Population Genetics Pt. B
1/35
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
36 Terms
Hybrid zone
A region in which genetically distinct parapatric populations (often species) interbreed. Because they’re right next to each other.
Cline
Gradual change in a character or allele frequencies over a geographic distance- often indicative of adaptive geographic variation.
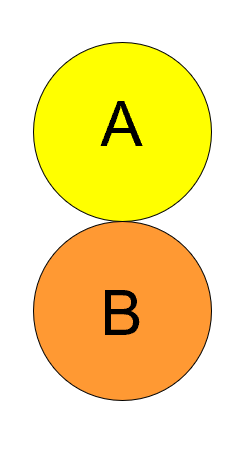
Allopatric
Populations that don’t overlap, and are often separated by a distance.
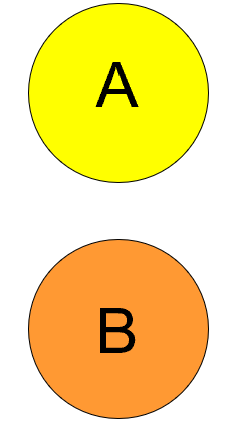
Sympatric
Populations that overlap each other
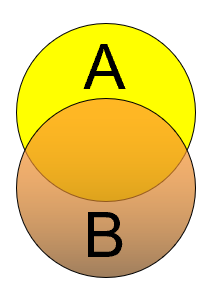
Parapatric
Populations that don’t overlap, but are just adjacent to each other (not separated by a distance.
Reinforcement
POTENTIAL CONSEQUENCE OF HYBRIDIZATION
Hybrids that are less fit than either purebred species. The species continue to diverge until hybridization can no longer occur.
Fusion
POTENTIAL CONSEQUENCE OF HYBRIDIZATION
Reproductive barriers weaken until the two species become one.
Stability
POTENTIAL CONSEQUENCE OF HYBRIDIZATION
Fit hybrids continue to be produced.

Hybridization
The process in which two complementary single-stranded DNA and/or RNA molecules bond together to form a double-stranded molecule.
Forward mutation (Violation of HW)
Assuming the population is large, and no selection (neutral) on new mutations.
A mutation from A to a. Rate is u
Reverse mutation (Violation of HW)
Assuming the population is large, and no selection (neutral) on new mutations.
A mutation from a to A. Rate is v
Genetic drift
Random fluctuations in the frequencies of alleles or haplotypes, often leading to some alleles being fixed (100% frequency). It is a form of nonadaptive evolution, consequence of chance.
Smaller the population, bigger the effect.
Bottleneck
A circumstance that decreases the size of a population.
Founder effect
A special type of bottleneck in which a small number of individuals establishes a new population. The new population will experience genetic drift because of small population size, and may lose some rare alleles from the original population.
Bottlenecks and founder effect consequences
Smaller the founder bottleneck, the faster heterozygosity decreases
After numerous generations, the population size increases, and mutation and recombination restore heterozygosity.
Heterozygosity
Condition of having two different alleles at a locus.
General indicator of the amount of genetic variability.
Coalescence
Concept that all gene copies in a population are derived from a common ancestor.
The smaller the population, the less time required for coalescence.
IMPORTANT: By chance, a FINITE pop (violation of HW) will eventually become monomorphic for one allele. The probability of a neutral allele (no selective advantage to phenotype) becoming fixed is equal to its initial frequency.
Monomorphic
DNA markers that cannot be used to differentiate between or among phenotypes.
If in the population only one allele occurs at a site or locus, we shall say it is monomorphic in that population.
If the jaguar has only one possible trait for that gene, it would be termed “monomorphic.”
Deme
Small independent populations of a species.
Metapopulation
Several proximate demes
Random walk
Equally likely consecutive changes that result in fixation or loss of an allele.
Some demes will either lose or become fixed for an allele- this probability increases with time.
Important Concepts of Genetic Drift 1
Allele or haplotype frequencies fluctuate randomly in a deme, and eventually one allele or haplotype will become fixed.
Due to #1, genetic variation at a locus declines with time, the frequency of heterozygotes [H=2p(1-p)] declines, and this decline can be a proxy for genetic drift in the population.
An allele’s probability of becoming fixed equals its frequency at that time, regardless of past changes in its frequency.
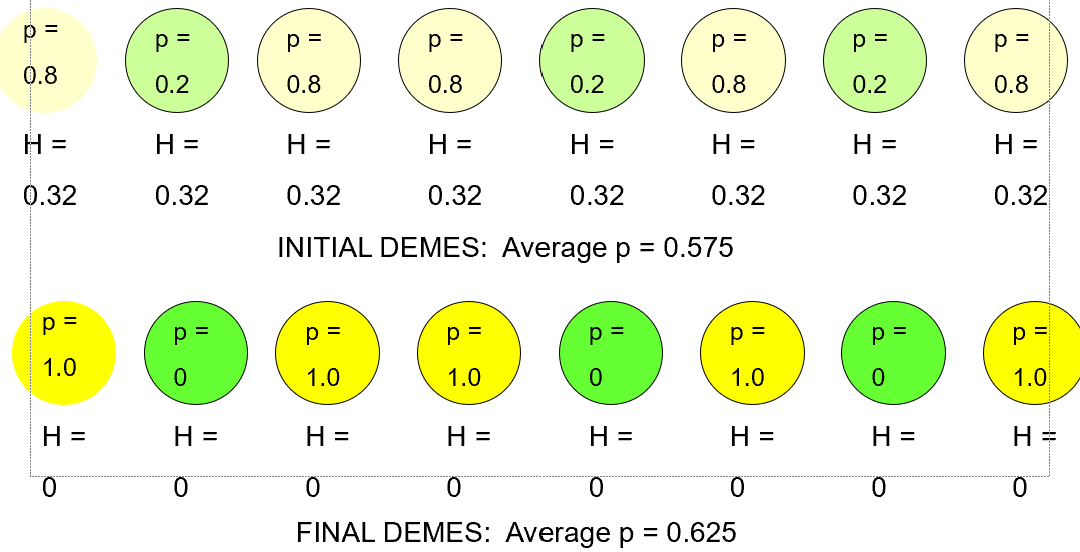
Important Concepts of Genetic Drift 2
Demes (populations) with the same initial allele frequency (p) undergo random walk, where a proportion (p) will become fixed for one form of the allele, and a proportion of others (1-p) become fixed for the alternate allele.
The frequency of a new mutation that is represented by one gene copy in a population with 2N gene copies is:
pt= ½ N= probability of p becoming fixed
NOTICE AS POPULATION SIZE (N) INCREASES, pt DECREASES
As seen in #5, genetic drift occurs faster in smaller populations. In a diploid population, the average time to fixation of a new, neutral mutant allele (assuming it becomes fixed) is 4N generations.
Important Concepts of Genetic Drift 3
Among demes in a metapopulation, the average allele frequency (p) changes little (small number of demes) or not at all (large number of demes), but as allele frequencies in each deme become 0 or 1 over time, the frequency of heterozygotes (H) declines to 0 in each deme, AND in the metapopulation as a whole.
Census size (aka absolute population size)
Actual number of adults in a population
Effective population size (Ne)
Number of individuals in an ideal population (where every adult mates) in which the rate of genetic drift (measured by decline in heterozygosity) would be the same as it is in the actual population.
E.G: 10,000 adults, but only 1,000 breed, then genetic drift proceeds at same rate as a population of 1,000 (Ne).
As Ne decreases….
Genetic drift and loss of heterozygosity increase.
Bad bc it can lead to decreased viability and/or inbreeding depression.
Effective Population Size can be reduced in 5 ways…
Variation in the offspring produced by females, males, or both
Sex ratio that differs from 1:1
Natural selection increases variation in offspring of certain phenotypes
If generations overlap, probability of inbreeding increases
Fluctuations in population size, especially when population sizes are small
Inbreeding depression
The reduction of fitness (survival and/or reproductive output) resulting from an increase in deleterious homozygous recessive alleles in inbred individuals- big problem with endangered species captive breeding programs
Genetic rescue
Introduction of new genes to inbred population
Inbreeding depression
Identical by descent
Descended from the same copy in a common ancestor.
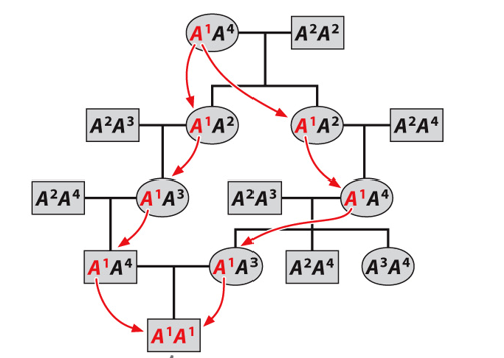
Inbreeding depression
Identical by state
Same in structure in function, but are descended from two dif. copies in ancestors.
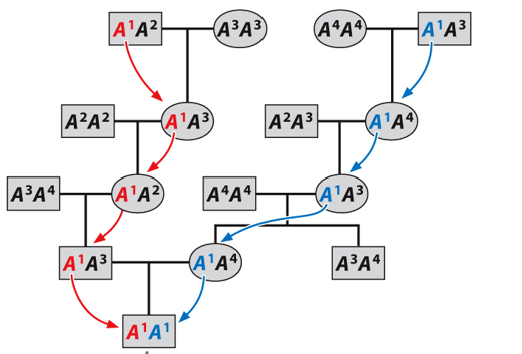
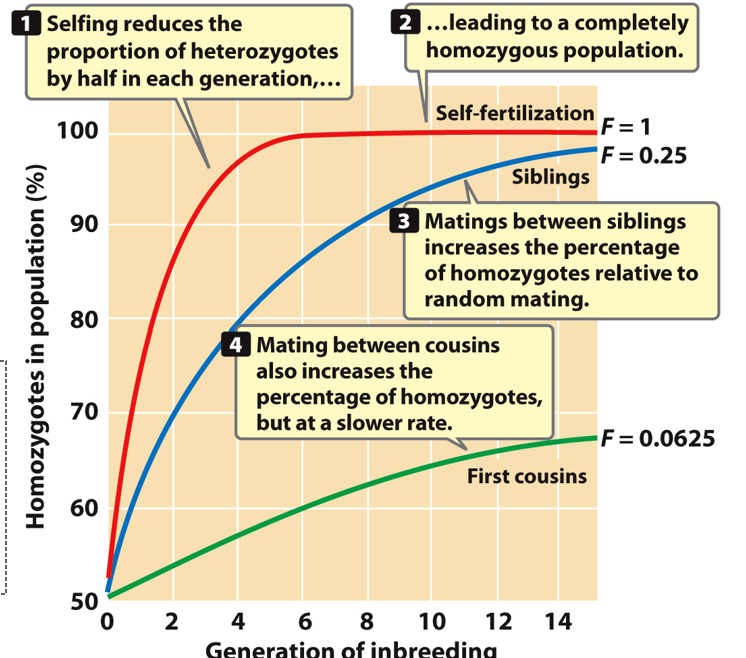
Inbreeding coefficient (F)
Measure of probability of two alleles being identical by descent. Ranges from 0 (random mating) to 1 (all alleles identical by descent)
Inbreeding Depression
Generations of Inbreeding
f(AA)= p²+Fqp
f(Aa)= 2pq-2Fpq
f(aa)= q²+Fpq
When inbreeding is occurring, heterozygote proportion decreases by 2Fpq, and half of this value is added to the proportion of each homozygote.
Self- Reduces heterozygotes by half each generation. Completely homozygous population
Siblings- Increases % of homozygotes relative to random mating
First cousins- Increases % of homozygotes, but at a slower rate
Mutation and Genetic Drift
As (N) increases, genetic drift has less of an effect.
Whereas mutation rate has more of an effect.
Heterozygosity increases with mutation rate and population size.
Fixation index (Fst)
Measures variation in a locus for two alleles among populations- ranges from 0 (no variation- lots of gene flow) to 1 (populations fixed for dif. alleles- no gene flow)
EVEN A LITTLE GENE FLOW KEEPS HETEROZYGOSITY HIGH