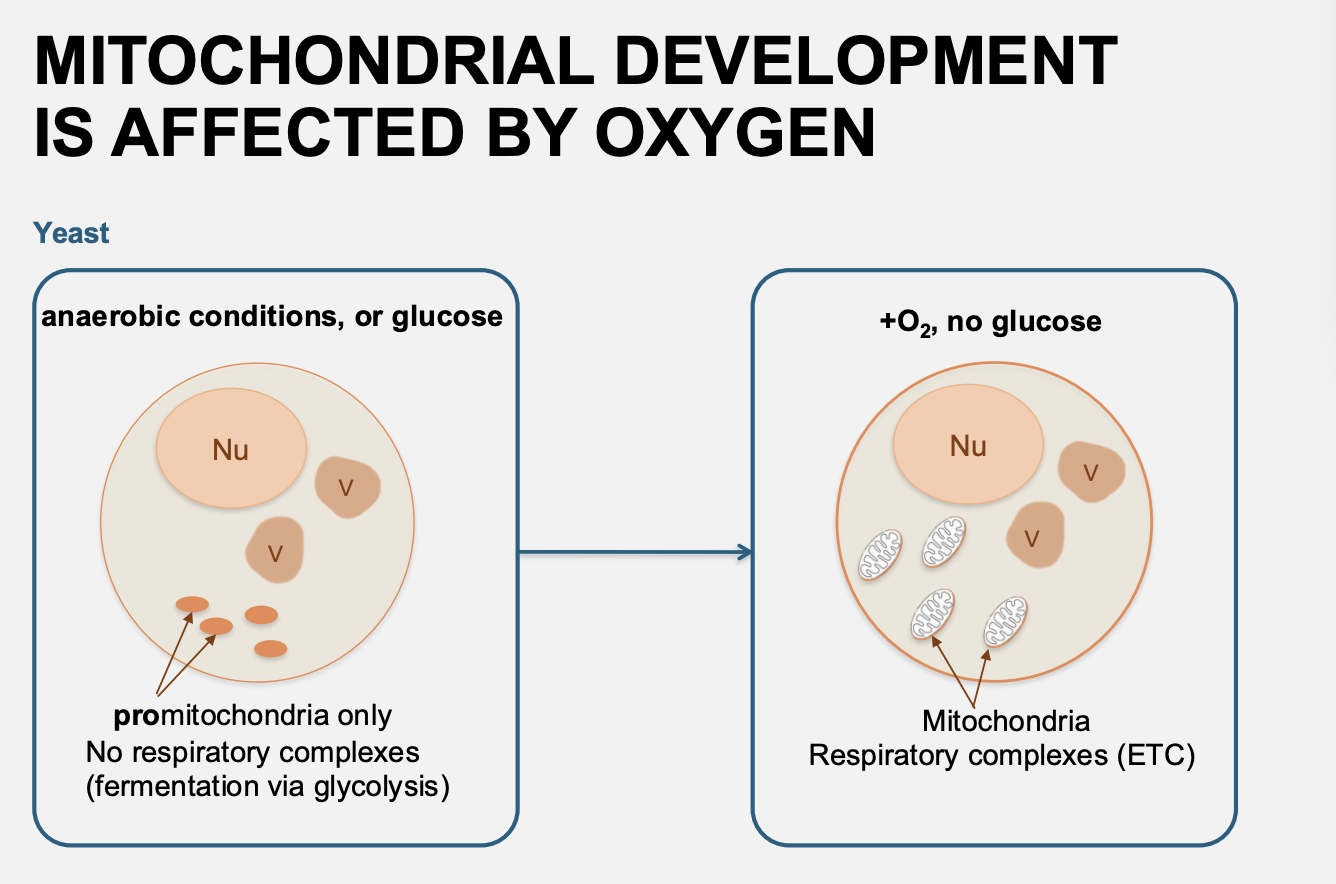
Not only is there regulation of haem (found in the mitochondria) but Oxygen regulates the nucleur encodes genes for mitochondria→ e.g CYC1 what is it and how regulated
Nuclear encodde CYC encoding cytochrome c
Regulated by → HAP1 (haem-activator protein 1)
How does it set up a kinda of feedback cycle, affect by oxygen
Oxygen→ makes Haem
Haem binds to HAP1
HAP1 activates Upstream Activating Sequences (UASs) in the CYC promoter
This promotes CYC expression and the making of cytochrome c WHICH Haem binds to for mitochondrial function
i.e: Overall need oxygen for haem and then the haem can be used for its actual function as well as transcibed the protein that it needs for its function (cytochrome C)
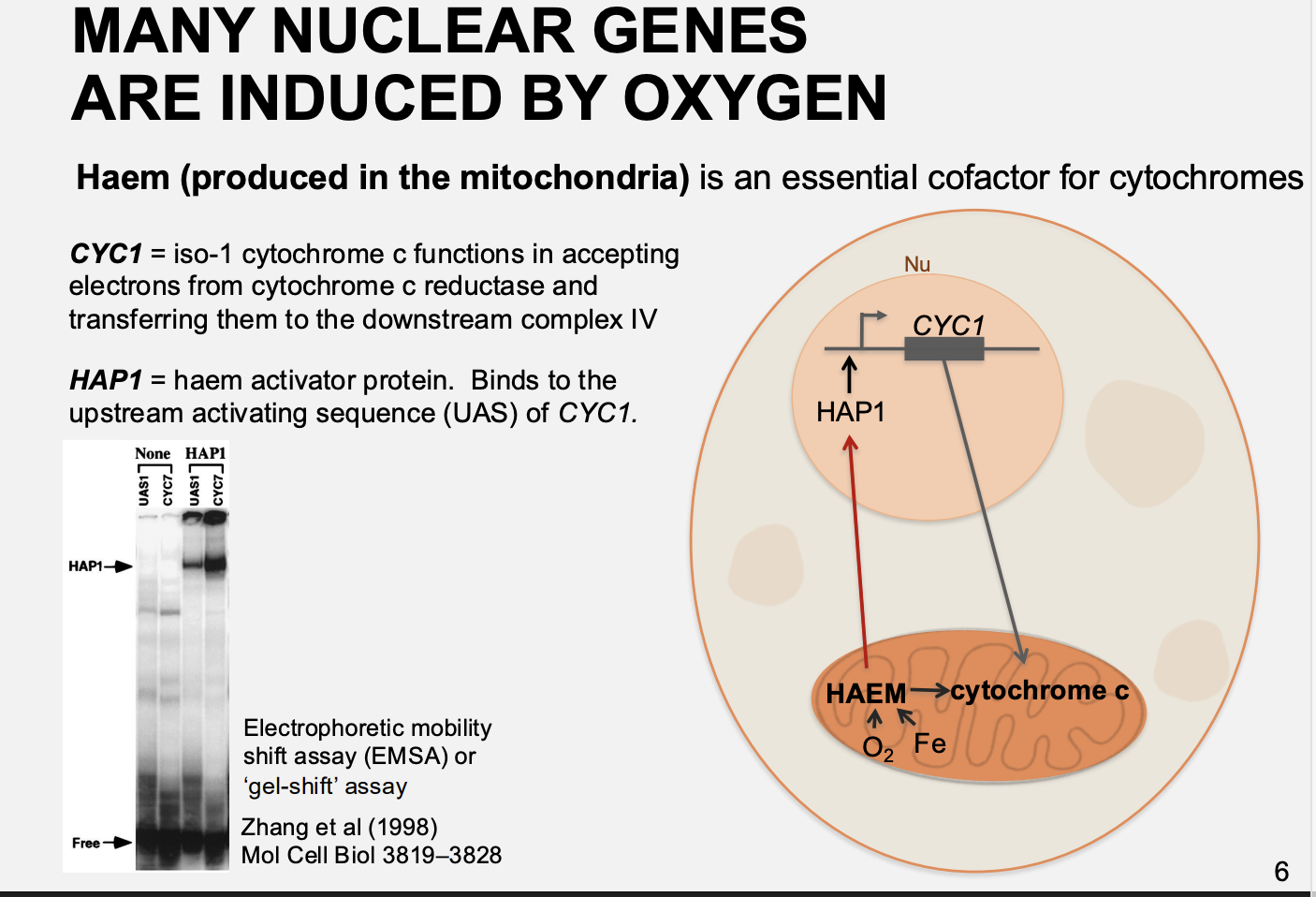
How does oxygen regulate genes that are not directly require Heam for regulation?
e.g COXIV, V, VI
haem stimulates HAP1
then the HAP1 activates HAP2/3/4/5 complex
which then goes to promote the COXIV, V, VI
makes cytochrome c oxidase/subunit IV
i.e→ does not bind directly to haem but still responds to it via the levels of HAP1 activation of gene for HAP4
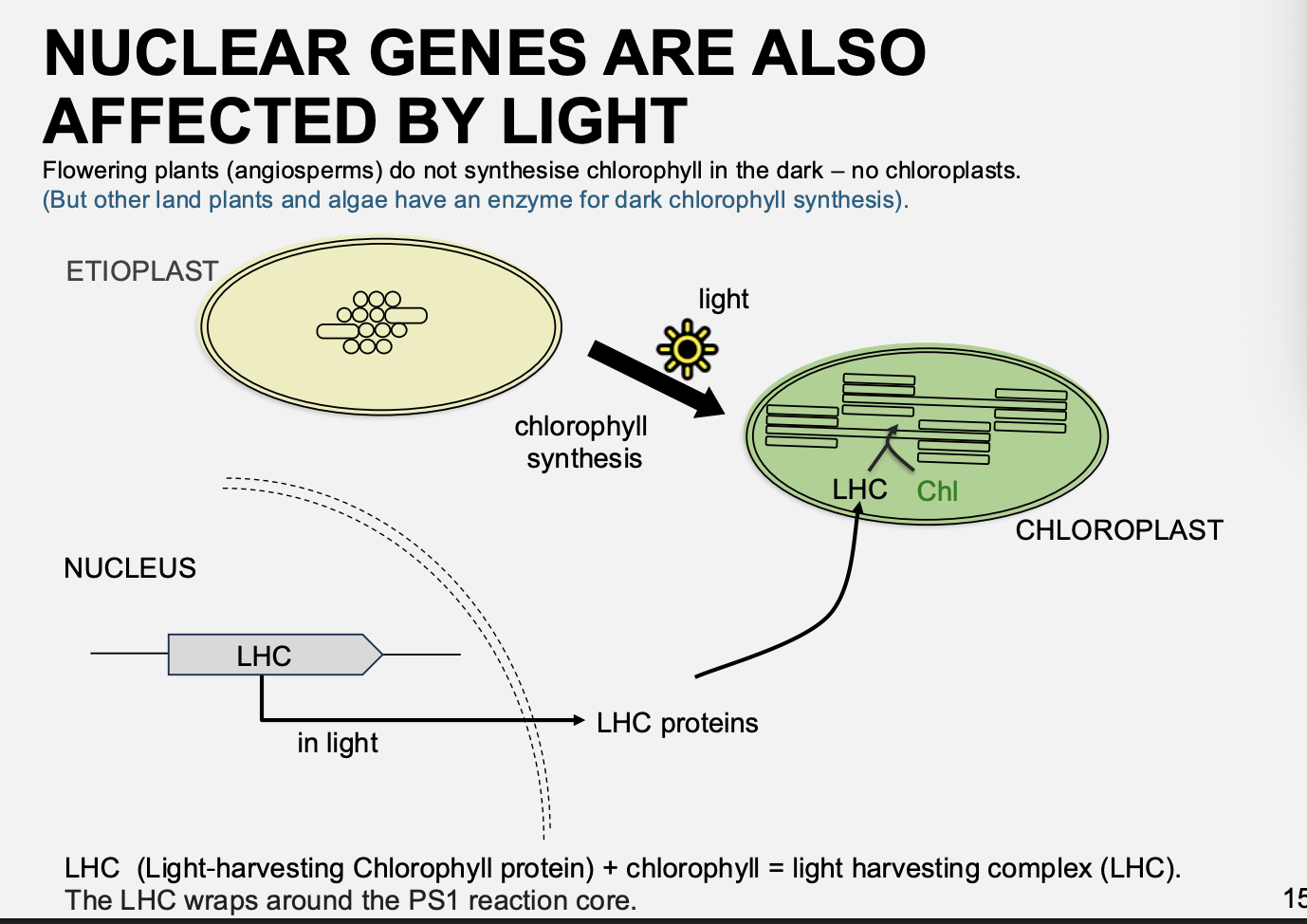
How does light change the function of chloroplasts
Flowering plants
Dark→ do not synthesise chlorophyll in the dark→ no chloroplasts→ etioplasts
Light→ make chlorophyll→ chloroplasts
Note: some land plants and algae in the dark…
have an enzyme for dark chlorophyll synthesis
dark-operative protochlorophyllide oxioreductase (DPOR)
So are green in the dark
i.e→ still shows how chloroplast function can be altered by the environment
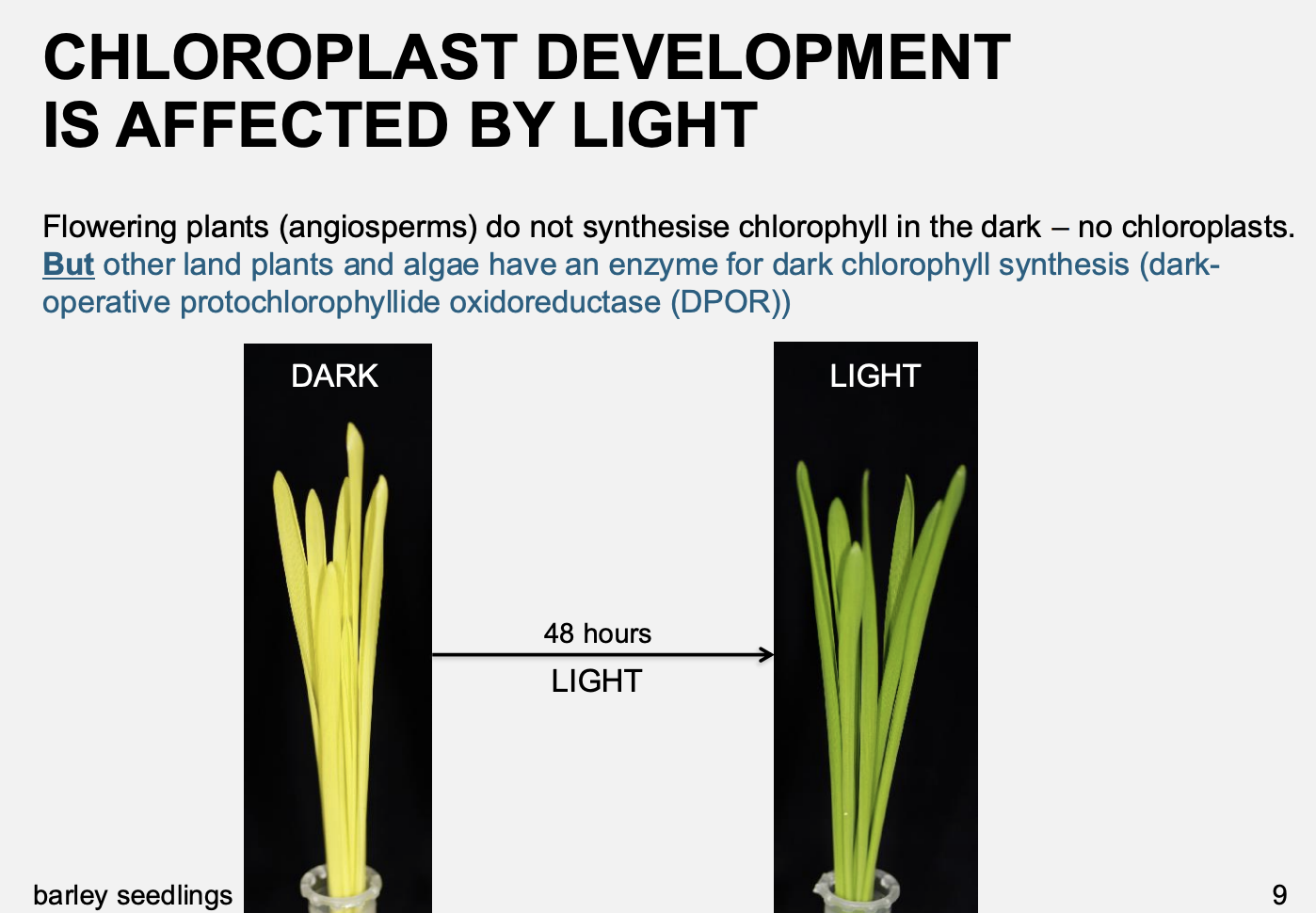
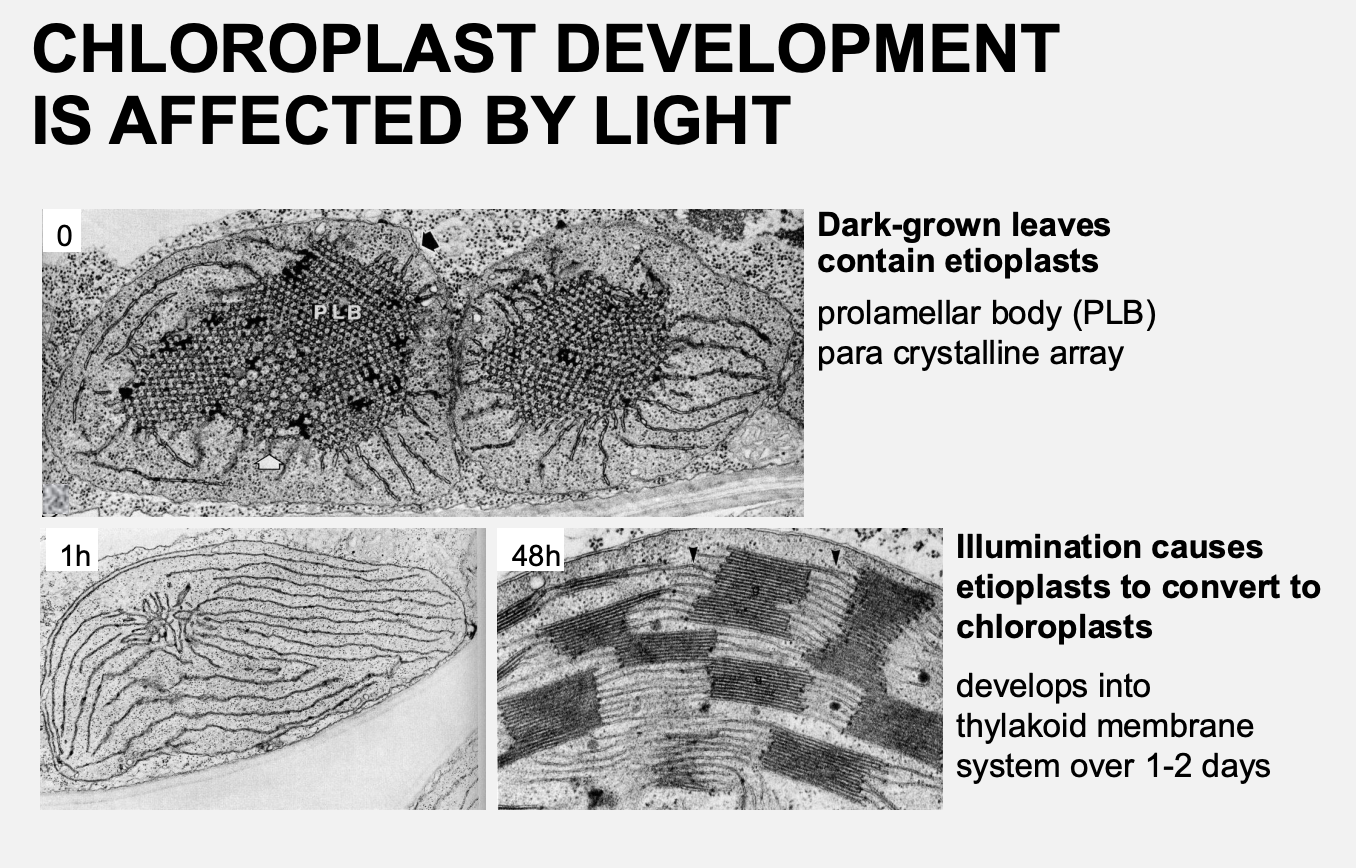
Chloroplast development from dark to light
Etioplast→ Crystalline structure
Prolamellar body
para crystalline array
all the components needed for chloroplast but just not in correct formation
Chlorolpast→ thylakoid membrane developed over 1-2 days
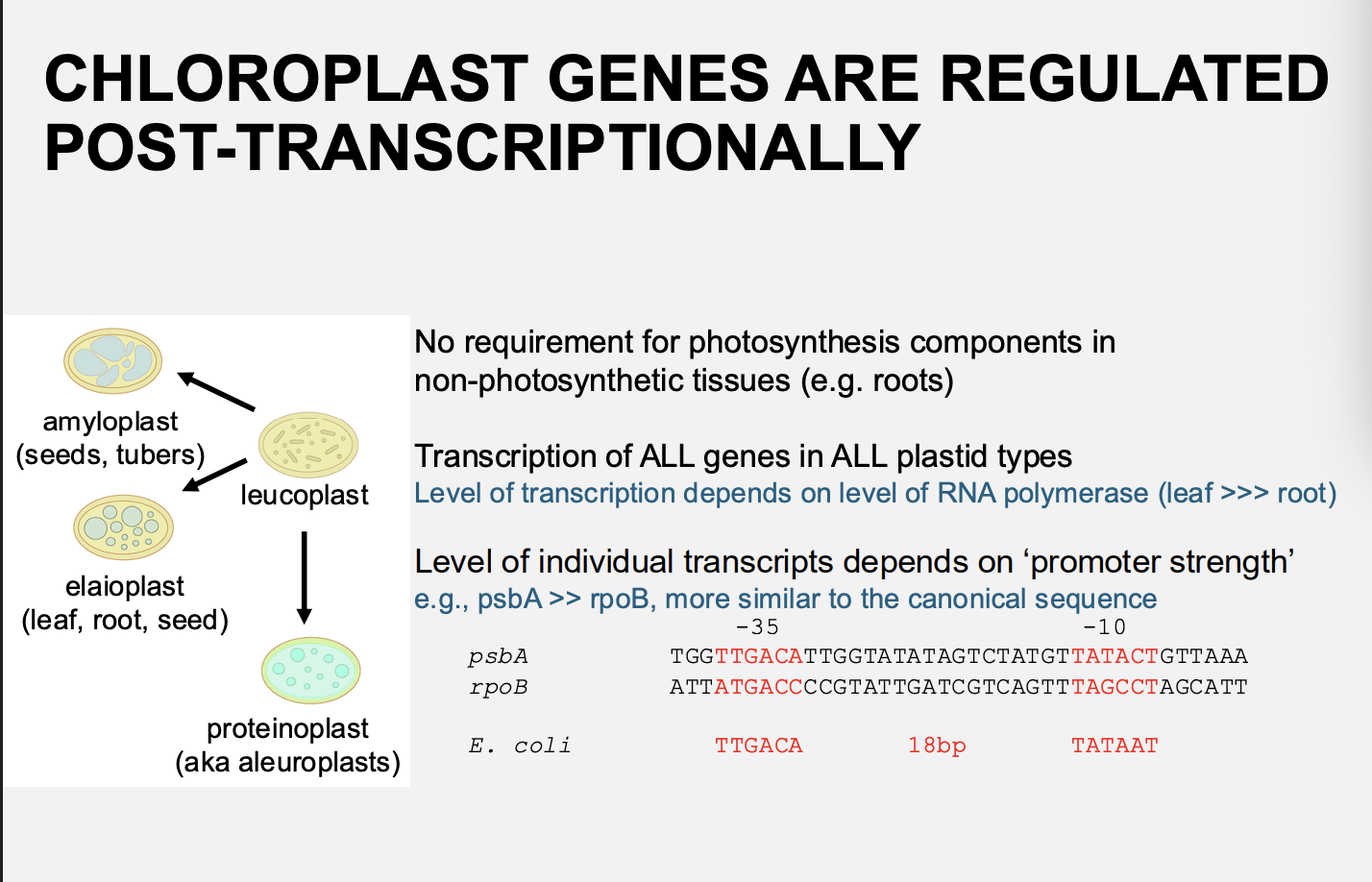
Even though there is no need for grene expression of phosotynethesis components in non plastids such as amyloplasts…
genes are transcibed in all tissues
there is no large differences in relative rates of transciption of individual genes in different tissues
and
no evidence for repressor or activator proteins for most individual genes
Then what are transcription levels dependent on?
Levels of RNA polymerase
The level of transcription of individual genes depends on promoter strength
But if transcription levels in amyloplasts and chloropasts are the same, why don’t amyloplasts contain photosynthetic machinery?
Chloroplast genes are regulated post-transciptionally:
I.e even through there is transcription of all genes in all plastid types
Level of transcription depends on level of RNA polymerase (leaf>>>root)
The level of individual transcripts depends on ‘promoter strength’
Extensive translational regulation→ what does this require
Positive regulation of translation:
i.e in the DARK→ there are transcripts but not translated
in the light→ the they get translated
There are many ways for this to happen
Example: Redox-regulated translation-initiation of psbA→ What is it
Forms the D subunit of the PSII core
Light→ translated
Dark→ not translated
As an mRNA transcript→ the psbA has a stem-loop structure in the 5’ UTR
Chloroplast polyadenylate-binding protein (cPABP) specifically binds this and is essential for translation
and a protein disulfided isomerase (cPDI) modulates the binding of this by chaning redoc status
Example: Redox-regulated translation-initiation of psbA→ How is its translation regulated: in the light
Light
PSI redox reactions cause FD reduced
This reduced FDTR
this Reduced protein disulfide isomerase (cPDI)
through redox potential or adenosine 5’-diphosphate-dependent phosphorylation
this allows the chloroplast polyadenylate-binding protein cPABP to bind to the 5’UTR step loop
binding to stem loop turns translation ON
PsbA translation is ON
(in the dark)
The cPDI is oxidised
so no cPABP binding
not attached to stem loop
translation off
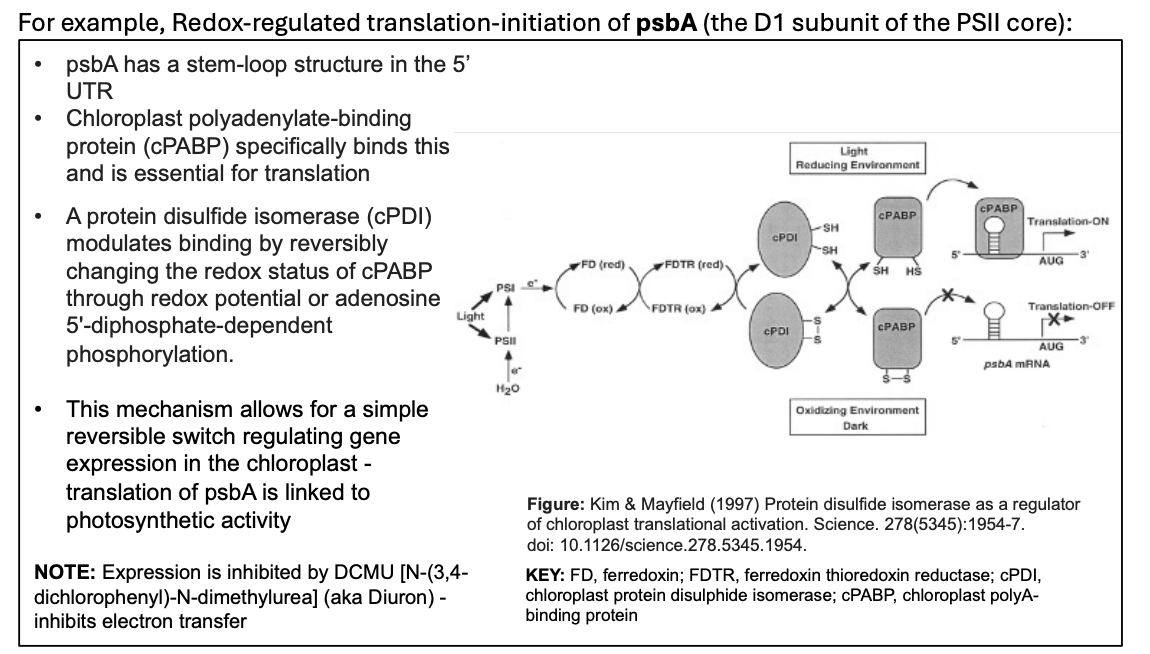
How we know that there is electron transfer/ redox reactions happening
Expression is inhibited by:
DCMU [N-(3,4- dichlorophenyl)-N-dimethylurea] (aka Diuron)
which inhibits electron transfer
Therefore what does this mechanism allows for
a simple reversible switch regulating gene expression in the chloroplast
translation of psbA is therefore linked to photosynthetic activty
overall: Shows how the environmental cue can change the function of chloroplast
Light can also alter chloroplast function/plastid type by regulating what
nuclear genes
instead of the chloroplast genes
→ e.g the transciption of LHC controlled by light
What is the light-harvesting complex
(antenna complex or LHC)
an array of protein and chlorophyll molecules
embedded in the thylakoid membrane of plants and cyanobacteria
transfer energy to one chlorophyll a molecules at the reaction centre of a photosystem
The transcription of LHC is controlled by light
note: this is not post-transciptionally like above!
its genes are encoded in the nucleus
and so light must regulate its transcription
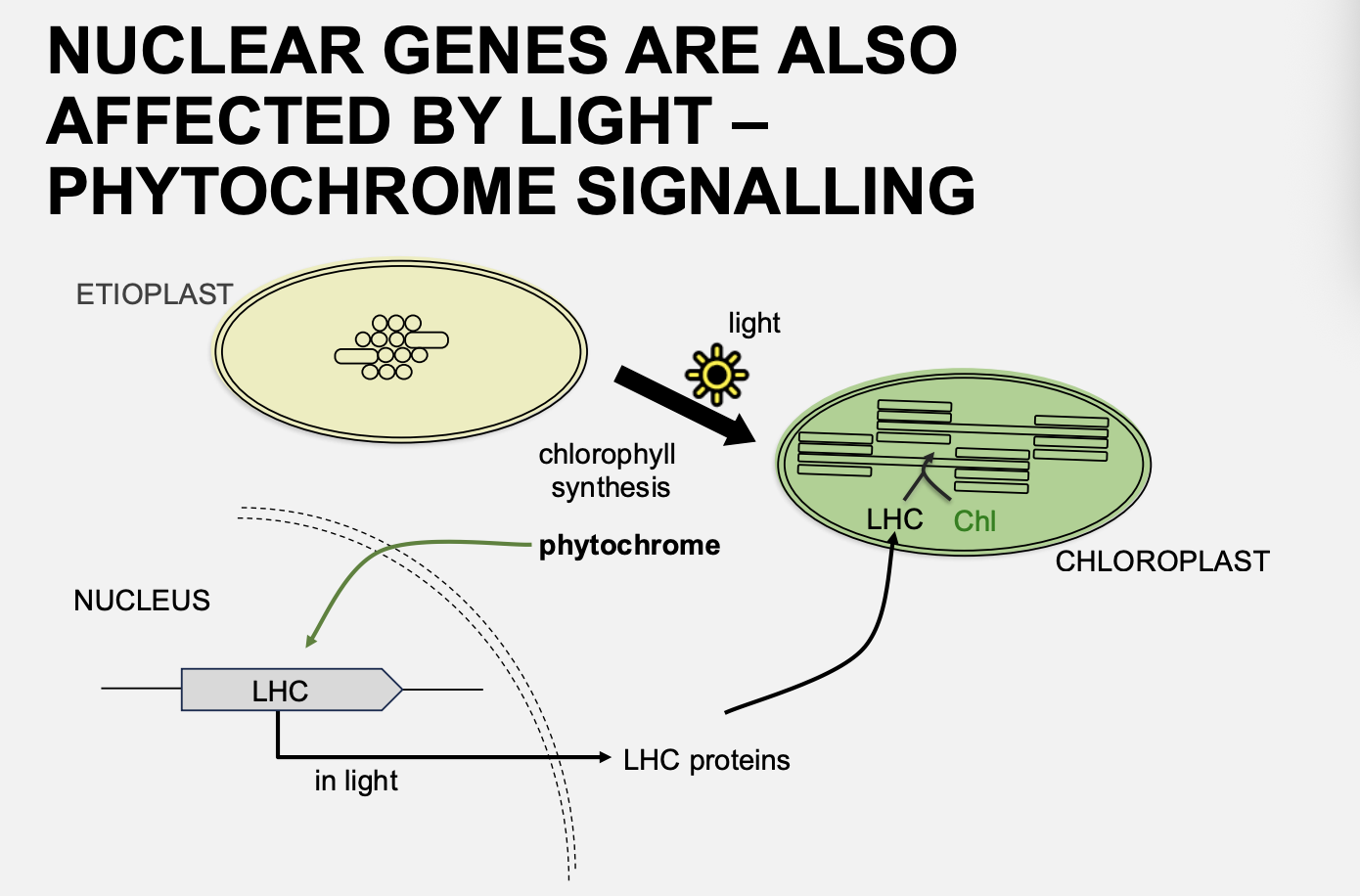
What aids this transciption regulation?
phytochrome signalling
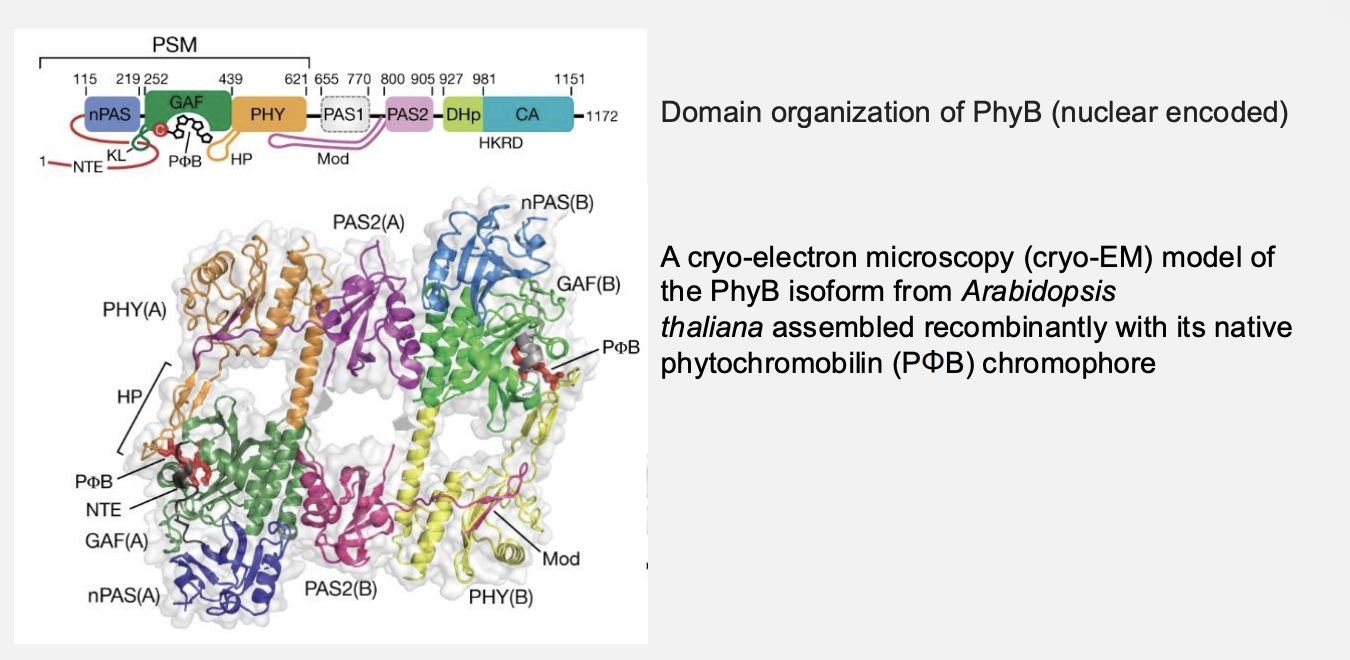
What are phytochromes
Photoreceptors autocatalytically bound to a bilin prosthetic group
they are made downstream of haem
note: in the dark when there is no chlorophyll made, haem feedback inhibits ALA synthesis
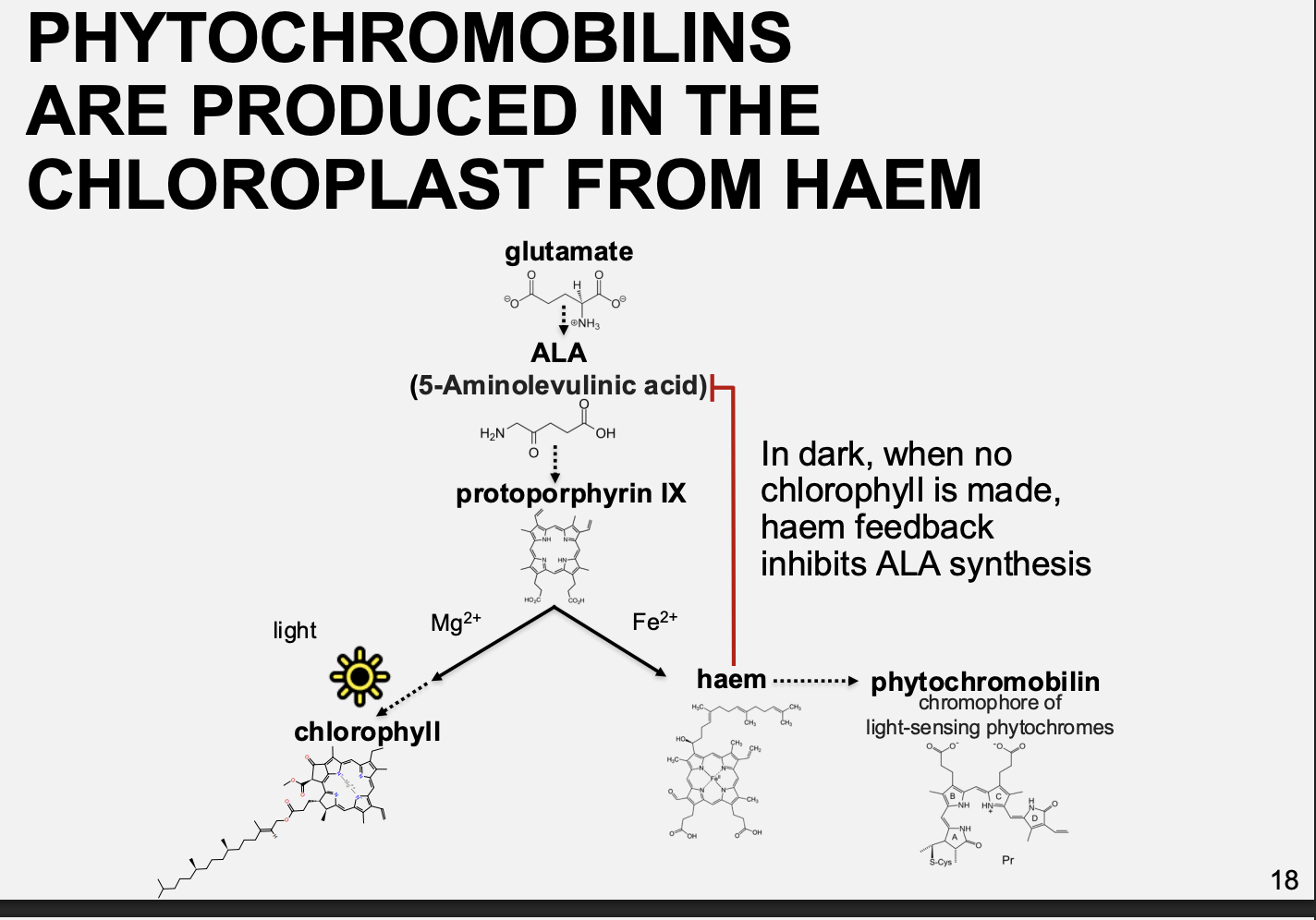
How do they act as photoreceptors
With light, changes the position of ring D
red light→ activation→ Pfr cis isomerisation
far red→ Pr→ inactivation→ transisolerisation?
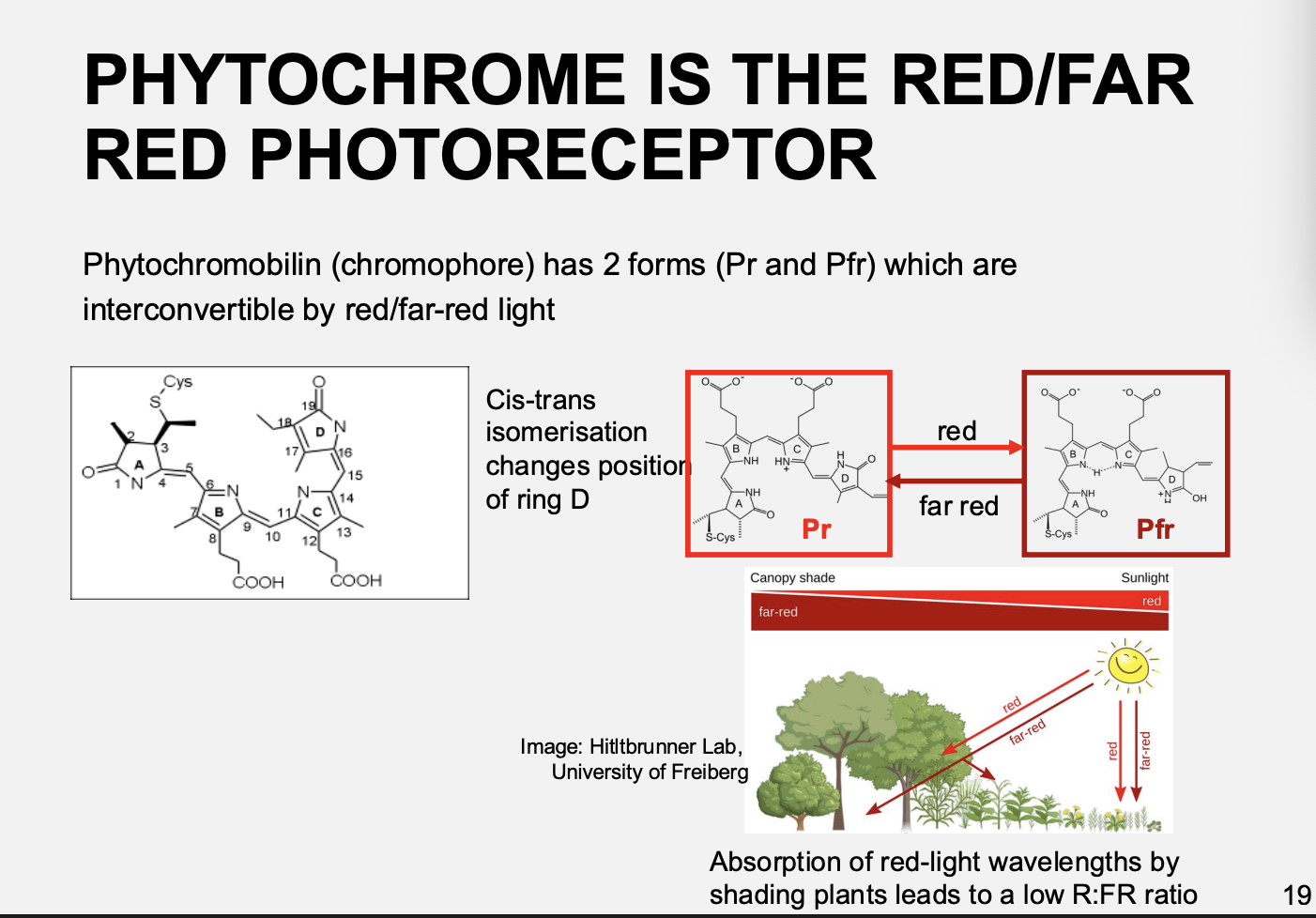
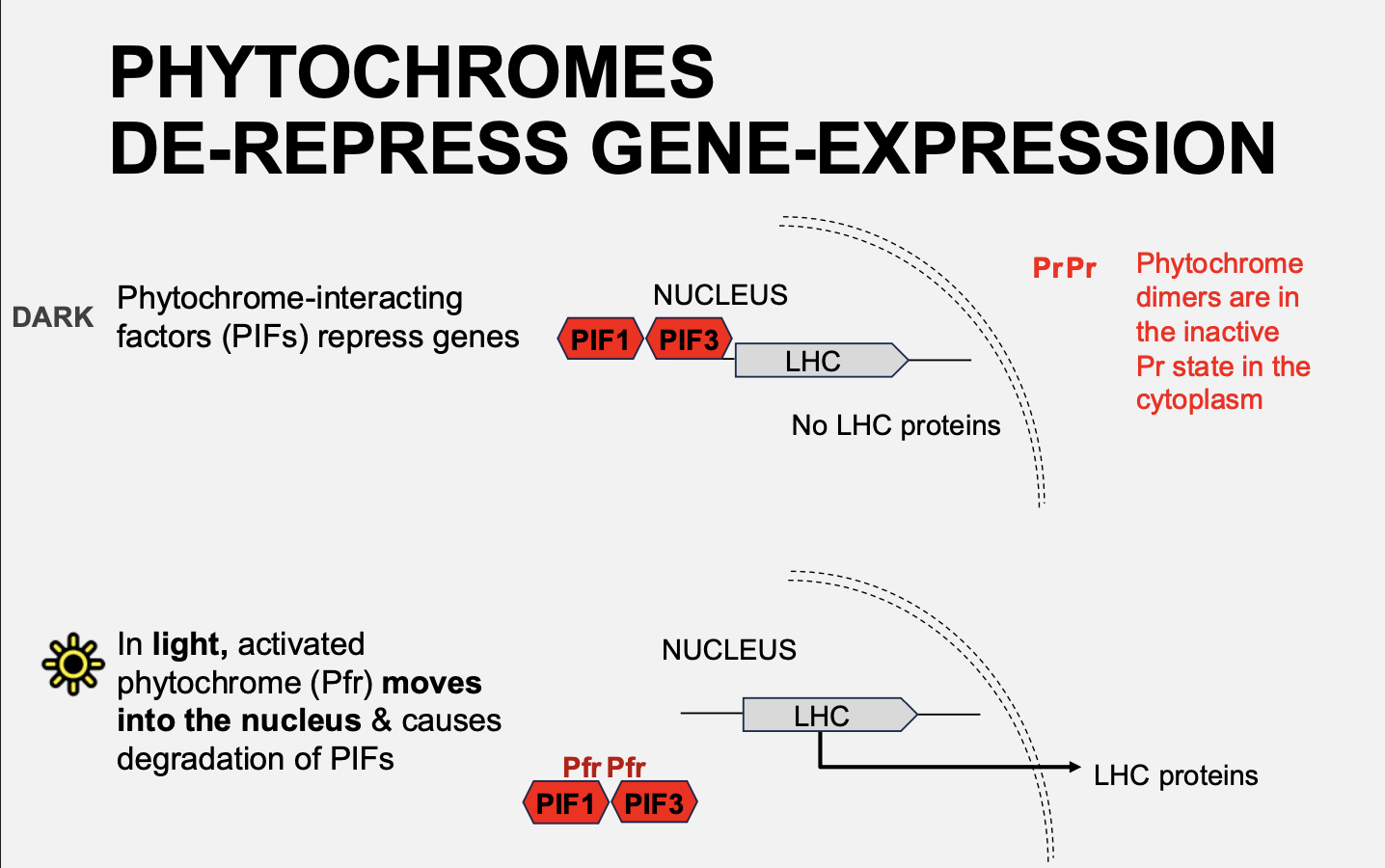
How does overall Light regulate LHC expression
Activation by red light
Pfr conformation
can translocate fro the cytosol to the nucleus
interact with the repressive transciption factors (PIFs) that bind the promoter of the genes including LHC
Allows LHC transciption
overall: phytochoromes de-repress gene-expression
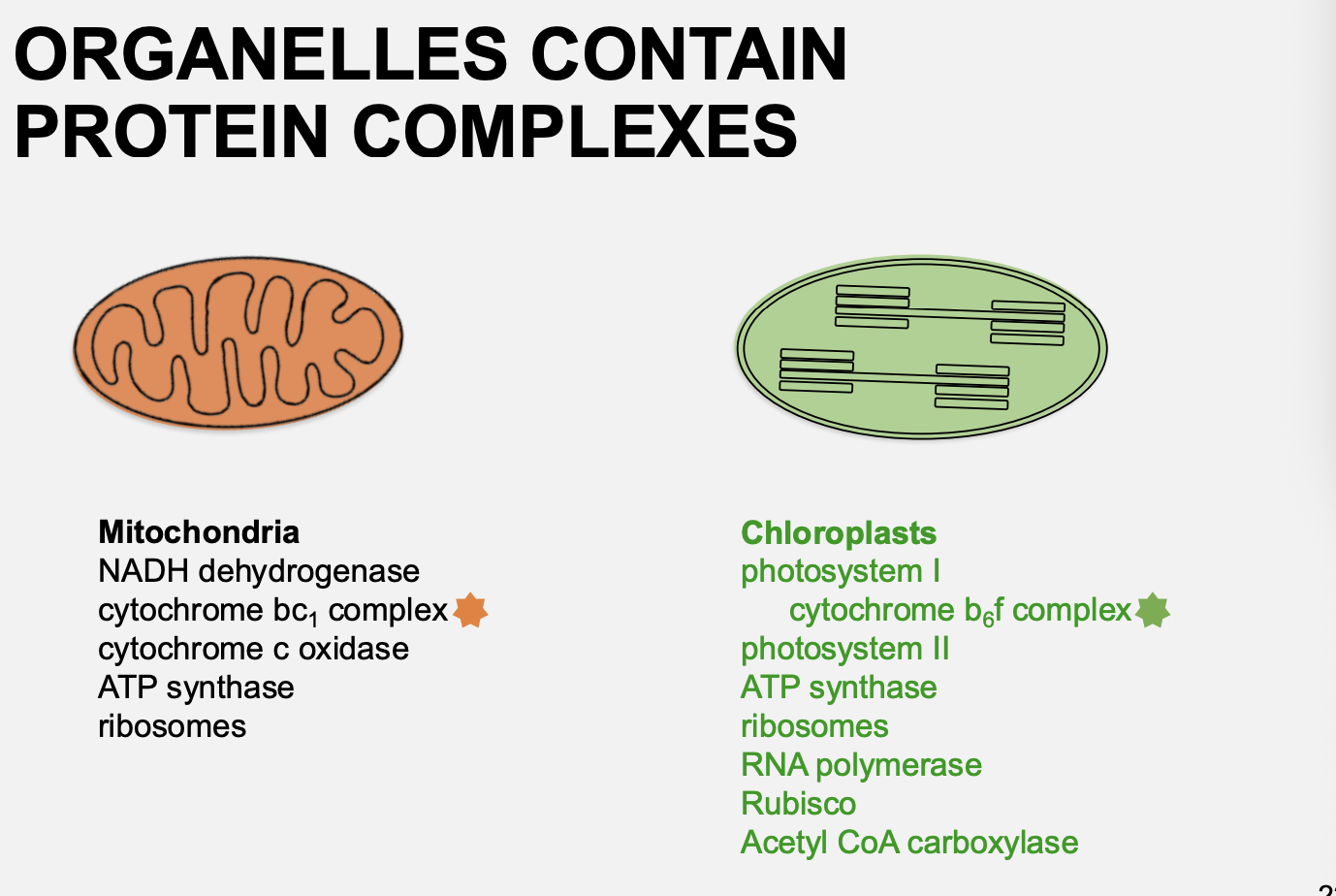
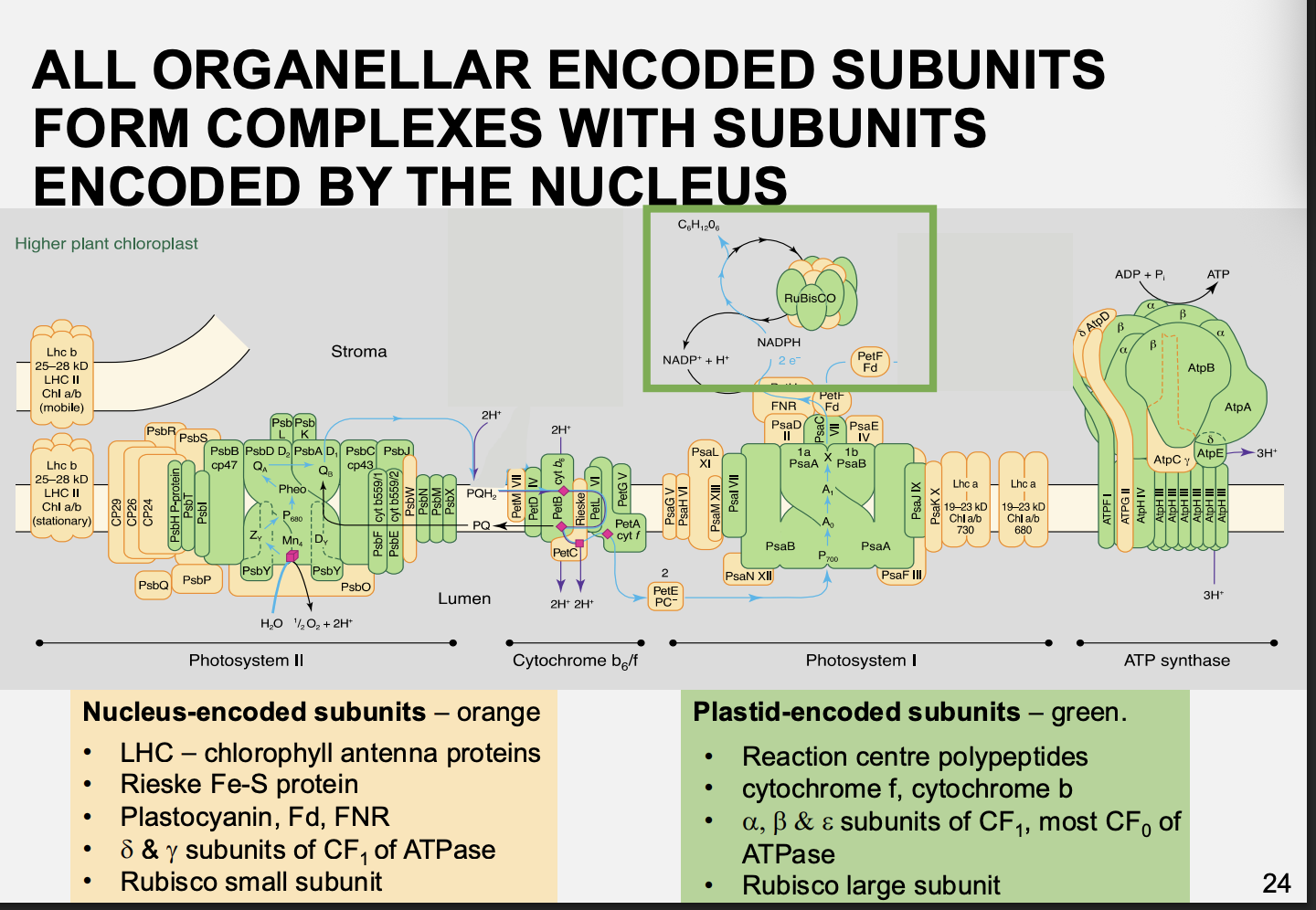
Coordintion of nuclear and organelle gene expression: complexes in oragnelles contain protein complexes that are encoed by
Both the organelles and the nucleus
e.g mitochondrial→ cytochrome c oxidase/subunit IV complex
e.g Chloroplast→ cytochrome b6f complex
Therefore: it is important that the ratio of nuclear to chloroplast proteins is correct and coordinated correctly
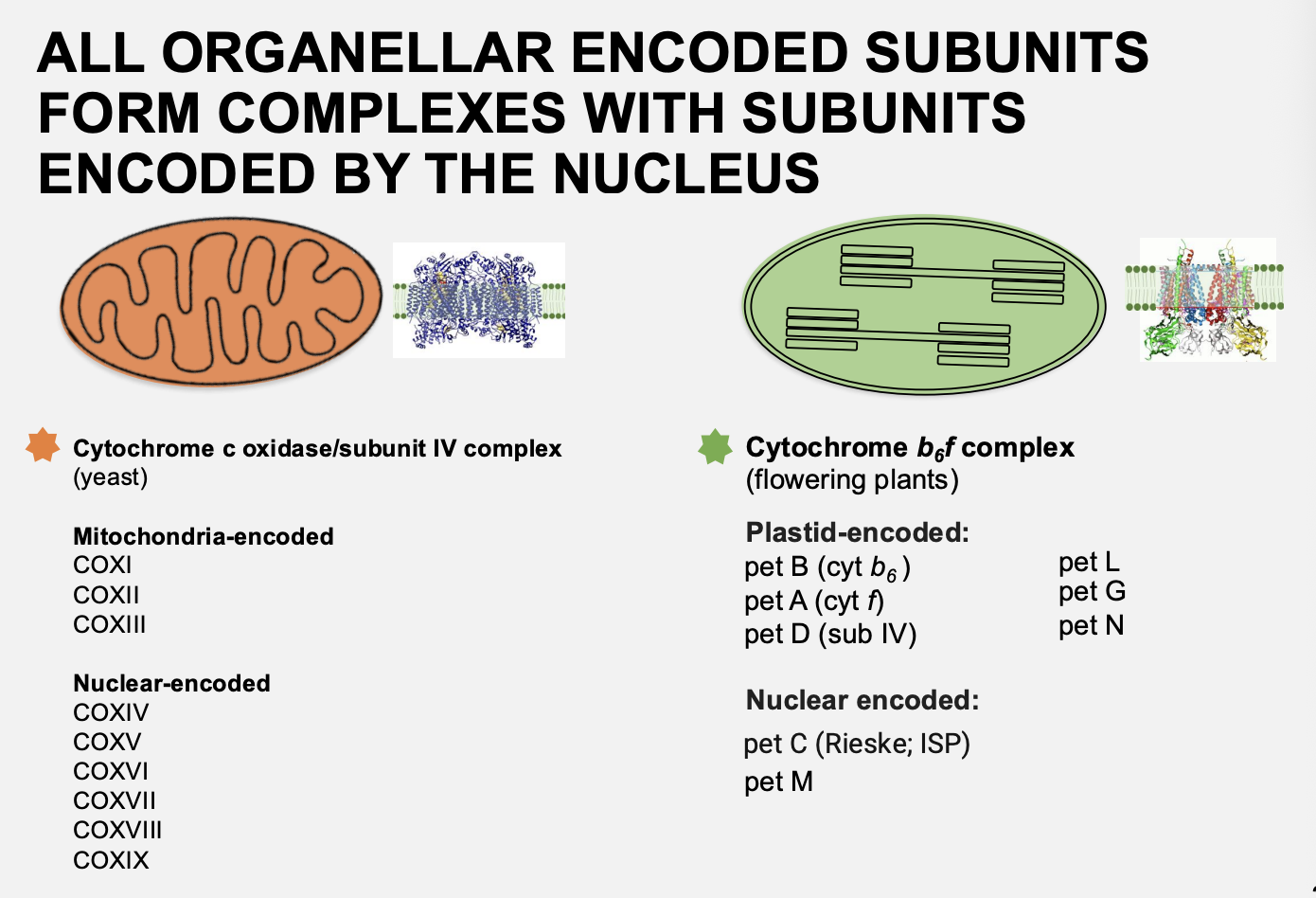
Picture of distribution of nuclear vs chloroplast encoded genes
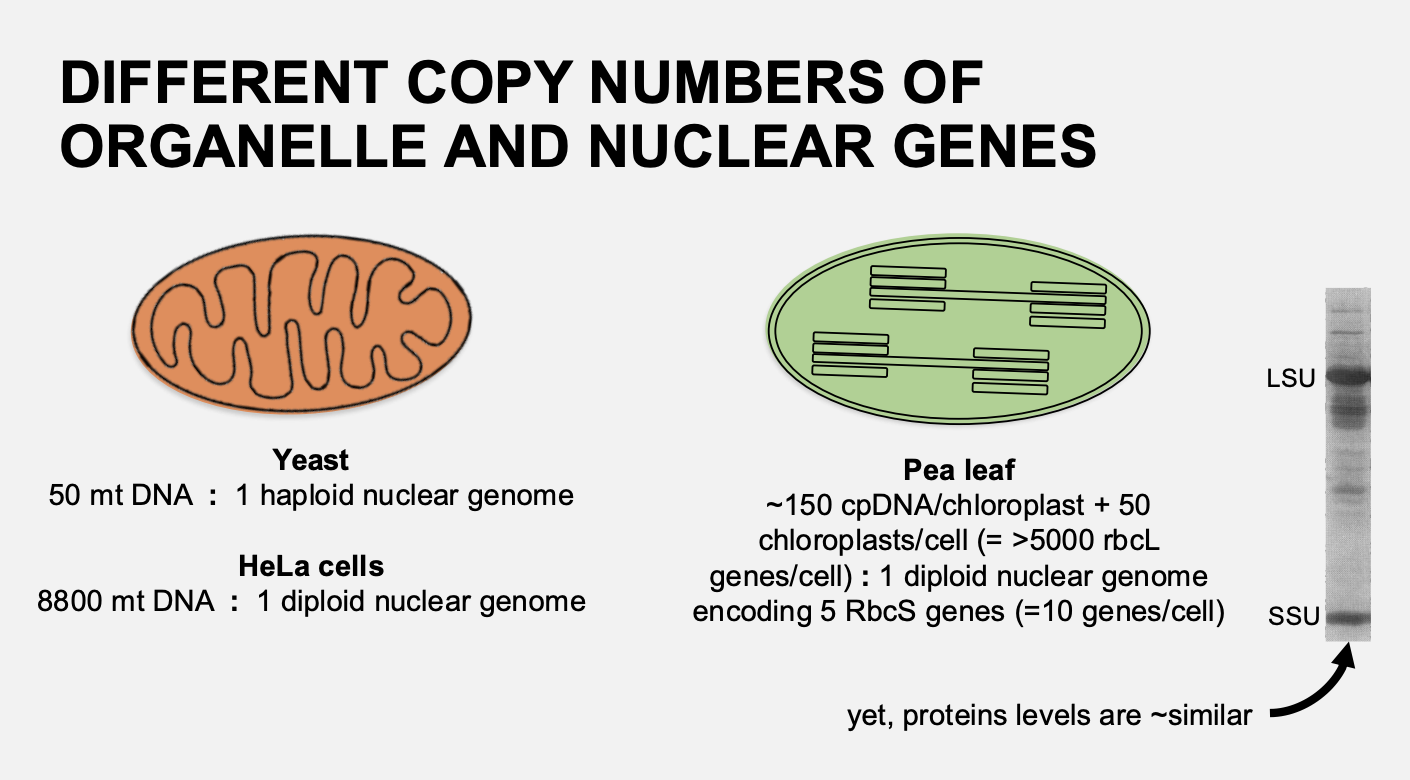
Problem with achieving this ratio
There are different copy numbers of organelles and nuclear genes
very large im blanace
Example:
Mitochondria→ Yeast 50 mit DNA molecules : 1 haploid genome (50:1)
Human HeLa cells 8800 mit DNA molecules : 1 diploid genome (8800:1)
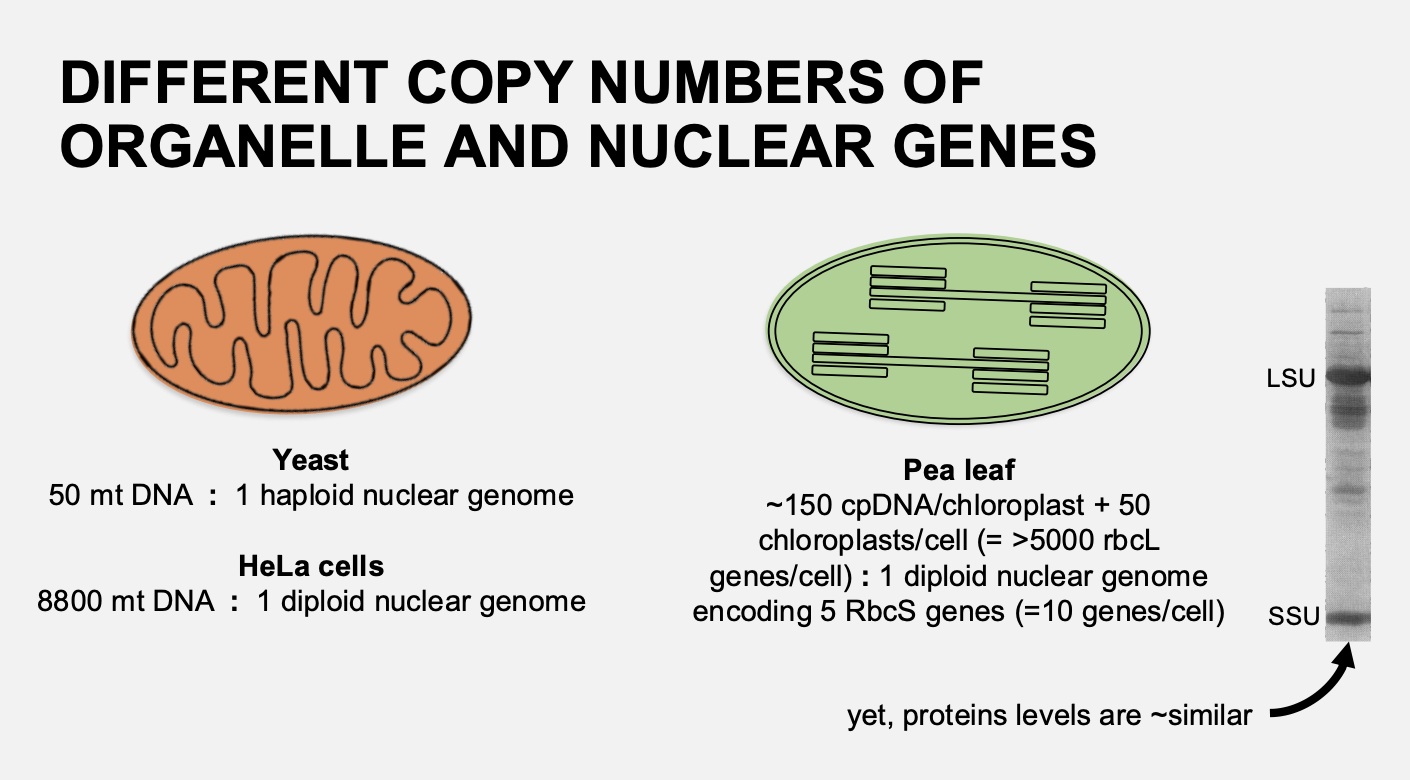
Why is the nuclear and mitochondrial expression genes not tightly coordinated in yeast?
synthesis of nuclear-encoded subunits can occur in absence of mitochondrial gene
e.g in petite p- mutatns (rho)
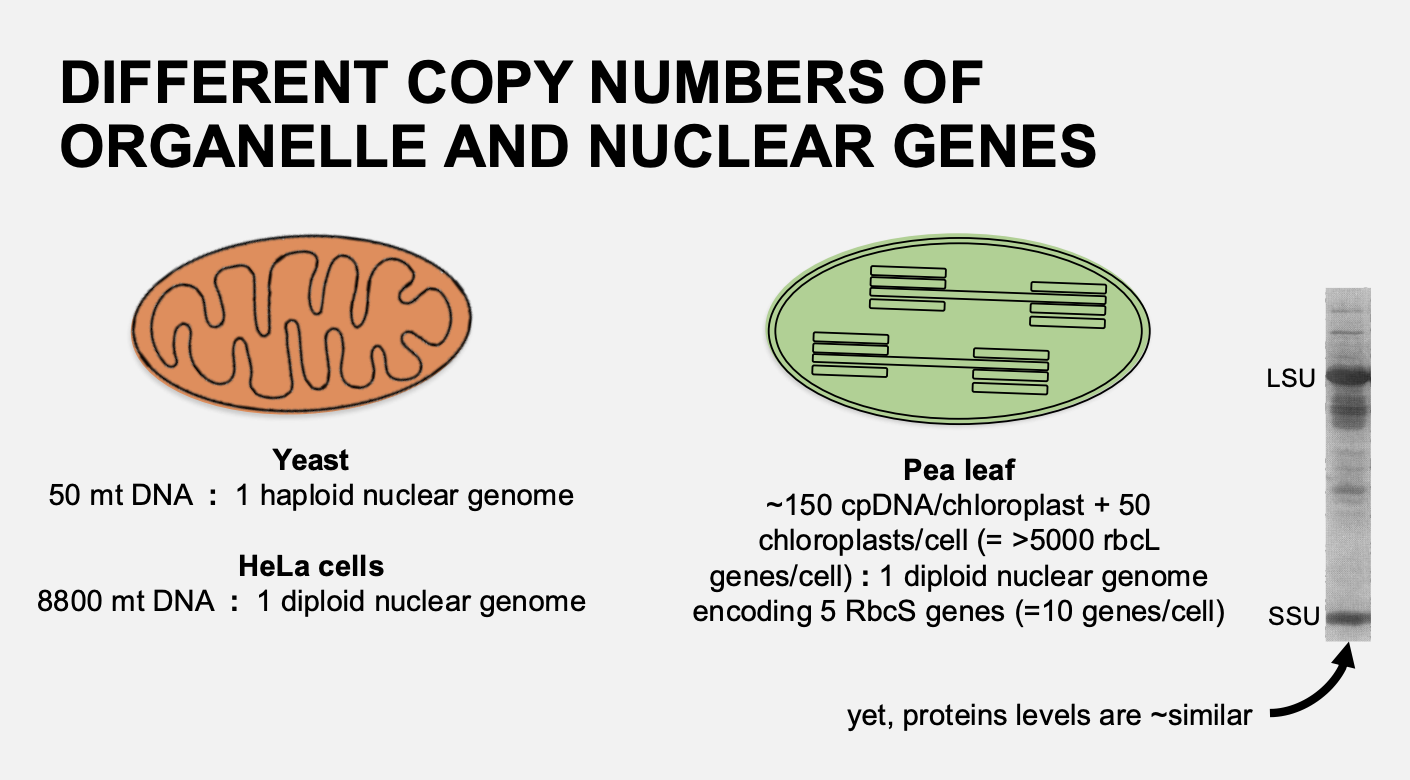
Example: Chloroplast protein Rubisco→ how is it encoded
Ribulose 1,5-bisphosphate carboxylase
L8S8 structure (large and small subunits)
Large subunit (LSU) (52 kDa; rbcL chloroplast gene: >5000 copies per leaf cell
Small subunit (SSU) (14 kDa); RbcS nuclear genes: 6-10 nuclear genes per cell
but→ see that in the northern blot→ the levels of LSU to SSU are similar
therefore→ there must be some kind of regulation of the translation
Evidence that the expression of protein subunits is not tightly coordinated
Protein synthesis inhibitors in vivo
Antisense RNA inhibition of S unit synthesis in transgenic plants
i.e→ there is coordination but it is not tight!
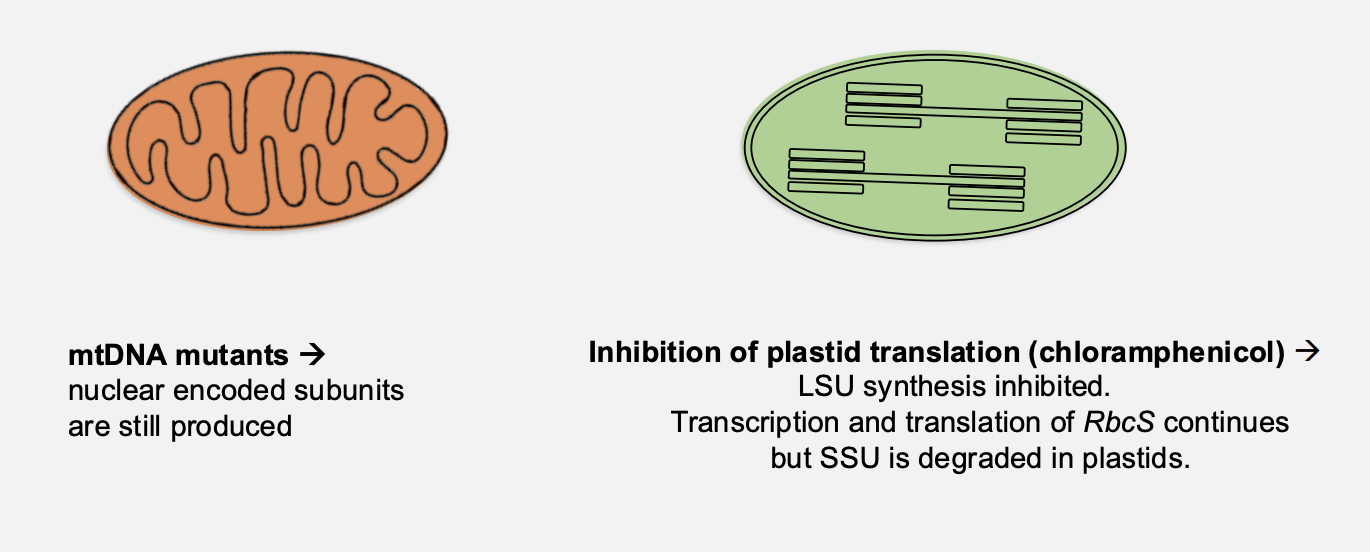
Protein synthesis inhibitors in vivo
cyclohexamide→ inhibits synthesis of SSUs (nucleus)
but rbcL transcription and LSU synthesis continues for some time
Chloramphenicol→ inhibits synthesis of LSUs (plastid)
but RbcS transcription and SSU synthesis continues for a short time
HOWEVER→ SSU des not accumulate and is degraded in the chloroplast
so there is no affect on the translation BUT there is still some degredation which might be involved in this regulation?
Antisense RNA inhibition of S subunit synthesis in transgenic plants
Tobacco plant produced containing antisense constrcut of RbcS cDNA
expressed from a consitutive promoter
result→ degredation of the mRNA and the decreased SSU synthesis
i.e it induced post-transciptional gene silencing
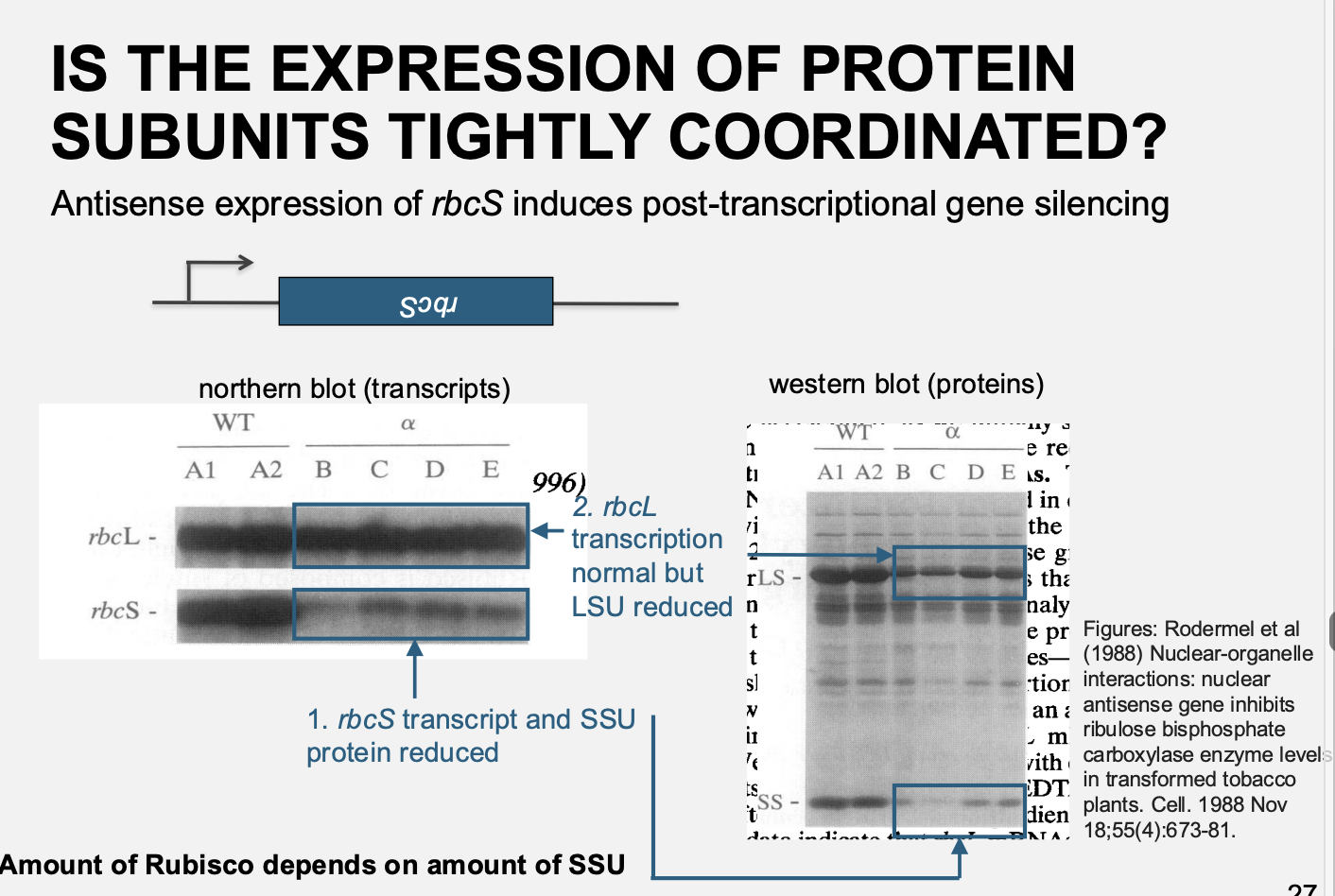
Summary of the Rubisco subunit reguation
the amount of rubsico correlates with the amount of SSU:
Reduction in SSU→ reduced accumulation os LSU, even if rbcL transcipt level is not affected
What does this suggest
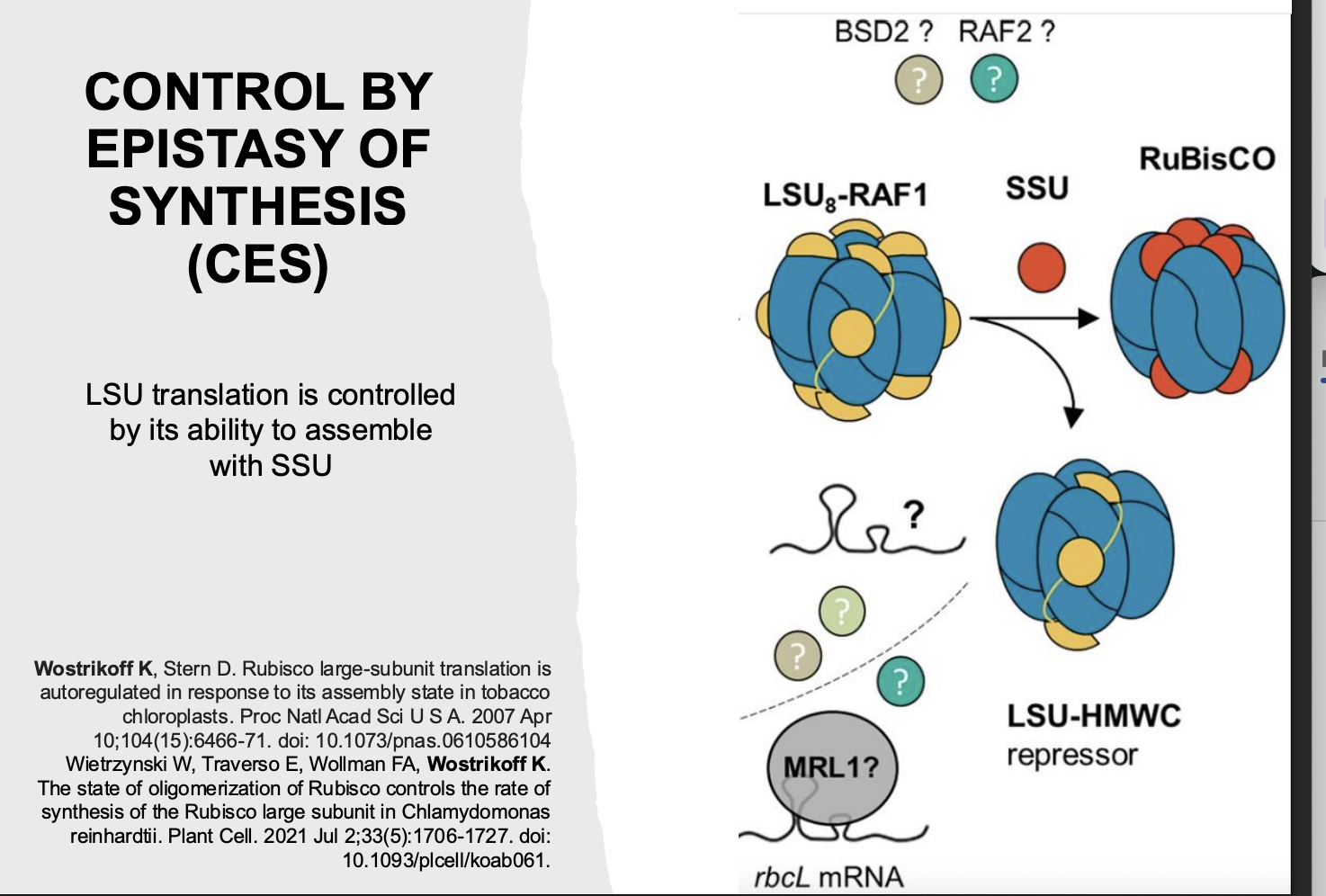
Assembly of Rubisco is driven by the availability of SSU and excess LSU subunits are degraded
How has this coordination been found to be achieved by
post-transciptional process→ ‘Control of Epistasy by synthesis’ CES:
LSU that is not complexed into Rubisco yet self- regulates by binding rbcL mRNA
i.e: In the absence of the SUU→ there is a repressor which causes the degredation of the LUU
This repressor system accounts for fluctuations in gene expression
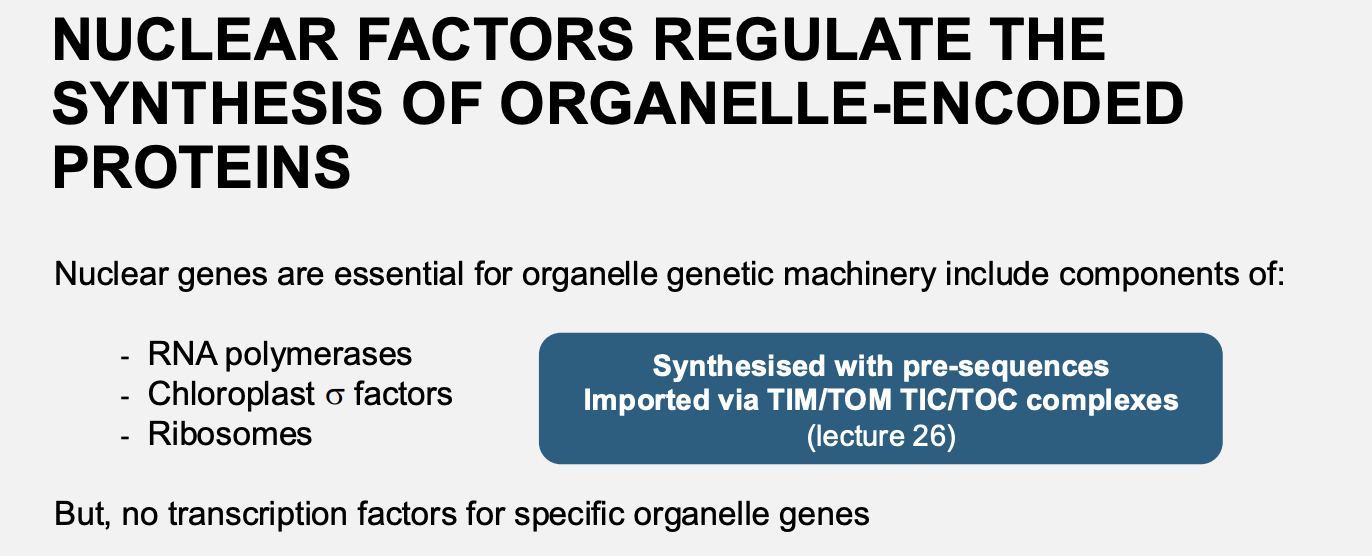
Nuclear factors for organelle biogenesis: Anterograde signalling→ nuclear genes are essential for organelle genetic machinery
The include components of
RNA polymerase
Chloroplast sigma factors
Ribosomes
They are synthesised with pre-sequences and imported via TIM/TOM or TIC/TOC
THEREFORE: the nucleus does
BUT they do not encode transcription factors for specific organelle genes
then how is post-transcitional regulation of mitochondrial genes done by the nucleus?
Major point of organelle gene expression control by the nucleus
Nucleus encoded proteins are also requred for the stability and translation of specific mRNAs in both mitochondria and chloroplasts
including editing
often, multilple nuclear genes are needed for a single organelle protein to be produced
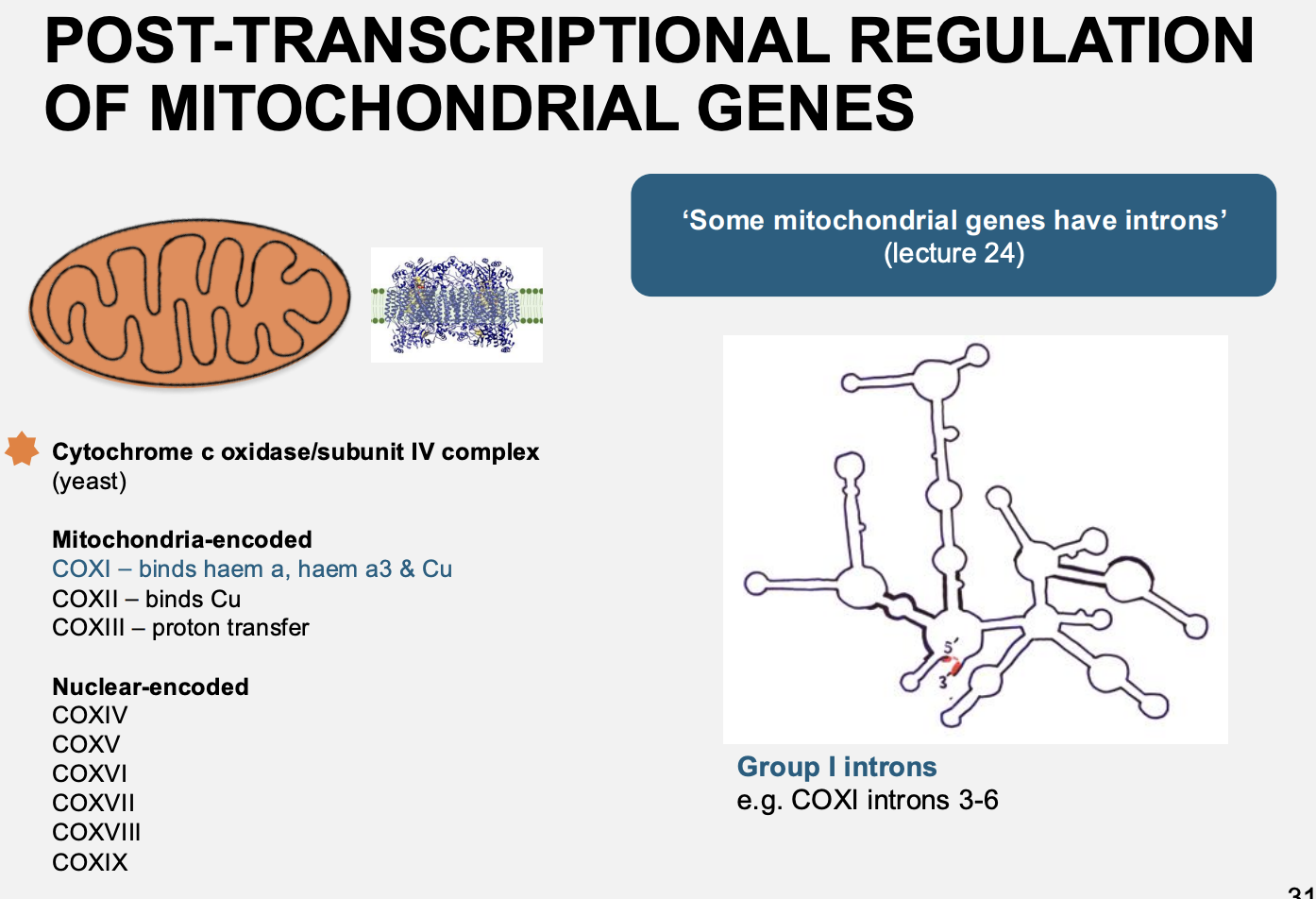
Example 1 → nucleuar factors regulate the splicing of mitochondrial cox genes
removal of intron 5b from COX1 requires 3 helicases
Northern blot shows the relative sizes of RNA moieties of COX
splice variances
different splices due to different nuclear mutations
therefore: nucleus has a role in ost-transiptional expression in the organelle
Found that:
in the nucleus? there are three helicase proteins for the removal of intron 5b in COX1
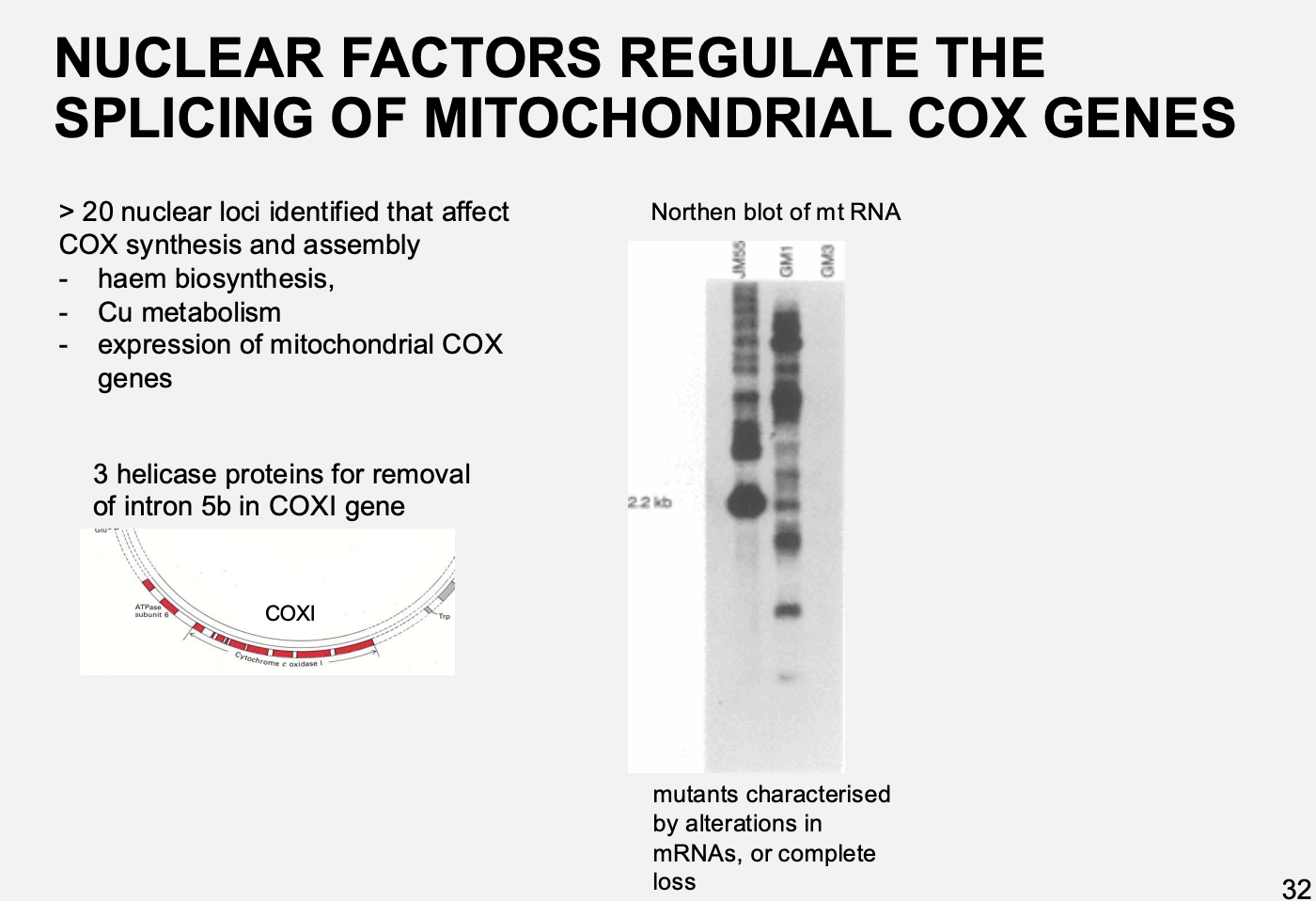
Example 2→ nuclear factors regulate the translation of mitochondrial cox genes
Procedure:
screen of yeast nuclear respiratory deficient mutants
result:
>20 additional genes identified that are trans-acting factors needed for COX assembly
Two types of trans-acting factors
Enzymes for haem biosynthesis and Cu-homeostasis and insertion into complex
proteins need for expression of mitochondrial COX genes:
stability, splicing and translation
PPRs
What are PPR proteins
bind and stabilise and help with splicing and translation
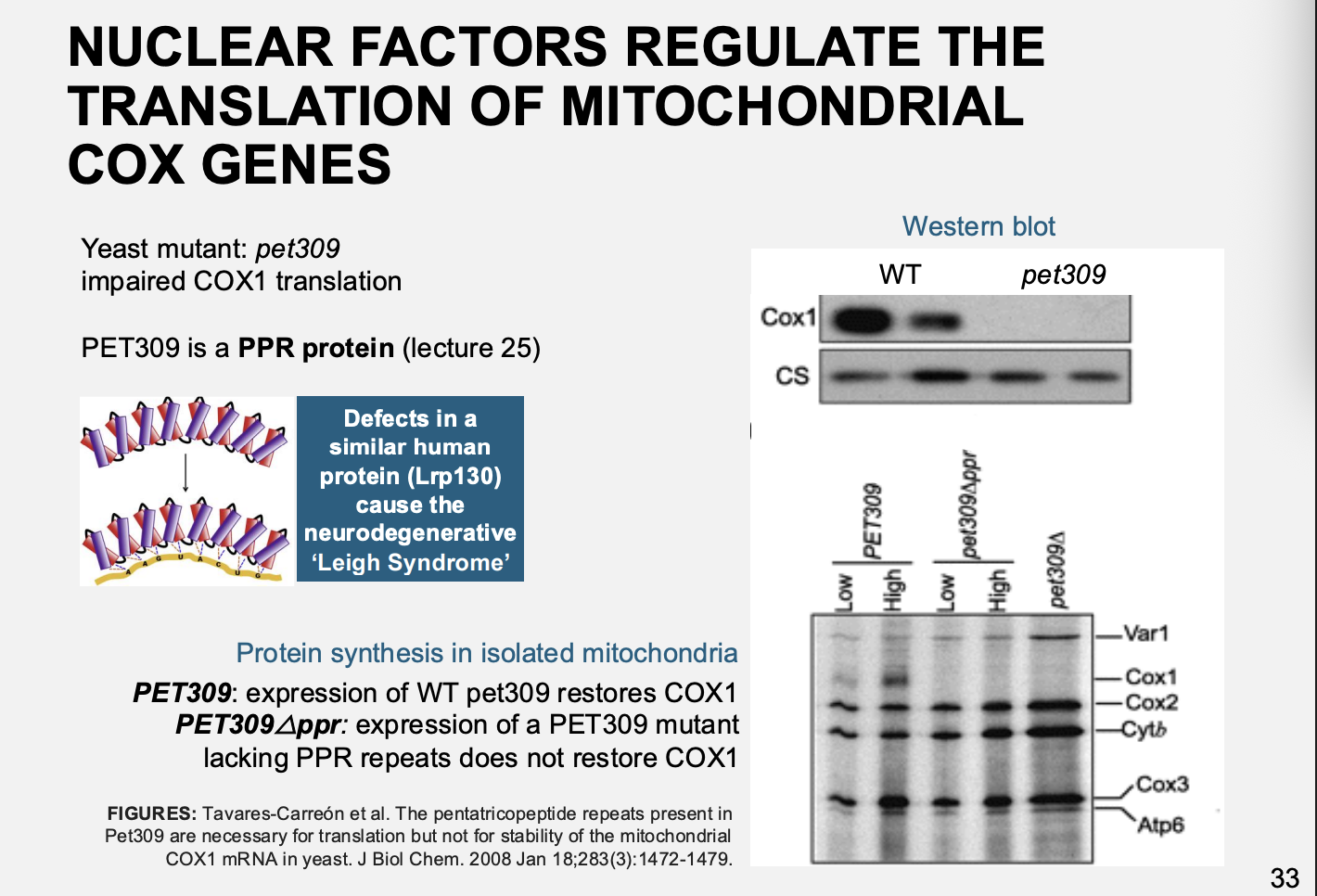
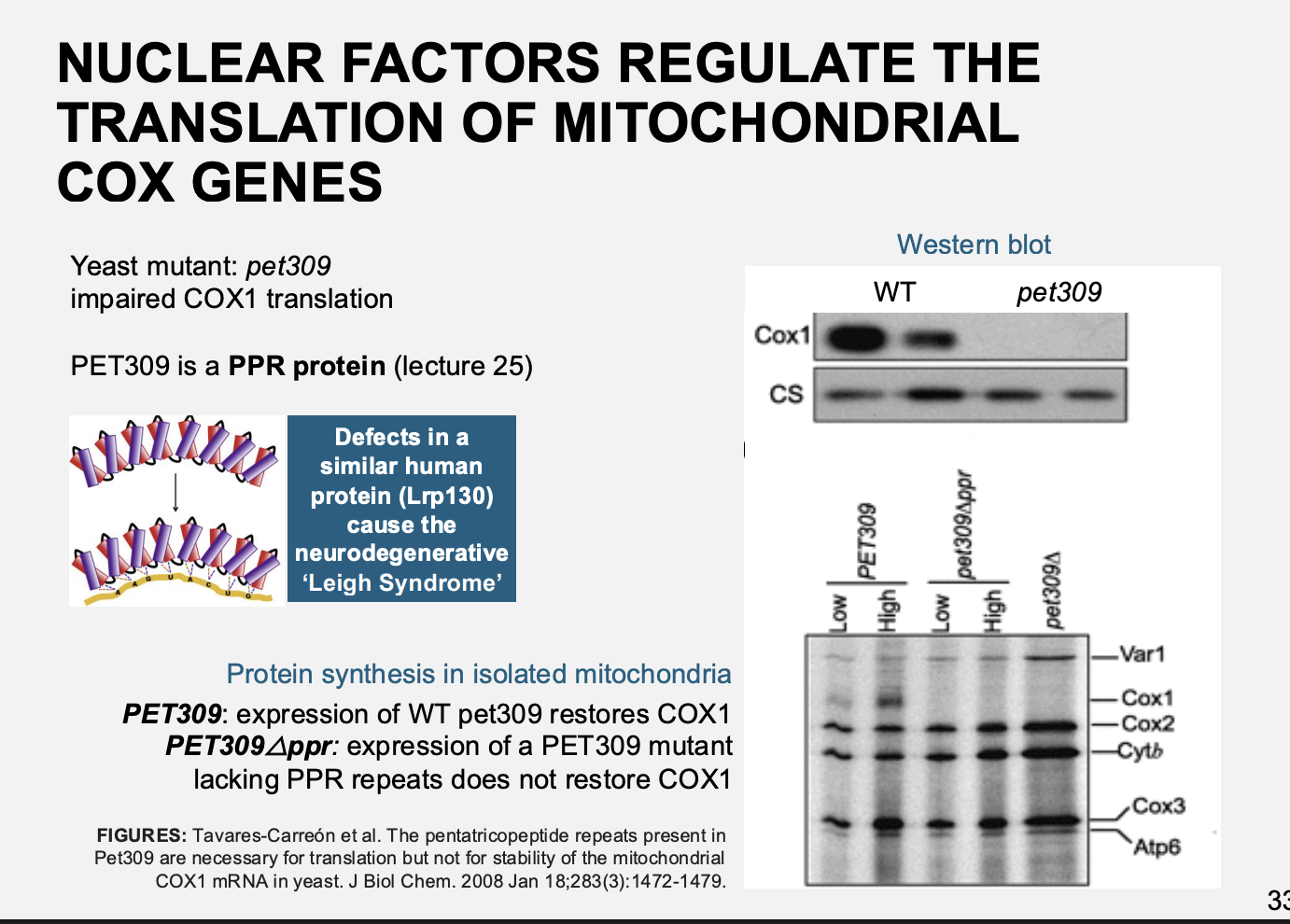
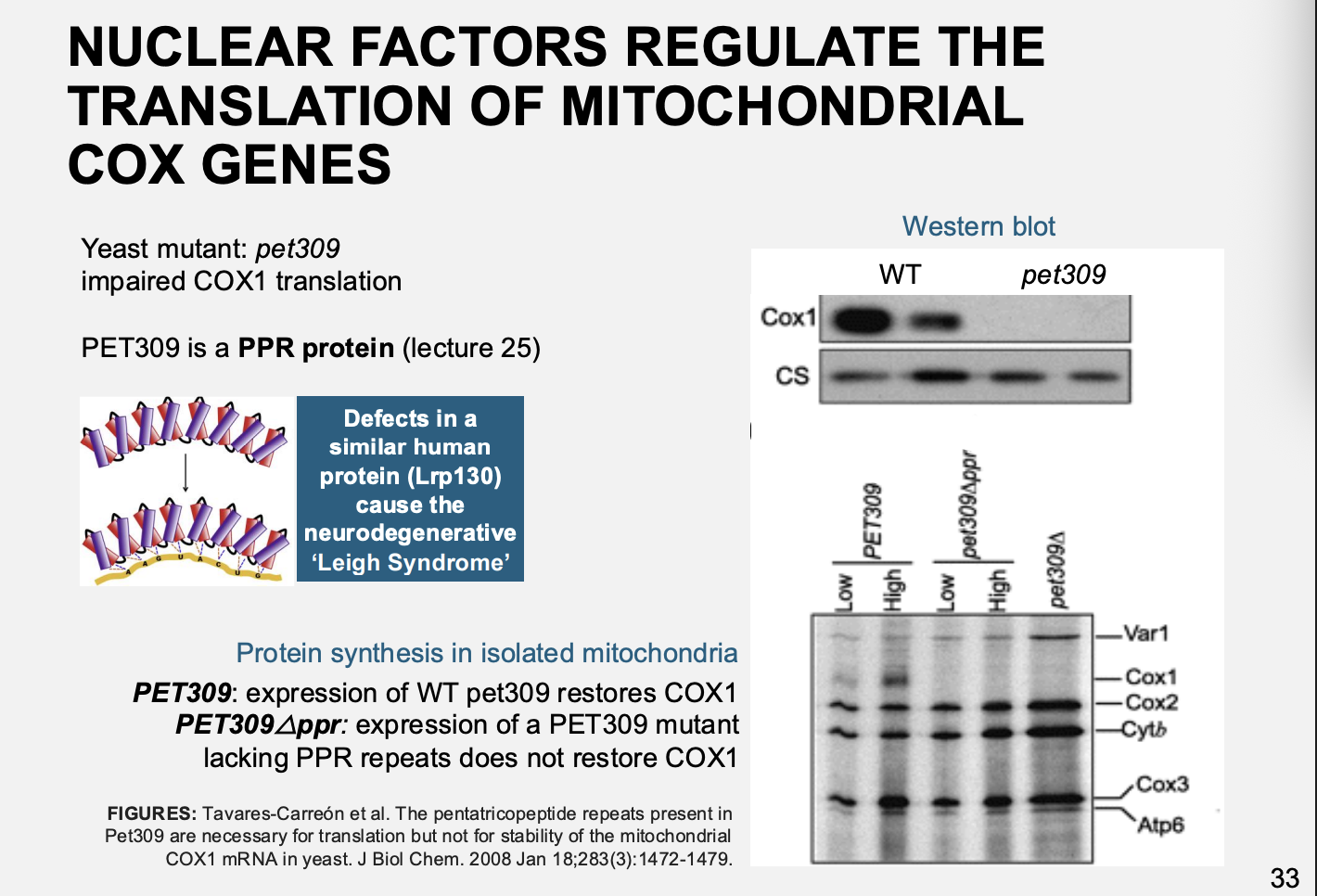
Example: petite mutant pet309
Pet is a PPR protein
results
Impaired in translation of Cox I mRNA
Complementation→ restored function
But→ complentation wtih proteins lacking the Huam nPet309→ neurodegenerative Leigh syndrome
due to inability to assemble COX
Therefore: show the need for nuclear regulation and the proteins it makes to regulate the organelle proteins
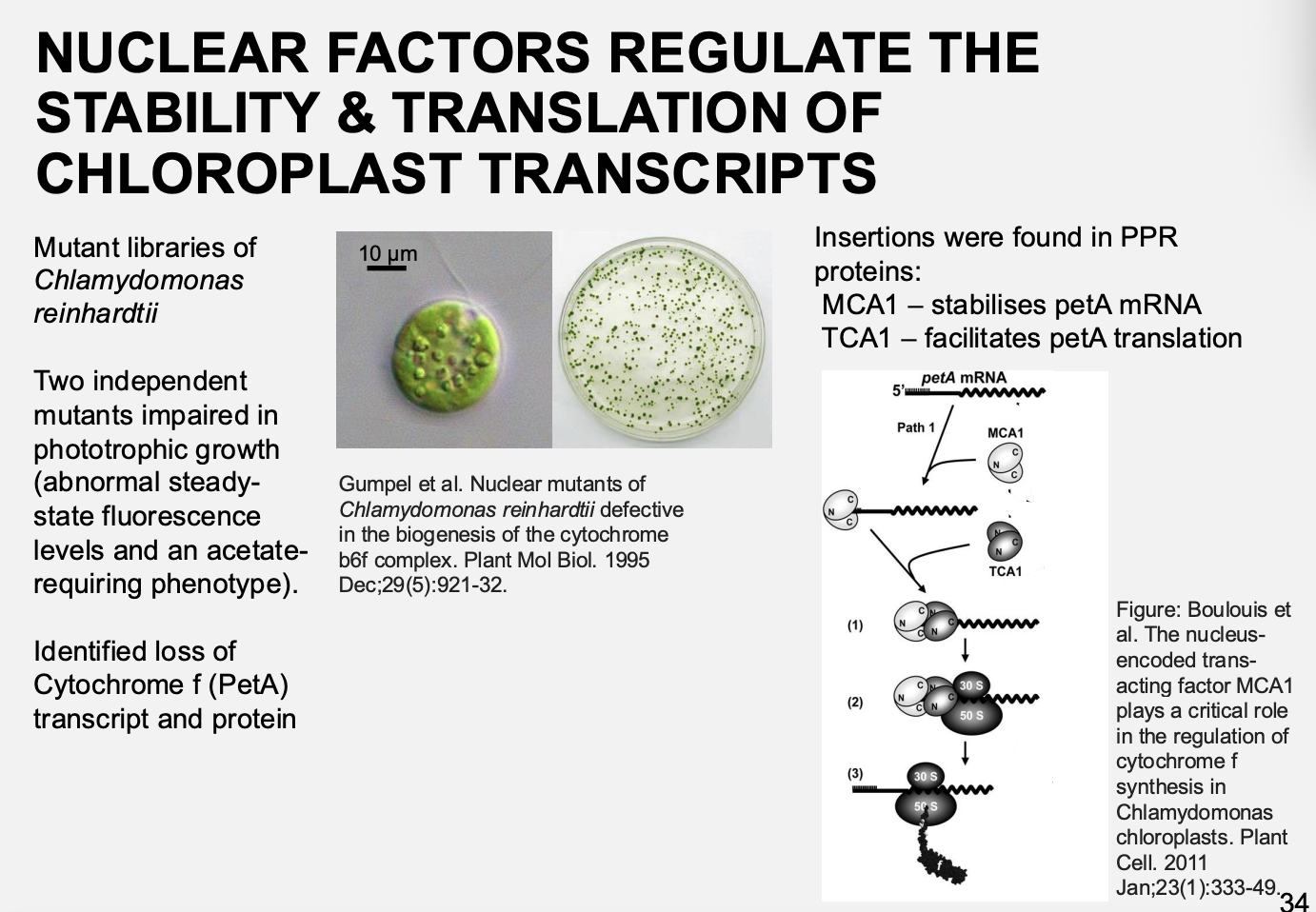
Nuclear factors regulate the stability and translation of chloroplast transcipts
mutant libraries of Chlamydomonas reinhardtii
Two independent mutants impaired in phototrophic growth:
Abnormal steady-state fluoresence levels
acetate-requiring phenotype
Result→ identifed loss of cytochrom f (PetA)
What was found to be the cause of these mutations
Insertions found in PPR proteins:
MCA1→ stabilised petA mRNA
TCA1→ Facilitates petA translation
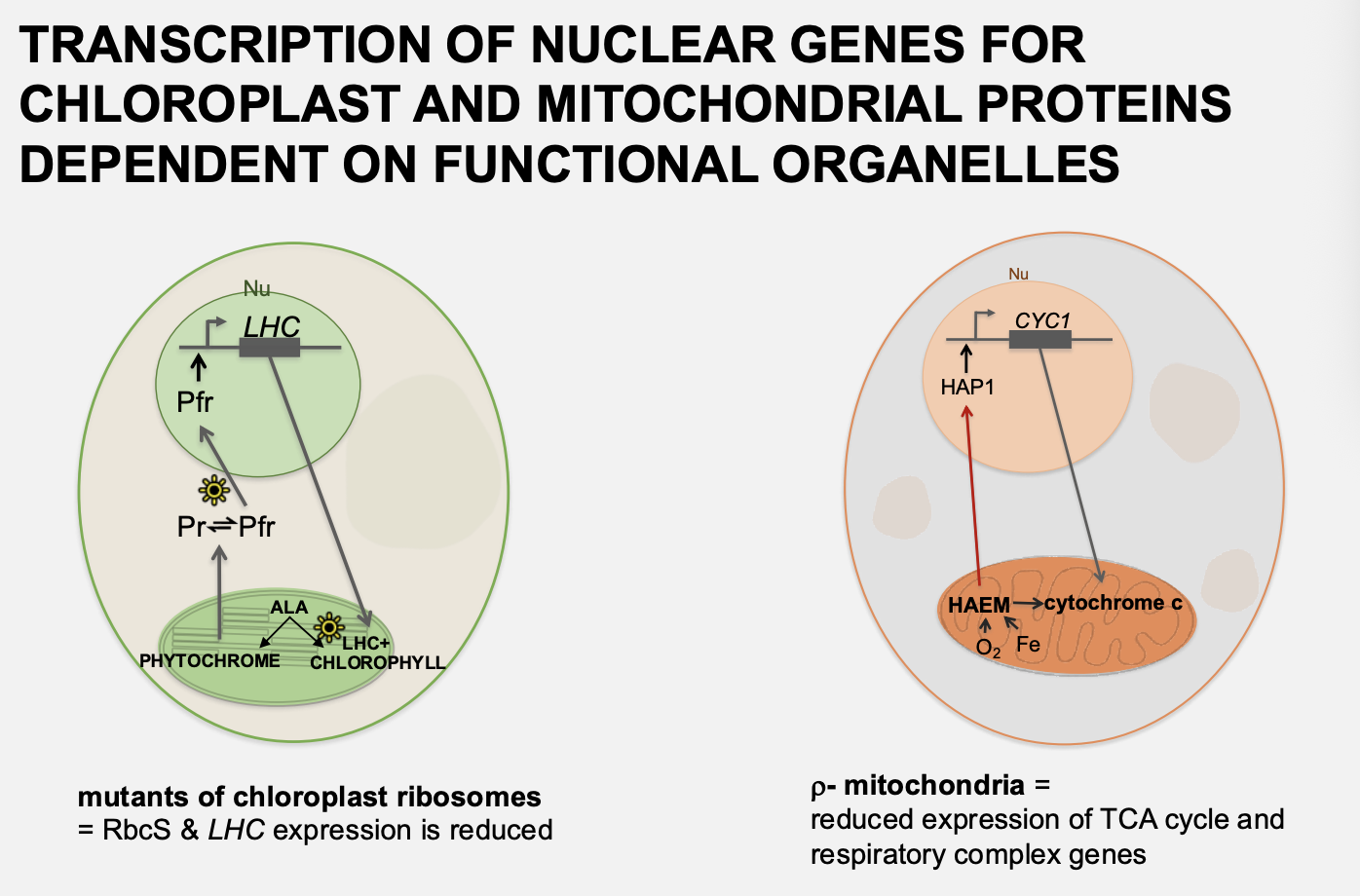
Signalling from organelles to the nucleus→ retrograde signalling compared to anterograde
Anterograde→ nucleus affecting organelle gene expression
retrograde→ signals from organelle to nucleus
transciption of nuclear genes for chloroplast and mitochondrial proteins is dependent on the functional state of the organelles
why do we know there is retrograde signalling
In the absence of functional chloropast or mitochondria
nuclear genes for organelle components are not transcibed
(or transcribed at basal levels)
Examples of this
Yeast p- mutants→ no mitochondria and so reduced expression of TCA cycle and respiratory complex genes
Barley mutants→ defects chloroplast ribosomes
→ reduced RbcS and LHC expression
In what ways can organelles communicate to the nucleus
Positive regulators→ from functional organelles
Repress nuclear gene expression→ Dysfunctional organelles
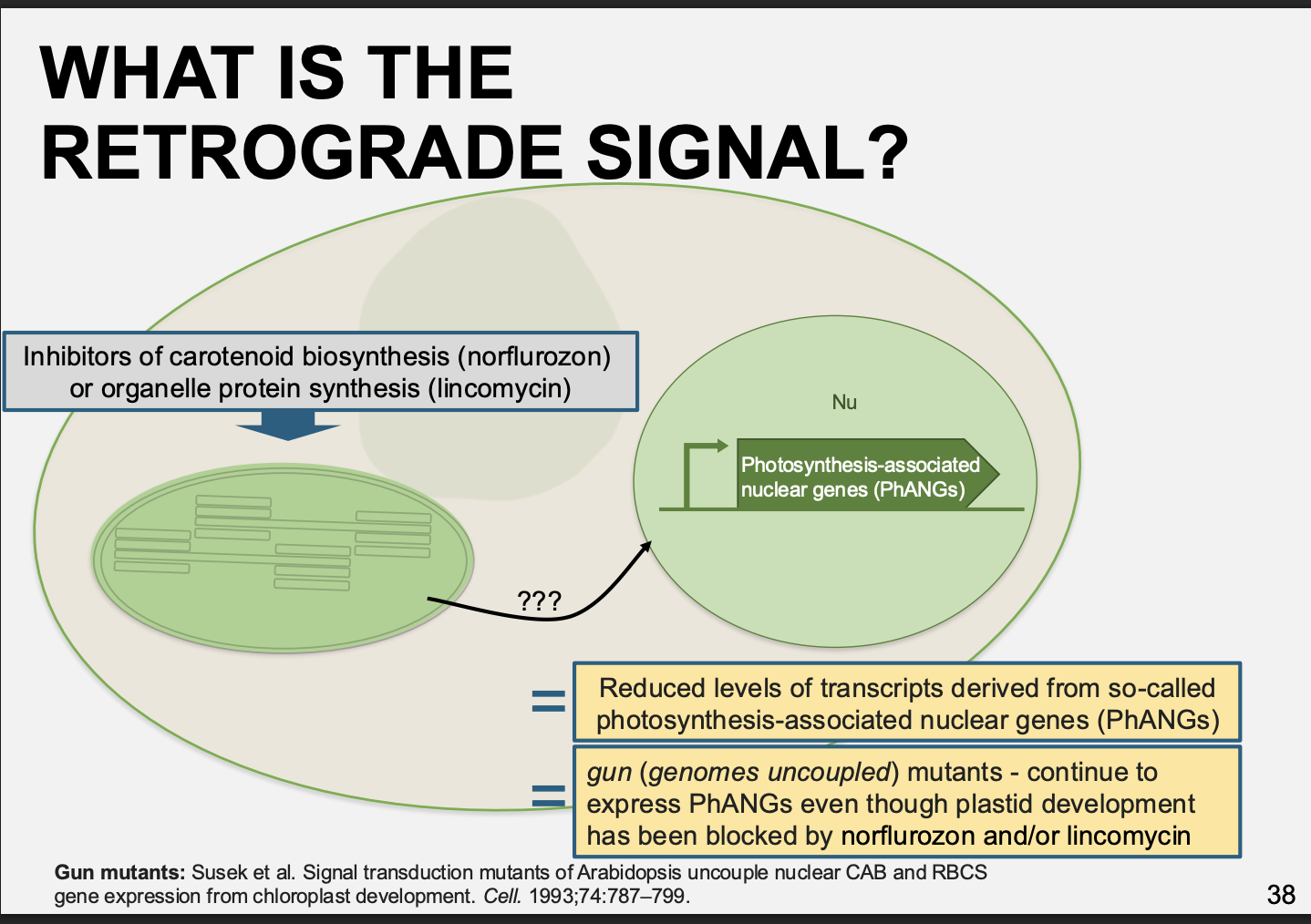
Example 1: of investigating nuclear transcription regulation by chloroplasts: way to cause chloroplast damage
Inhibitors of chloroplast transcription and translation
e.g Tagetitoxin→ inhibits chloroplast RNA polymerase
e.g Linomycin and chloramphenicol→ chloroplast protein synthesis inhibitors
Results:
prevent nuclear gene expression early in seedling development
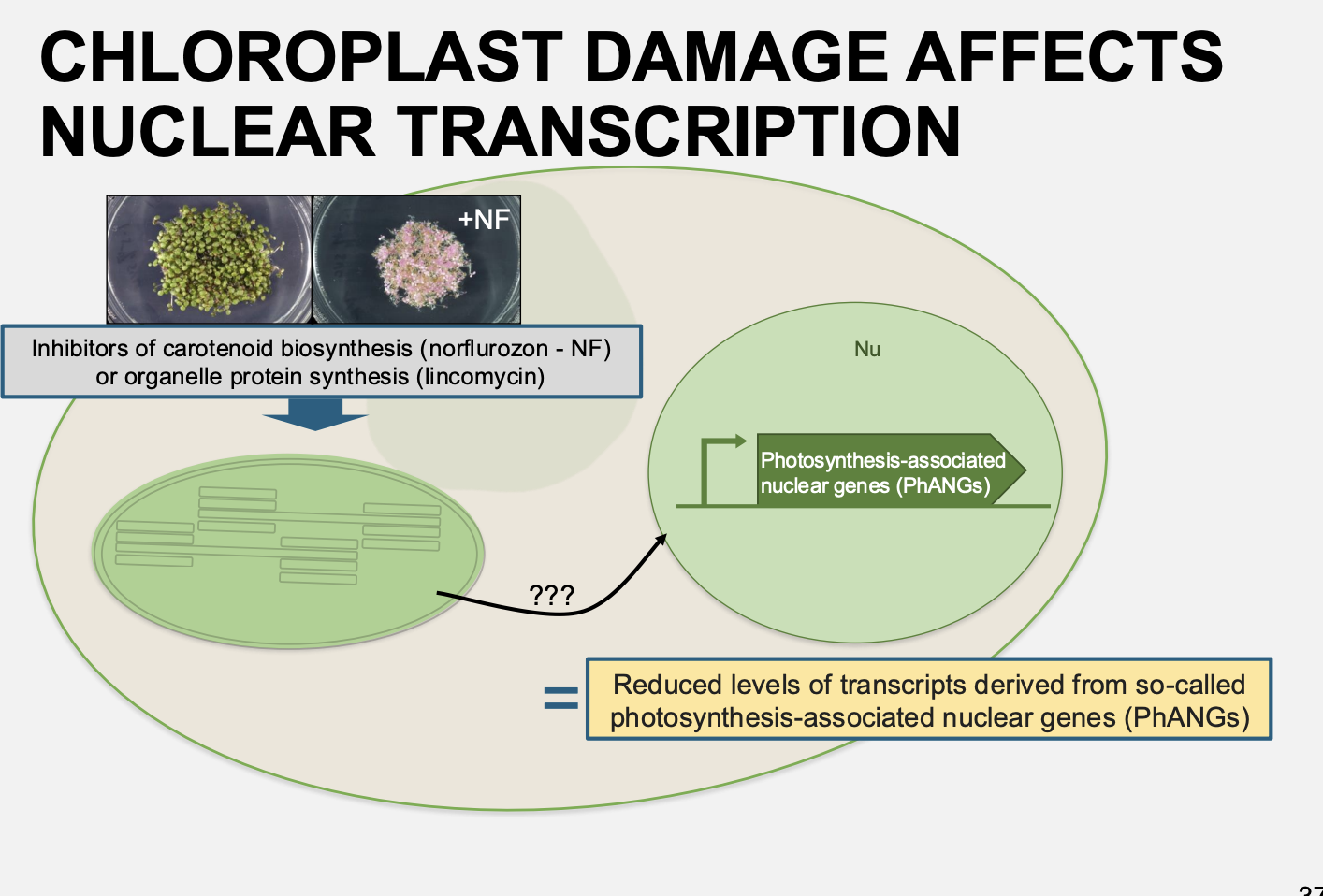
Example 2: of investigating nuclear transcription regulation by chloroplasts→ way to cause chloroplast damage
Photooxidation of chloroplasts
in carotenoid-deficient mutant plants or in plants treated with norflurazon
inhibitor of carotenoid biosynthesis
Results:
Absence of carotenoids→ photobleaching of chlorophyll and destruction of normal chloroplast function
In these two examples→ the treatments lead to what
reduced levels of transcipts derived from so-called photosyntheis-associated nuclear genes
PhANGs
Such as those for light-harvesting chlorophyll protein (Lhc) or Rubisco small subunit→ RbcS
Is the genes encoding products for mitochondrial and cytosolic componenets affected?
no
How was the actual retrograde signal found
mutant libraries
identified → ‘gun’ mutants
gun= genomes uncoupled mutants
These contiued to express PhANGs even thorugh plastid development has been blocked by the treatments
What did this identfify:
these genes are involved in tetrapyrrol biosynthesis→ which include haem
i.e: found mutatns that allowed genes to be expressed even thorugh the chloroplast was dyring
→ note: hard to work with→ have to work with seedlings coz can’t phoyosynthesise
so these mutants mean the nucleus encoded the chloroplast genes anyway, even though there was no retrograde signal from the chloroplast coz they were dying
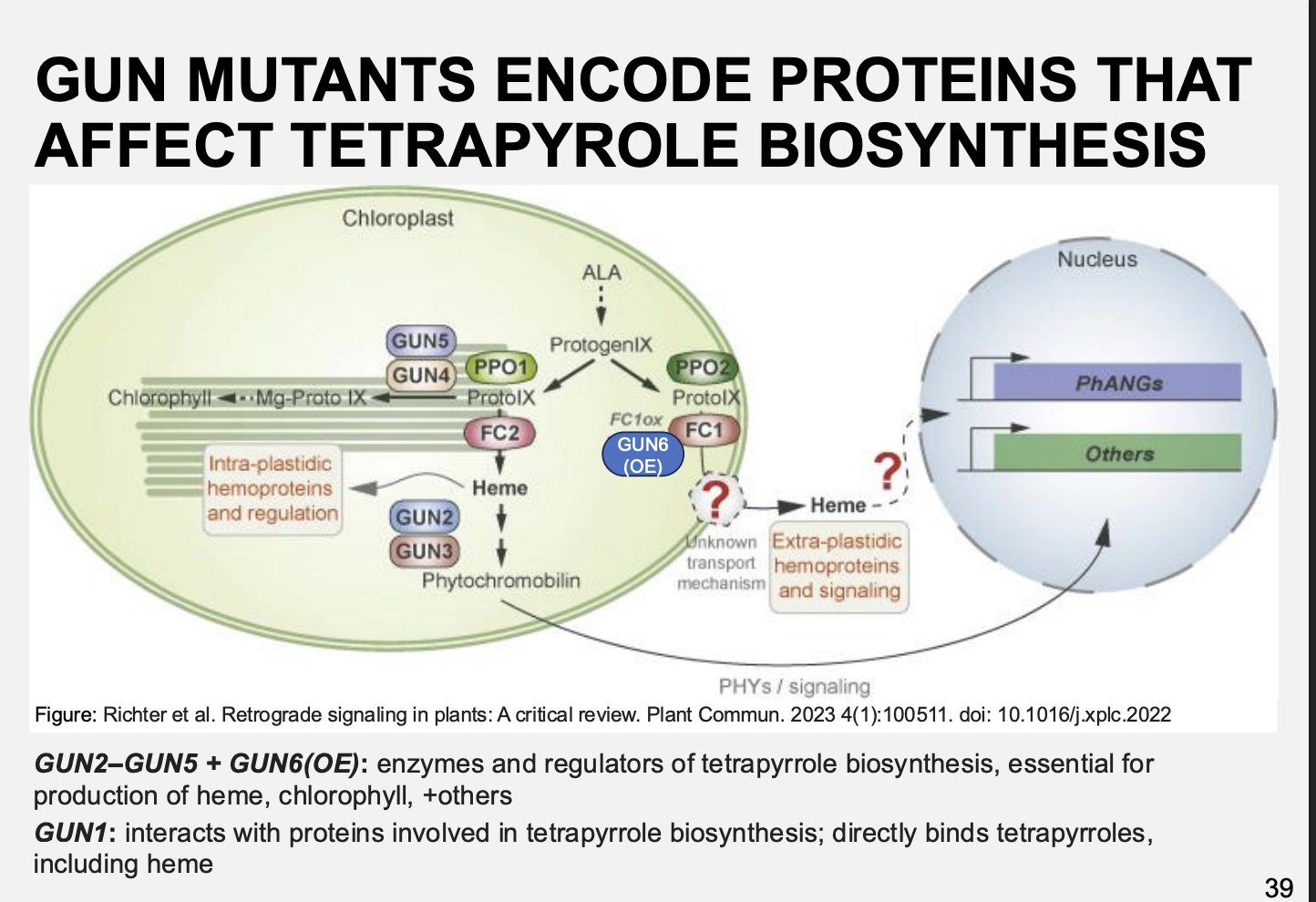
What go gun mutants do
encode proteins that affect tetrapyrole biosynthesis
examples:
GUN2-GUN5 + GUN6(OE)→ enzymes and regulators of tetrapyrrole biosynthesis→ essential for production of heme, chlorophyll and others
GUN1→ interacts with proteins involved in tetrapyrrole biosynthesis→ directly binds tetrapyrroles including heme
Summary
O2 supply impacts mitochondrial biogenesis
Light impacts chlorophyll biogenesis
• Functional organelles are required for eeicient nuclear gene expression
Anterograde signals: nuclear-encoded genes are required for organelle function, including PPR proteins for RNA stability and translation
Retrograde signals: - mitochondria: heme produced in mitochondria required for expression of nuclear-encoded cytochrome c - chloroplasts: tetrapyrrole biosynthesis (possibly heme) required for expression of photosynthesis-associated nuclear genes (PhANGs)
Signals identified by perturbing the expression of: - nuclear genes (e.g., antisense, overexpression, mutant libraries) - organelle genes (e.g., mitochondrial mutations; treating plants with norflurozon/lincomycin)
Organelle inheritance - coadaptation of nuclear and organelle genomes
Defects in mitochondria - linked with many diseases, and ageing, in humans - cytoplasmic male sterility in plants