2. control of transcription termination of prokaryotic cells
1/22
Earn XP
Description and Tags
part II of prokaryotic gene expression regulation
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
23 Terms
How is transcription termination regulated = attenuation control
By different positions of terminators
if first terminator at the beginning/first —> small mRNA as RNA pol stops at term1
if terminator father down at the end the coin sequence —> mRNA coding for gene is transcribed
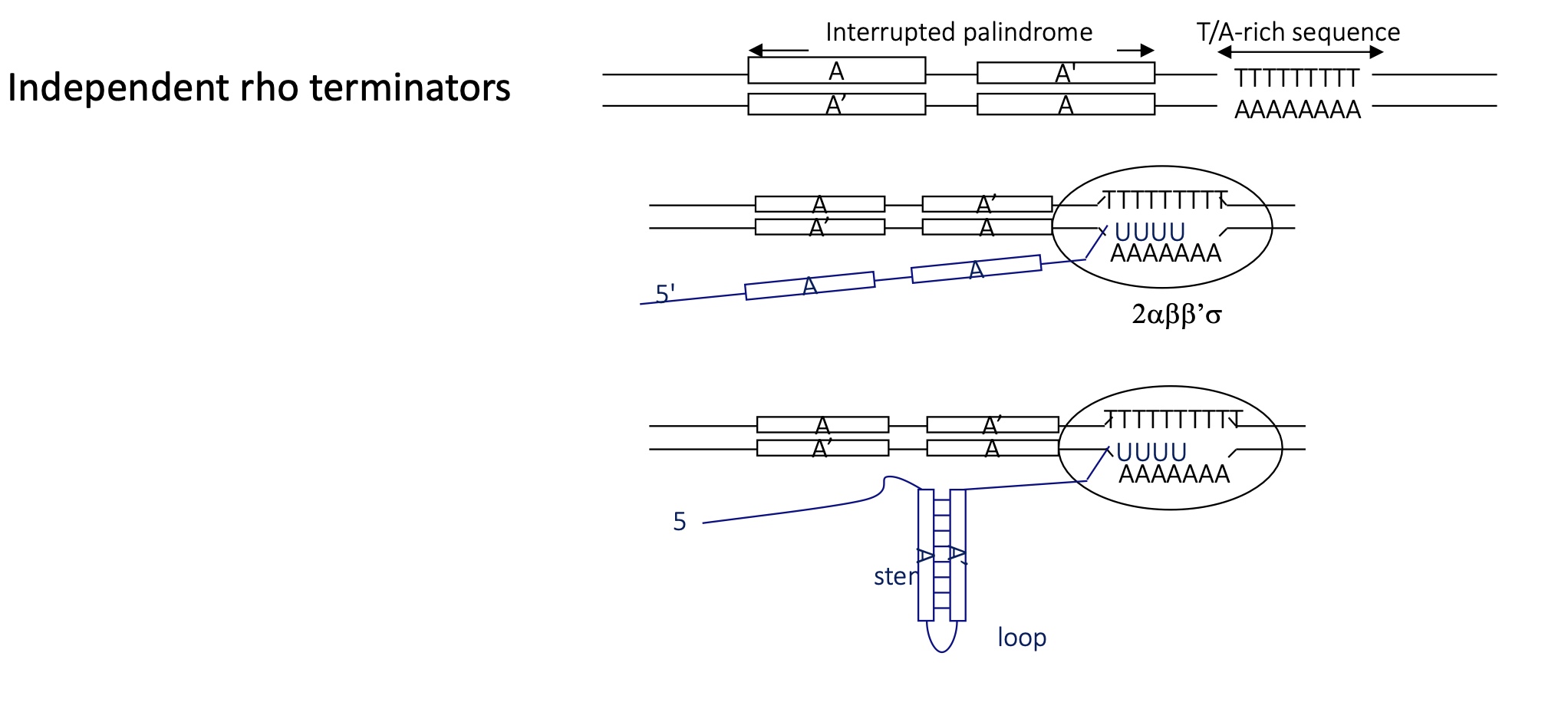
what is the stricture of an independent Rho terminator
interrupted palindrome (sequence above is the same as complementary sequence), followed by a T/A rich sequence
RNA pol transcribes palindrome sequence
complementary sequence hybridise to form a stem loop sequence upstream of T/A rich sequence
RNA pol pause due to this structure at the T/A rich sequence
RNA pol has a weak interaction w/ T/A rich sequence (no C/G) —> immutable interaction
termination os transcription
how can termination be prevented ?
by preventing the formation of the stem loop structure upstream of the T/A rich sequence, which can be blocked by :
translation of a leader protein (ribosome mediated attenuation )
w/ a protein
w/ an tRNA = box riboswitch
by binding a metabolite = riboswitch
2.1 control of termination by a leader protein, give an e.g
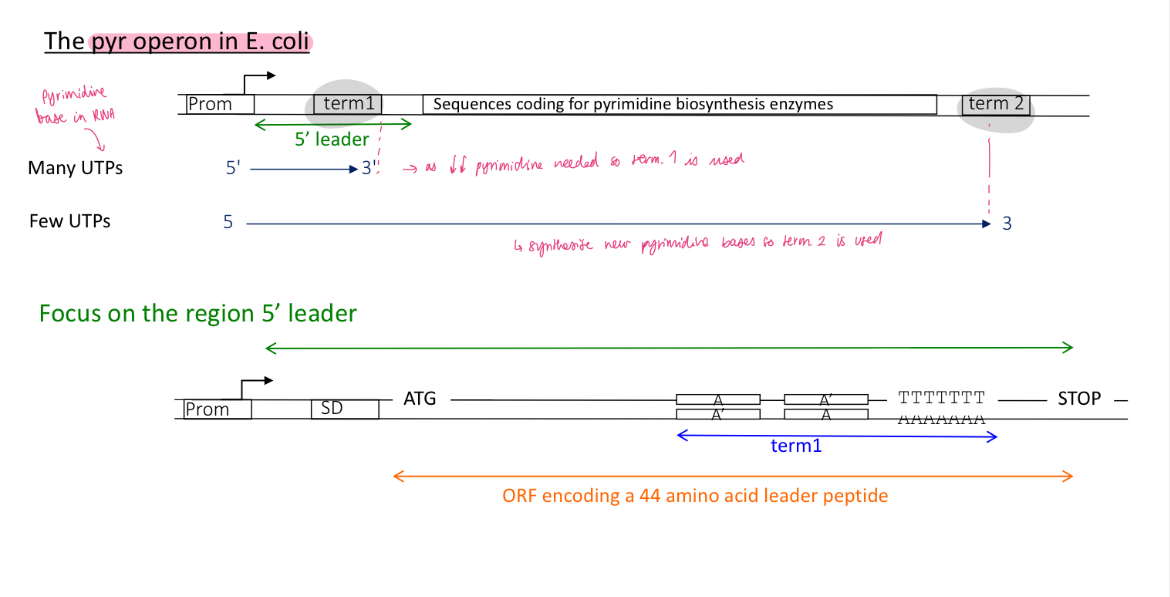
pyr operon in e.coli
the operon codes for pyrimidine bases (UTP) in RNA
the operon had 2 terminators Term1 and Term2
at the beginning of the operon there is a sequence that’s codes for
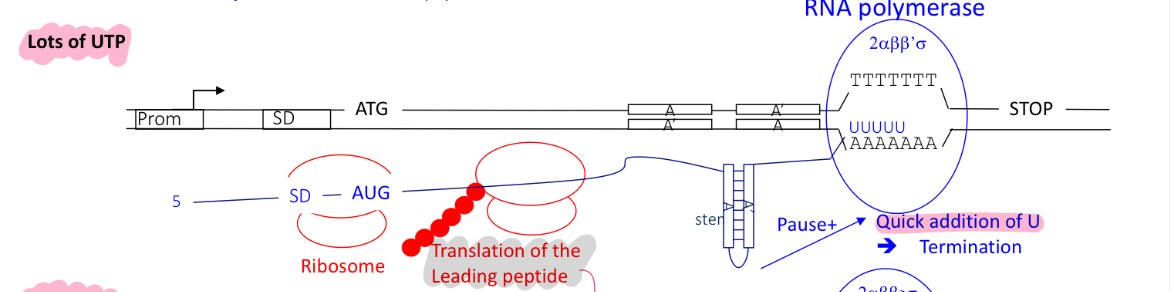
2.1 how is the control of termination of the pyr operon regulated in the presence of many UTPs
term 1 is used when there’s lots of UTPs (as less pyr bases are needed)
translation of leading protein before RNA is complete w/ transcription
RNA pol adds quickly the complementary U bases to the T/A rich sequences
RNA forms a terminator stem loop structure upstream of U rich region
termination occurs at term1 —> RNA pol falls off and theres no expression of genes encoding for pyr bases
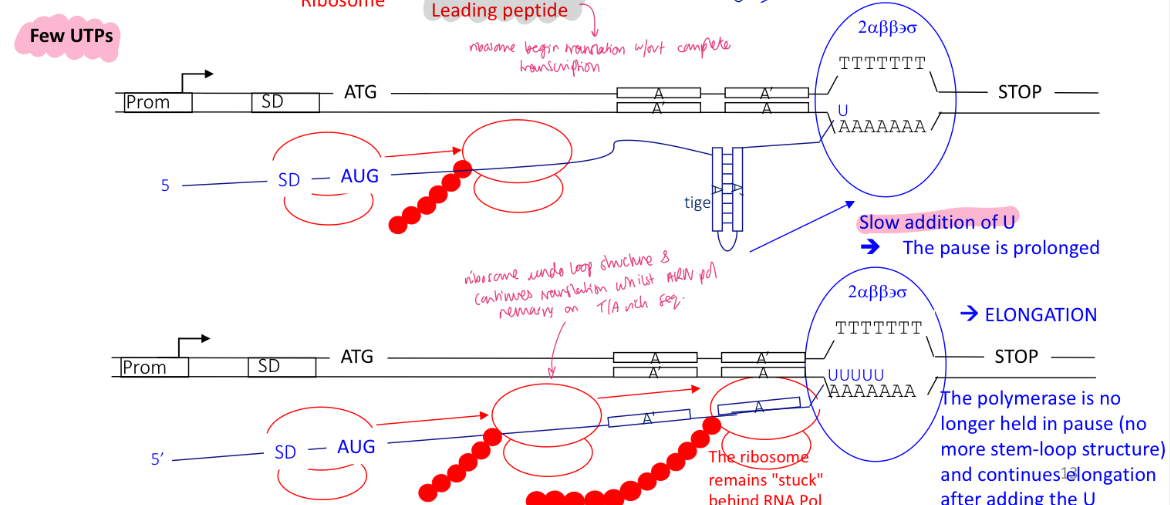
2.1 how is the control of termination of the pyr operon regulated in the presence of little UTPs
term 2 is utilised as more pyr bases are needed : UTP affects transcription efficiency
the ribosomes translating the leading proteins remains stuck behind the RNA pol
no stem loops structure
RNA pol is slowly adding U complementary bases to the T/A rich sequences
RNA pol is not paused and continues transcription —> gene encoding for pyr bases are expressed
2.1 give me a quick summary of how pyr operon termination is controlled by a leader protein
High UTP → efficient transcription/translation → Term1 stem-loop forms → transcription terminated → no pyr gene expression.
Low UTP → stalled ribosome + slow polymerase → no Term1 → transcription continues → pyr genes ON.
2.2 control of termination w/ a protein, give me an e.g
the hut operon in B.subtilis
hut = histdine utilisation ( codes for enzymes that break down His and produce ATP)
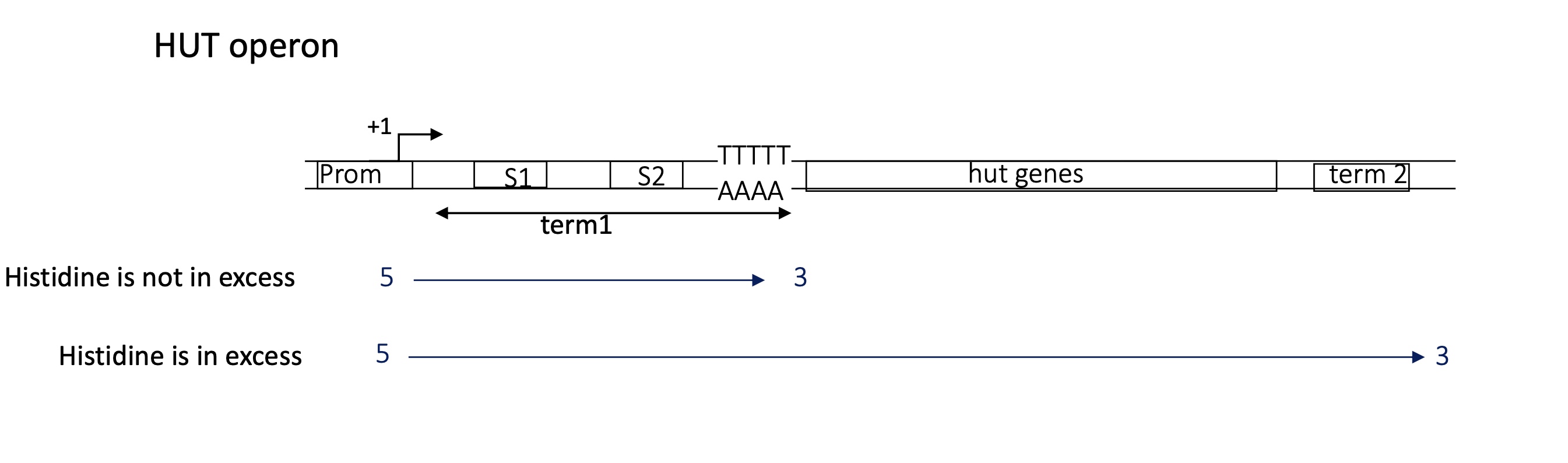
2.2 what protein regulates the termination of hut operon
HutP = RNA binding protein = regulatory protein
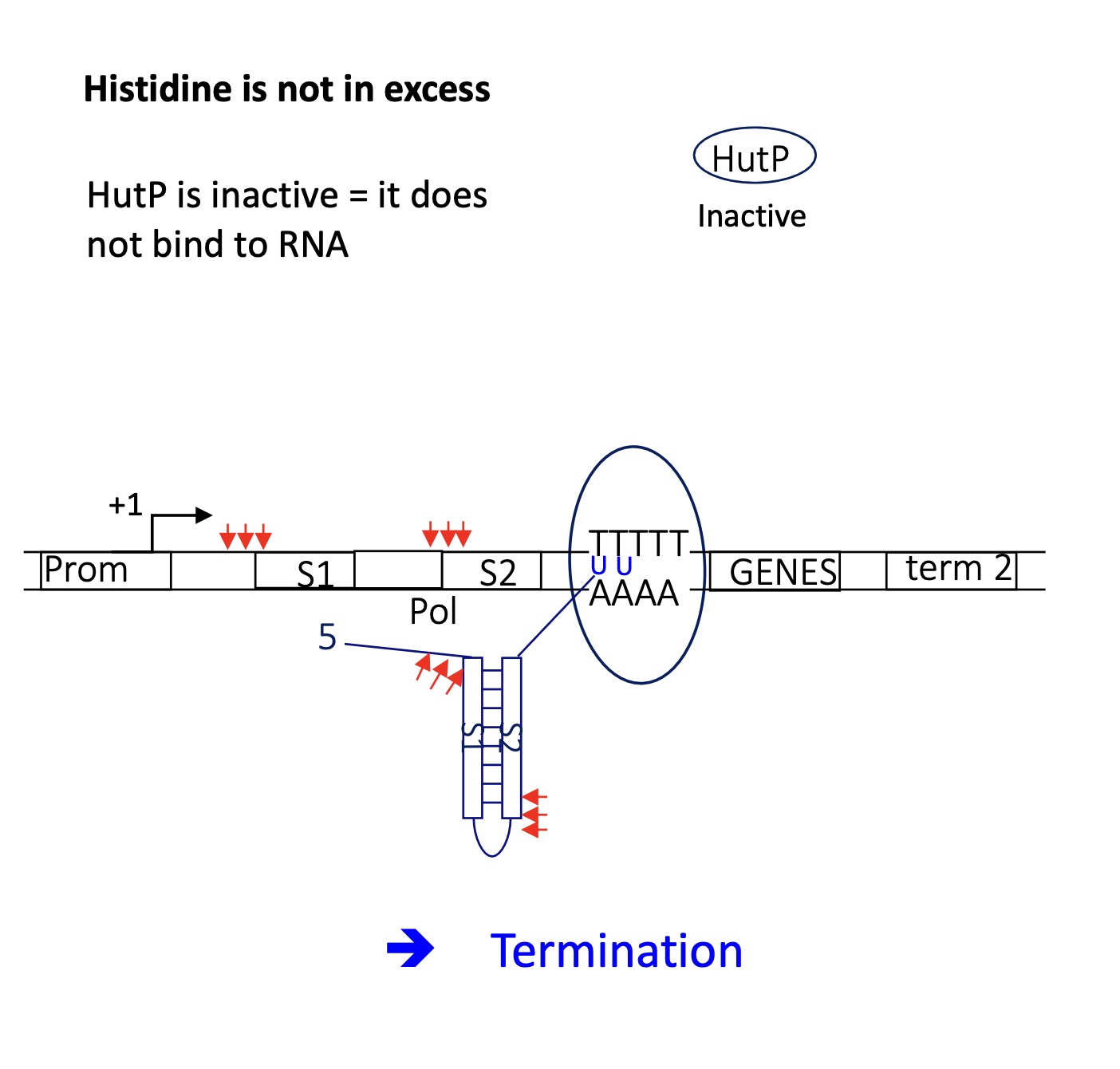
2.2 what happens to the termination of the hut operon in the presence of little His
HutP is inactive —> doesn’t bind to RNA
termi ator stem loop structure is formed upstream of the U rich region
detachment of RNA pol
termination of transcription —> no gene expressed
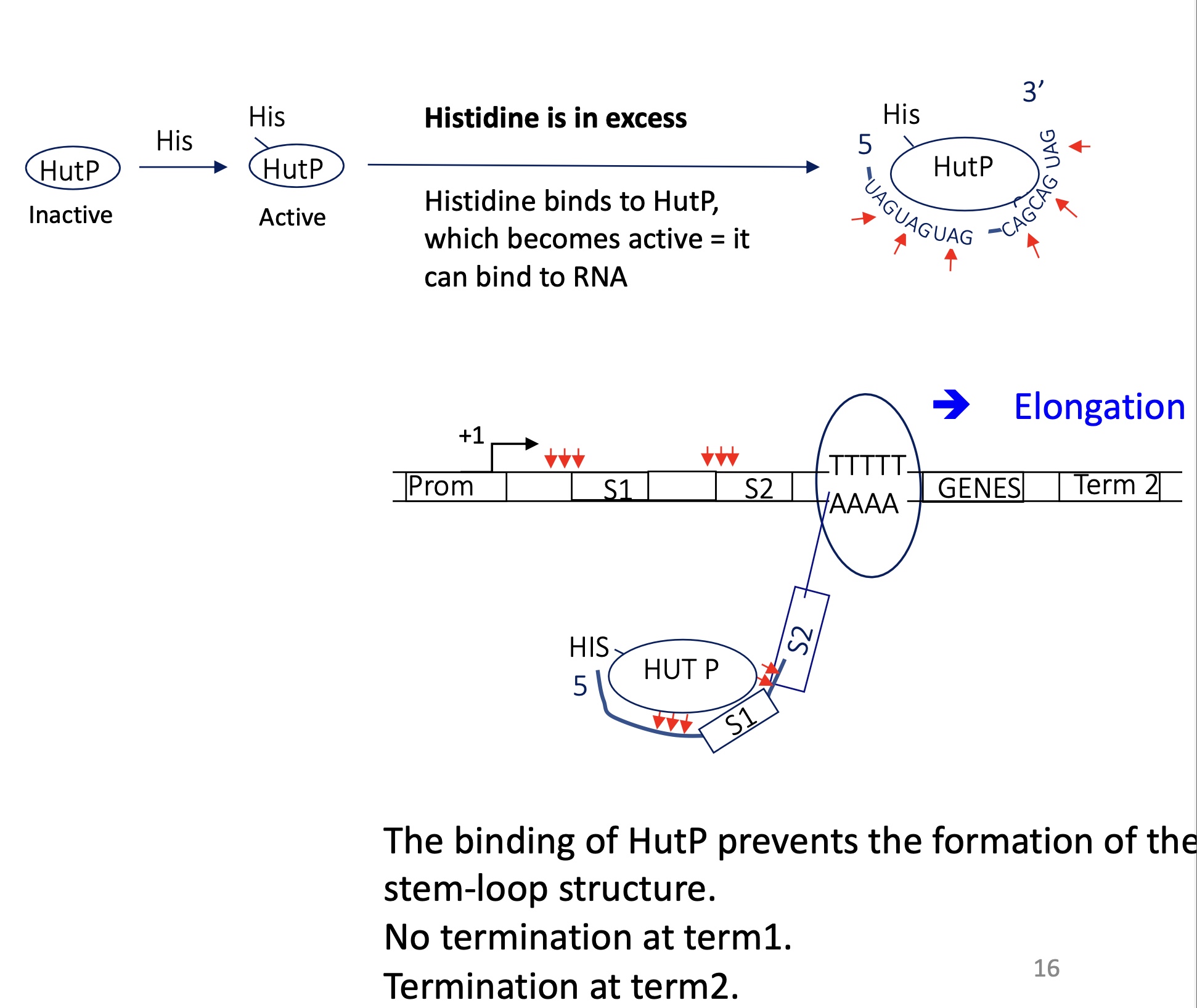
2.2 what happens to the termination of the hut operon in the presence of many His
His binds to HutP
HutP can bind to specific sequence CAG/UAG on RNA
HutP binds upstream/overlapping palindromic sequences
Inhibition of the formation of terminator stem loop structure
RNA pol doesn’t terminate at term 1 —> expression of genes
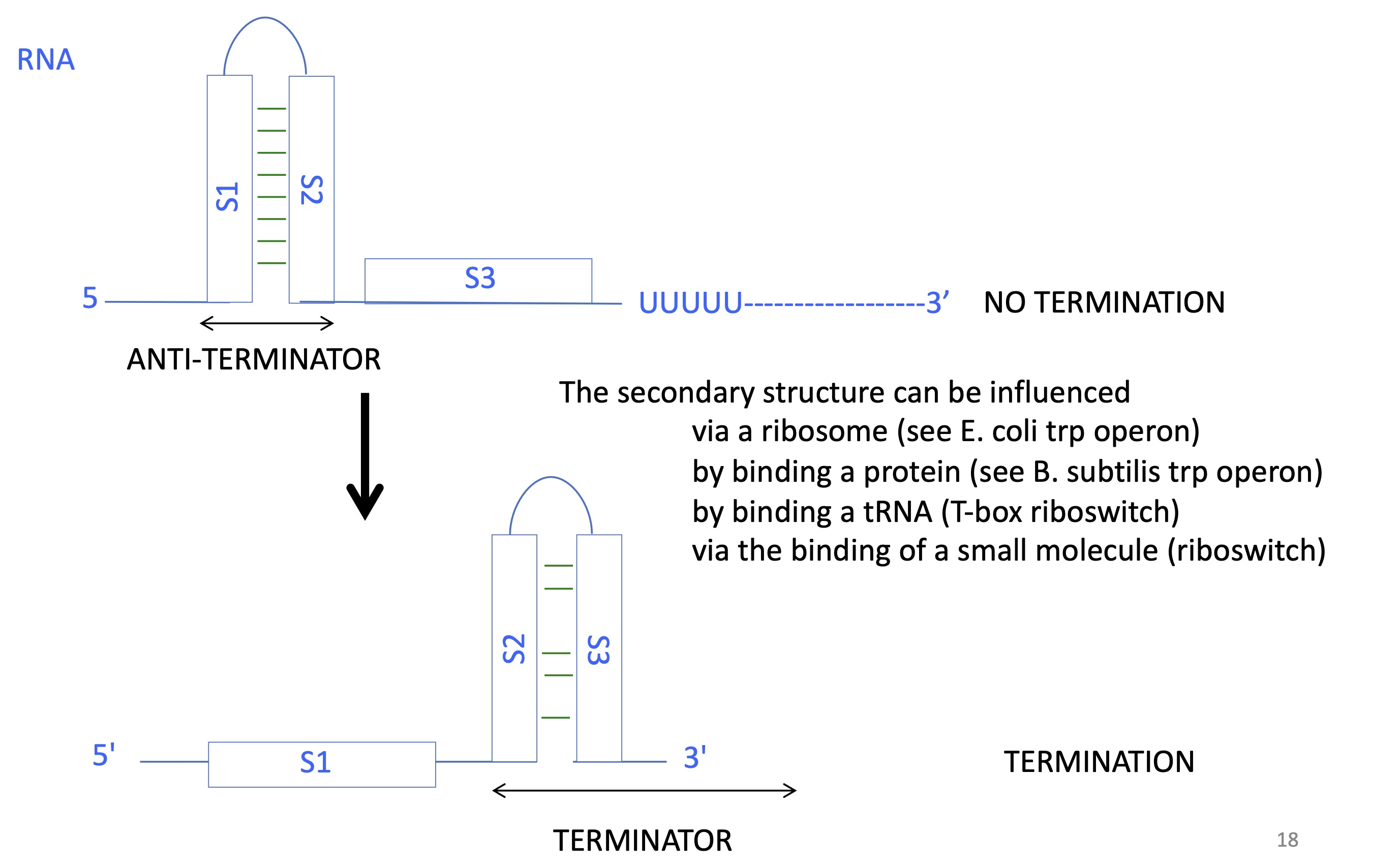
2.2. what are the alternative RNA structures to control termination
terminator stem loop structure —> two palindromic sequences hybridise upstream of the U rich region
anti-terminator stem loop structure —> two palindromic sequences hybridise before a sequence upstream of U rich region
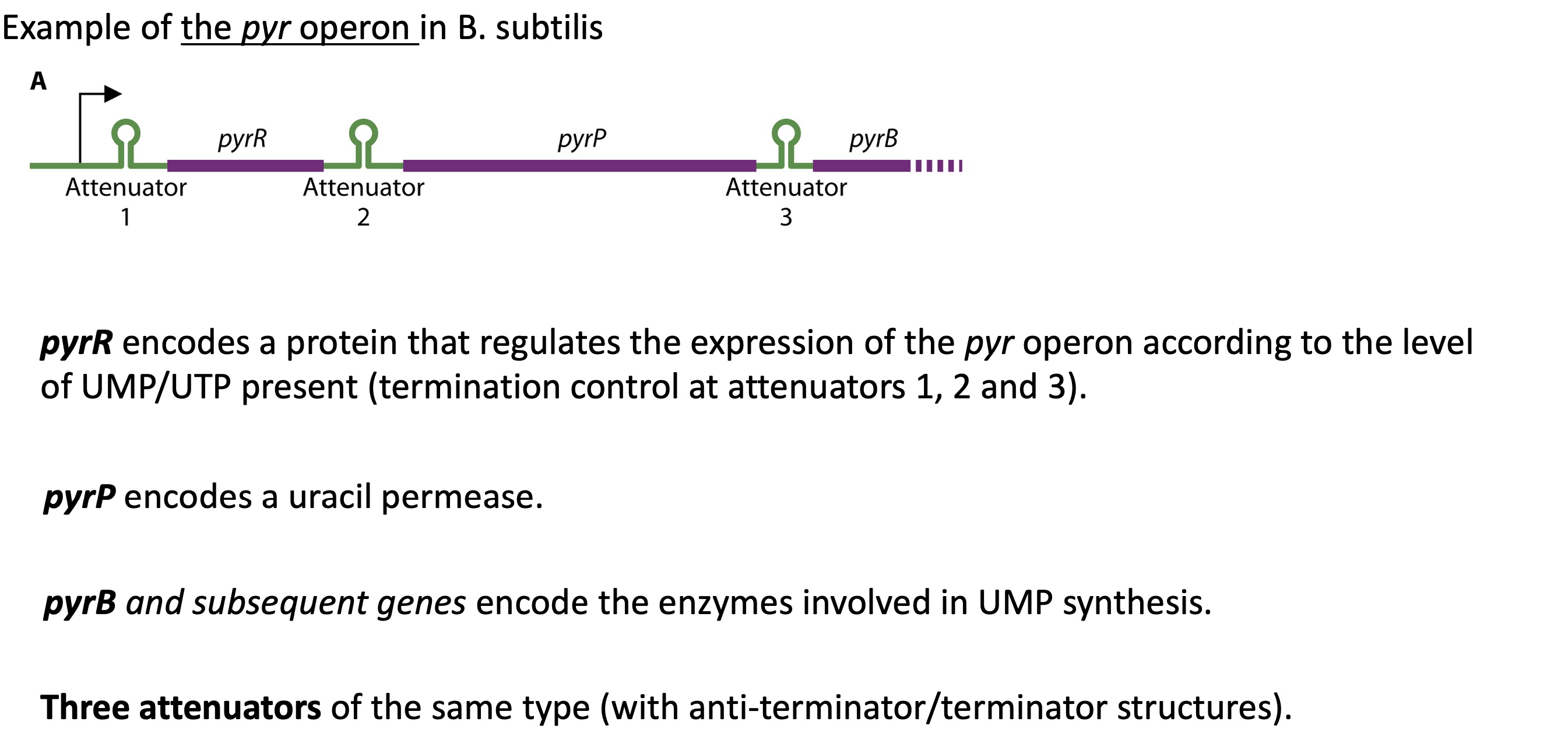
2.2. what’s another example of termination control w/ a protein (also controlled by leader protein)
pyr operon that encodes for different pyr genes :
pyrR : codes for protein that regulates pyr operon expression according to the level of UTPs
pyrP : encodes for uracil permeate
pyrB & following genes : codes for enzymes involved in UMP (pyrimidine nucleotide) synthase
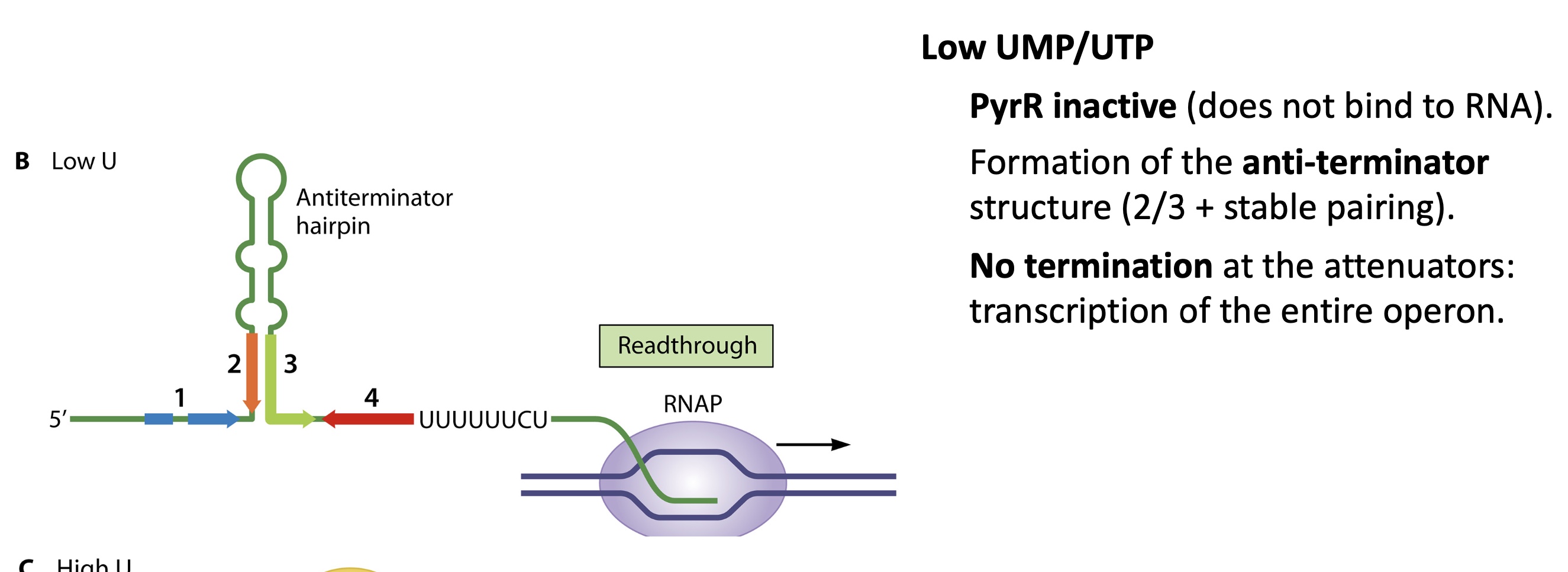
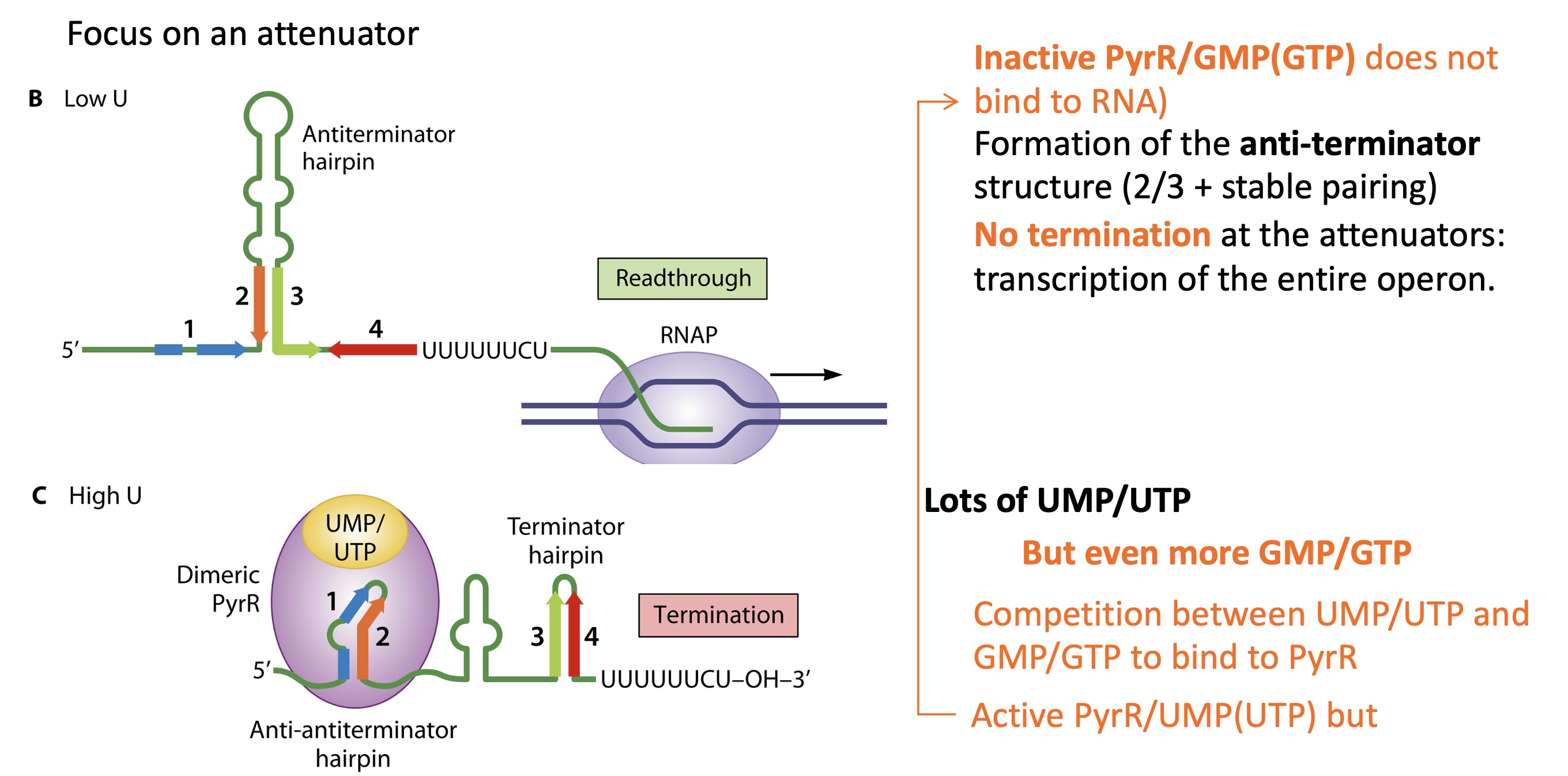
2.2 how is termination of pyr operon controlled in low levels of UTP/UMP
PyrR is inactive and doesn’t bind to RNA
anti terminator structure (S2-S3) is very stable (secondary structure of RNA )
RNA pol doesn’t terminate and whole pyr operon is expressed
2.2 how is termination of pyr operon controlled in high levels of UTP/UMP
UMP/UTP binds to PyrR —> dimerisation —> active
PyrR binds to RNA and induces stem loops structure of S1-S2 = anti-anti terminator stem loops
S3-S4 hybridise and forms a terminator stem loops
Termination at the attenuators
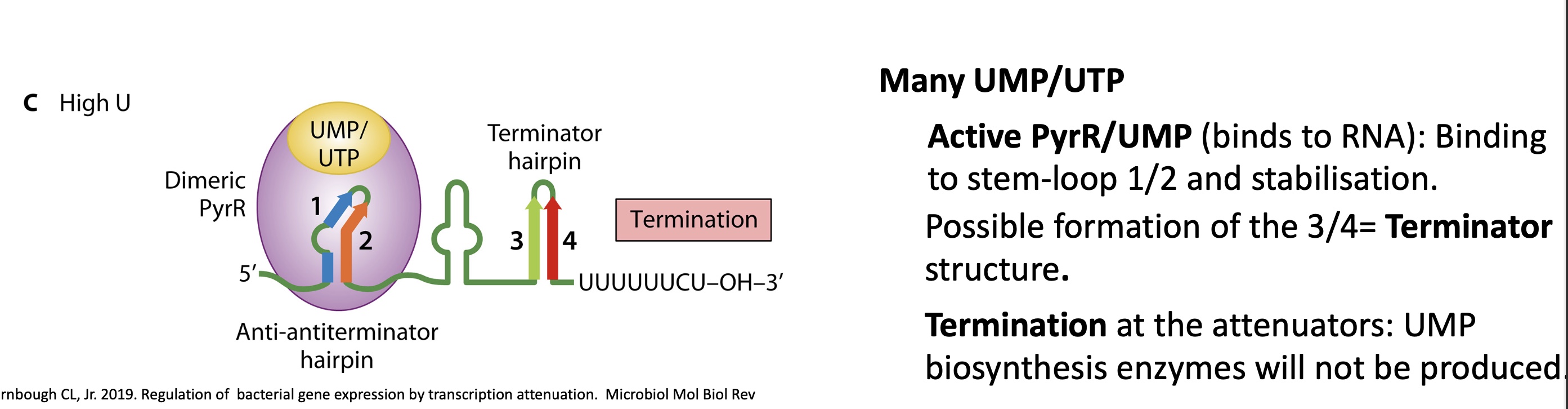
2.2 what happens the termination control if there’s lots of UMP/UTP but even more GMP/GTP
competition between GT/MP and UT/MP to bind to PyrR
when UT/MP binds to PyrR —> active form
when GT/MP binds to PyrR —> inactive form
PyrR doesn’t bind to RNA, formation of anti terminator stem loop structure
no termination and pyr operon expressed
TO KEEP EQUILIBRIUM OF PYRIMIDINE/PURINE
2.3 what is the definition of a riboswitch
cis regulatory system (regulation from same sequence/DNA) of an mRNA that directly senses physiological signals causing a change in the RNA structure
terminator stem loop
anti terminator stem loop
riboswitch can be a :
metabolite e.g sugar, vitamin
t box : tRNA
2.3 what is an example of a box riboswitch
gene encoding for tyr synthetase tRNA
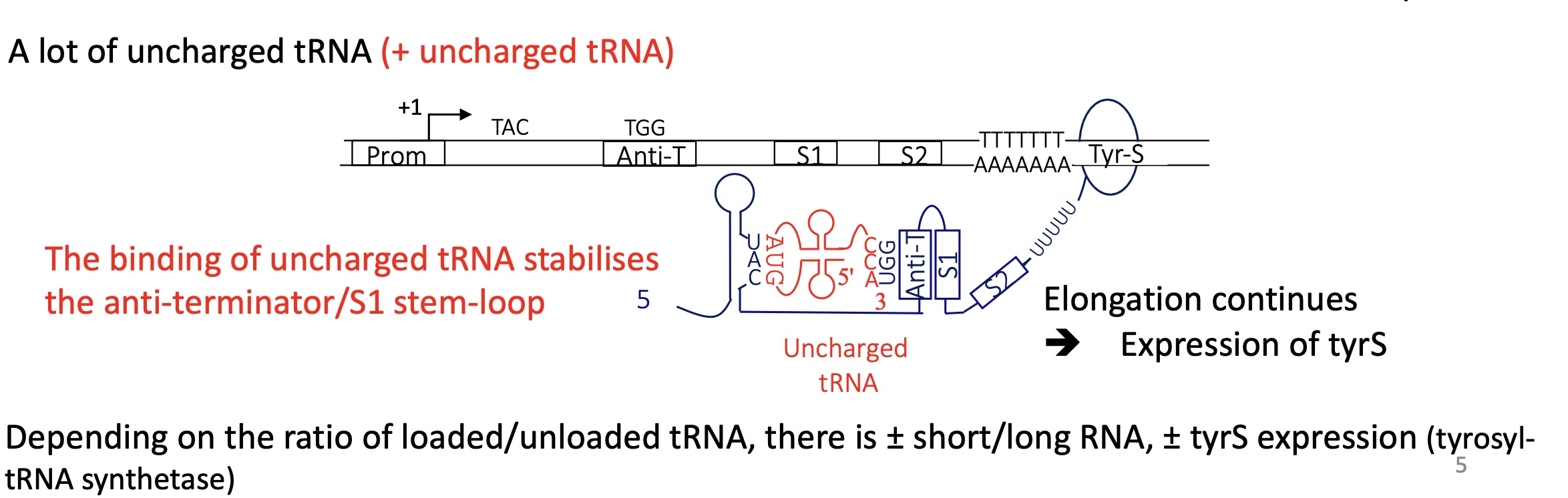
2.3 how does the ration of loaded to unloaded tRNA influence the regulation of certain genes
if there is an excess of uncharged tRNA :
increased expression of genes involved in the biosynthesis of the corresponding AA
increased expression of the gene encoding for the corresponding aminoacyl transferase
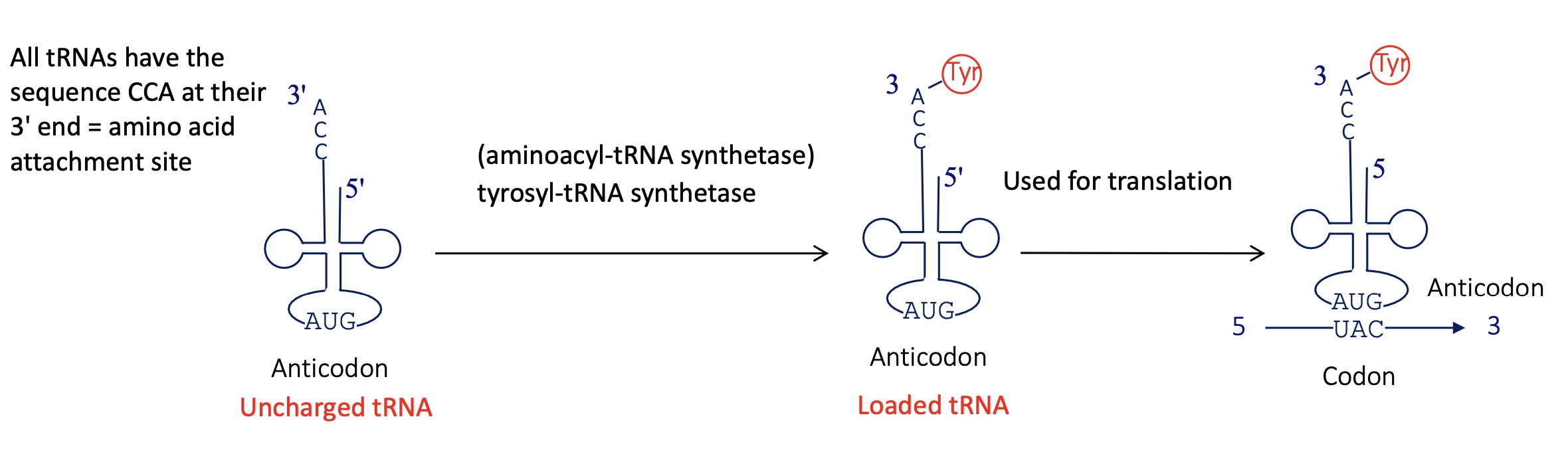
2.3 using the example of tyrosine how is the tRNA loaded with tyr
All tRNA have a CCA sequence at their 3’ = AA attachment site
tyrosyl-tRNA synthetase (aminoacyl tRNA synthetase) will charge the tRNA w/ tyr at the CCA sequence
loaded tRNA can bind to codon and be used for translation
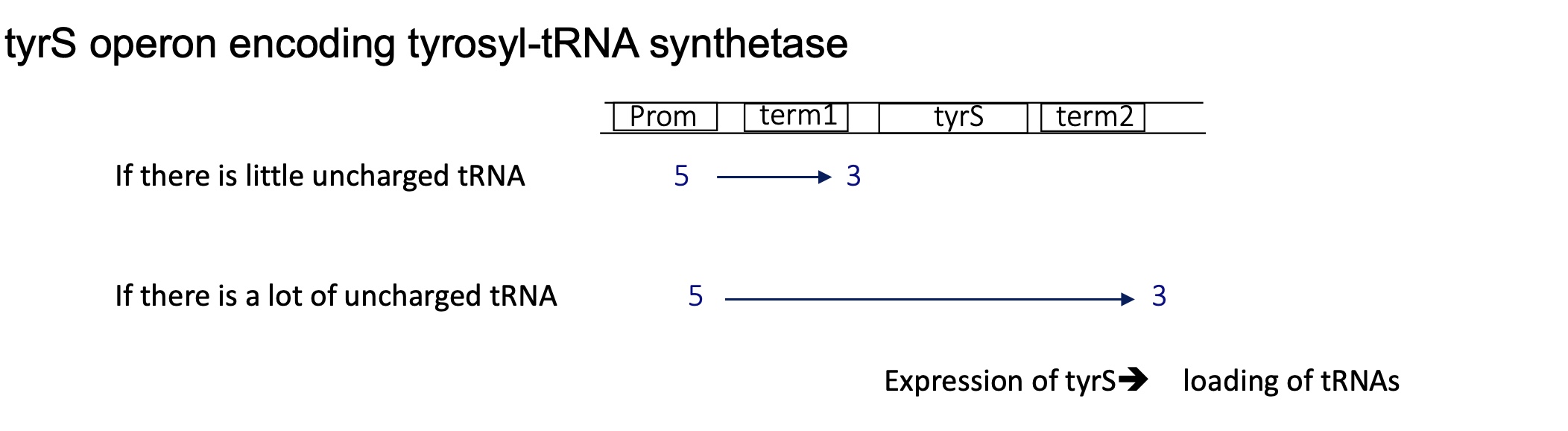
2.3 what does the tyre operon encoding for tyrosyl-tRNA synthetase look like
contains 2 terminators :
term1 after the promoter —> used when there’s high ratio of charges tRNA so less tyrosyl-tRNA synthetase needed
term2 at the end of the tyrS gene —> used when there’s a high ratio of uncharged tRNA so more tyrosyl-tRNA synthetase is needed
2.3 if there’s few uncharged tRNA/many charged tRNA what is the termination control for tyrS
mRNA transcribed has a:
stem loop structure w/ codon recognised by corresponding tRNA ( UAC for tyr codon) —> remains unbound to tRNA
T box which contains an anti terminator + UGG triplet doesn’t bind to S1 —. no anti terminator loop structure formed
Terminator stem loop of S1-S2 formed at the U rich region
—> termination at term1 as there’s no uncharged tRNA to bind to t-box
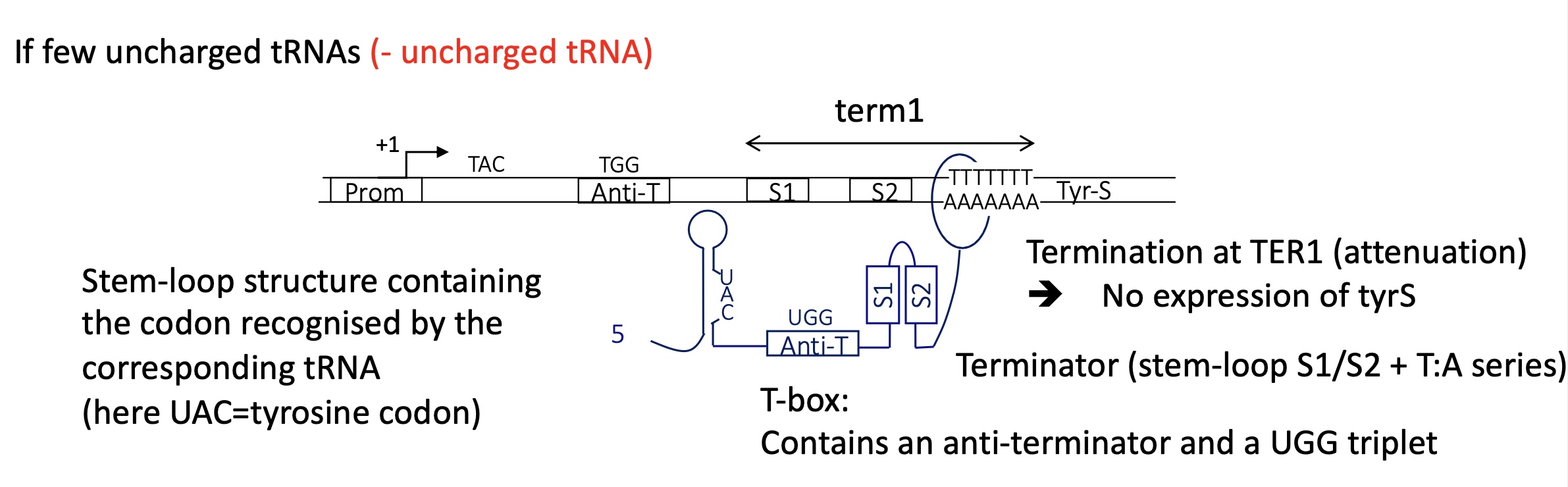
2.3 if there’s many uncharged tRNA/few charged tRNA what is the termination control for tyrS
mRNA transcribed :
the uncharged tRNA binds to codon (UAC) on the stem loop structure and the UGG triplet on the T-box
stabilises the formation of an anti terminator team loop structure between T box and S1
no terminator stem loops structure
elongation continues
—> termination at tem 2 and expressie of tyrS