Molec Cell Ch 4- Protein structure and function
1/77
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
78 Terms
The shape of a protein is specified by its ______
amino acid sequence
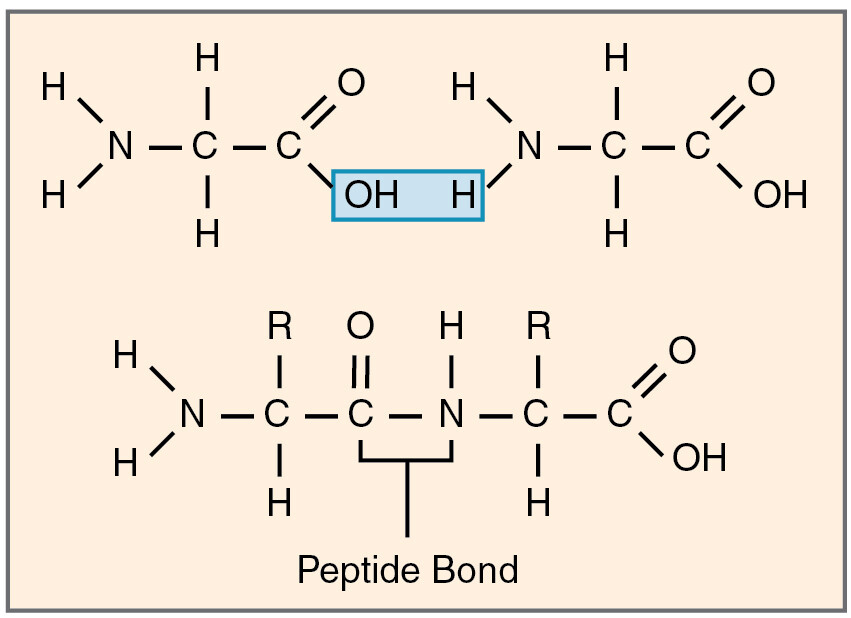
Where is the peptide bond located in the polypeptide sequence ?
at the C-N bond. NOT at the N-C bond.
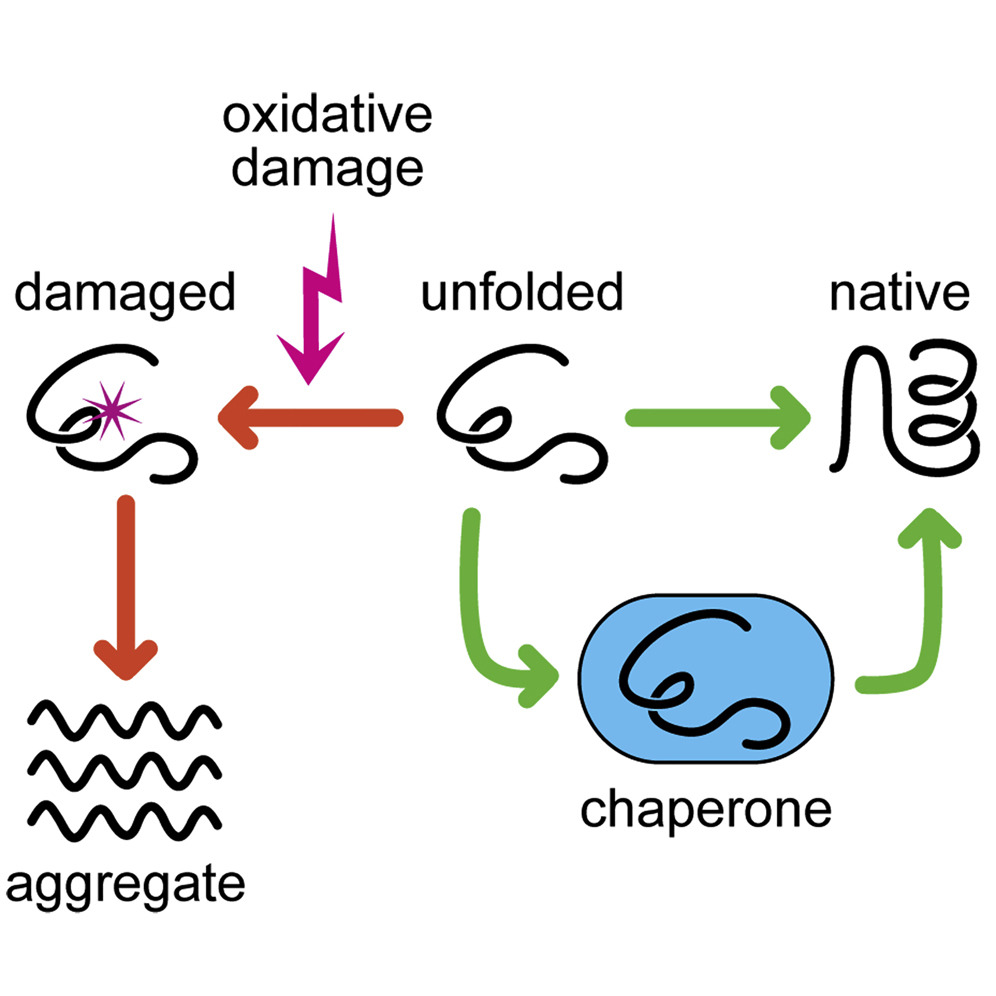
Noncovalent, (Electrostatic attractions, hydrogen bonds, and van der waals attractions) weak interactions within the polypeptide ultimately lead to the overall 3D conformation. Proteins fold into a conformation of lowest energy.
True.
Some proteins can spontaneously remold into their proper shape but most need the assistance of chaperone proteins to fold properly.
True
Hydrogen bonds within the polypeptide backbone produce the secondary structure.
> This is the alpha helices and beta sheets.
True
_______ are often found as transmembrane regions of proteins (exposed hydrophobic amino acids)
Alpha helices - These regions are also used to interact with other polypeptides.
{The helices wrap around each other to minimize exposure of hydrophobic amino acid side chains to aqueous environment}
________ form rigid structures at the core of many proteins.
-can be parallel or antiparallel.
-can form large pores called B-barrels.
- can stack to form damaging amyloid structures (secondary structure; insoluble fibrous protein aggregates)
Beta sheets
Prions
cause normal proteins to misfold and produce amyloid structures.
The interaction of ______ between secondary structures produces regions of tertiary structure with function; protein domains
R groups (side chains)
Can different domains be found across multiple proteins?
Yes. Most proteins contain multiple domains.
Proteins are organized into ______ based upon ______ domains.
Families; homologous
quaternary structure.
Large protein molecules often contain more than one polypeptide chain
-subunits can be the same or different.
Outside cells Collagen and Elastin are common constituents of extracellular matrix and form fibers in tendons and ligaments.
Collagen polypeptides are organized as a _____
Triple coiled coil.
Elastin is composed of?
Multiple polypeptides cross-linked to each other (via di-sulfide bonds that form between cysteine residues)
Inside cells, tubulin forms long, stiff microtubules.
> Actin forms filaments that support the plasma membrane.
> Keratin forms fibers that reinforce epithelial cells- it is the major protein in hair and horn.
Proteins bond to other molecules via noncovalent interactions between the surface of the protein and ______.
Ligand
_____ catalyze covalent bond breakage or formation
Enzymes
Enzymes chemically transform the ligands to which they bind, thus speeding up chemical reactions (aka: catalysts)
True
Proteins are regulated through the mechanisms:
Allosteric regulation, feedback inhibition, positive feedback, and feed-forward regulation.
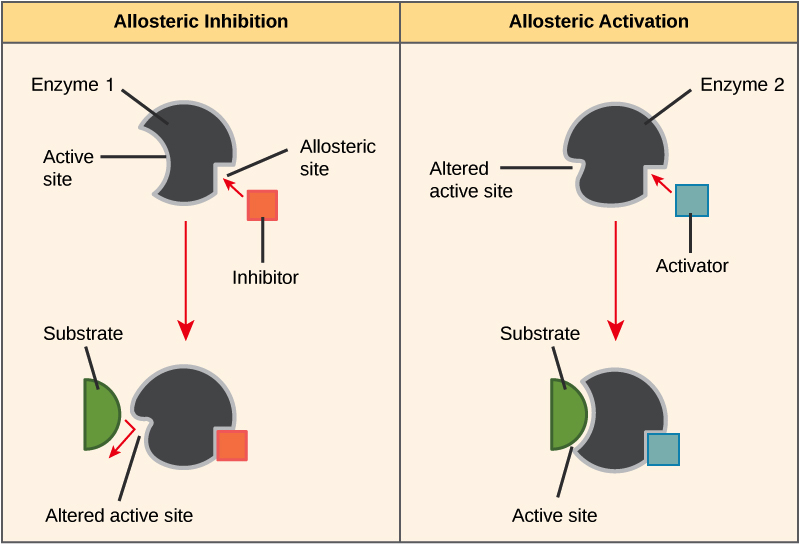
allosteric regulation
The binding of a regulatory molecule to a protein at one site (outside of the catalytic site) that affects the function of the protein at a different site.
Allosteric regulators are often products of other chemical reactions in the same biochemical pathway.
True
Proteins have multiple binding sites for _______ and _______.
Catalytic activity and regulation.
Many proteins utilize multiple regulatory sites in order to integrate multiple pieces of information from the environment.
True
Regulation via phosphorylation:
Phosphorylation is the process by which a phosphate group is added to a molecule.
Covalent modifications can regulate where a protein is located, regulate its function, and regulate its degradation.
True
G-proteins are regulated by_______________.
Binding and hydrolysis of GTP.
Interacting with GAP and GEF proteins.
GAP - GTPase activating proteins
GEF - Guanine nucleotide exchange factors
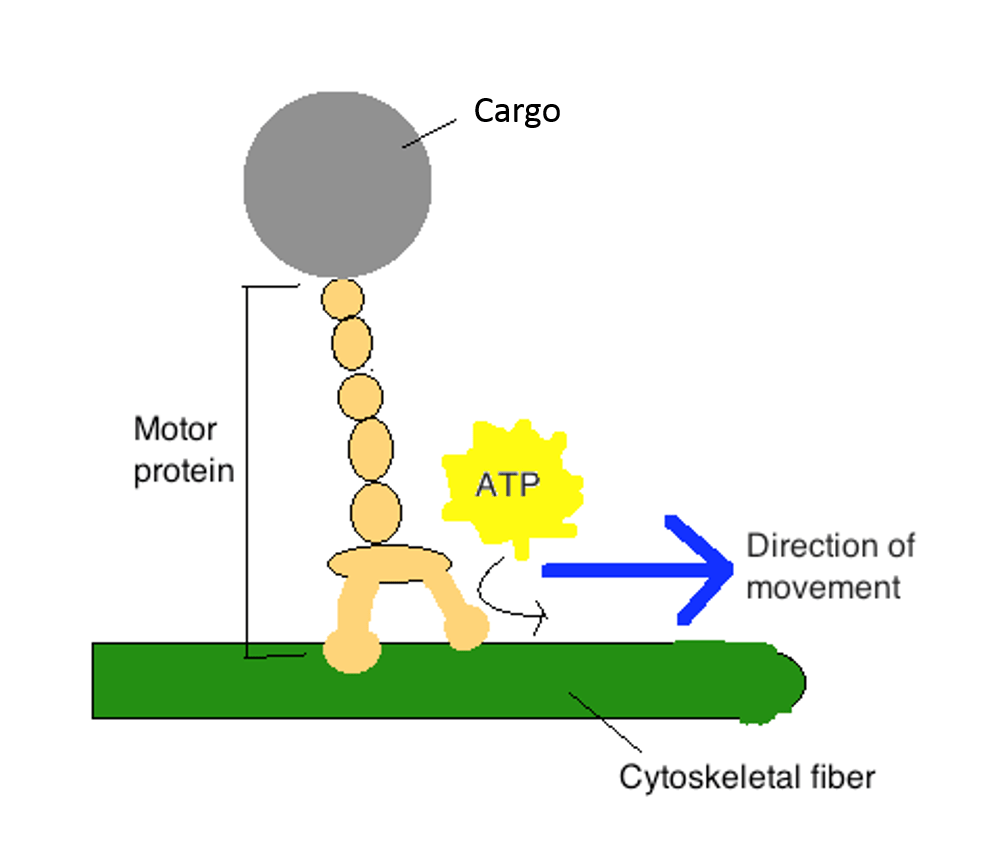
_________ allows motor proteins to produce directed movements in cells.
ATP hydrolysis (irreversible)
Proteins can be tagged for destruction. > Proteins tagged with polyubiquitin will be brought to the proteasome for degradation.
A large number of noncovalent interactions is required to hold two regions of a polypeptide chain together in a stable conformation.
The amino acid sequence = the primary structure of a protein.
True
primary structure
amino acid sequence
what leads to the overall 3D conformation of proteins?
non-covalent weak interactions w/in the polypeptide
chaperone proteins
assist in the correct folding, assembly, stabilization, and degradation of other proteins
alpha helices
Spiral-shaped
Found in cell membranes (transmembrane regions) (hydrophobic amino acids exposed)
alpha helical regions used to interact w/ other polypeptides
formed by hydrogen bonds
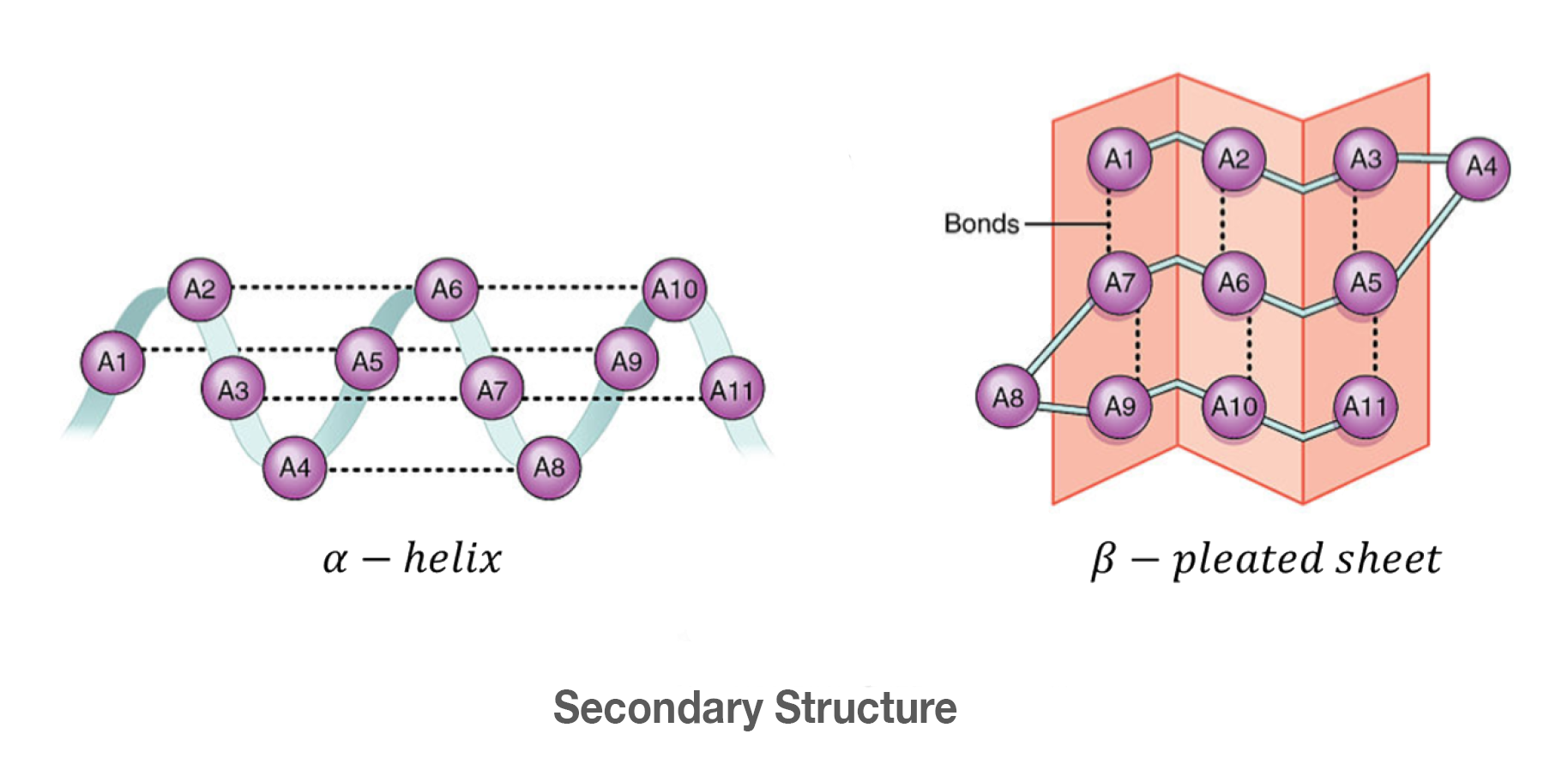
B-sheets
Flat, rigid structures
Can be:
Parallel
Antiparallel
Functions:
Form protein cores
Can create β-barrels (large pores)
⚠ Important pathology
β-sheets can stack into amyloids (harmful)
Prions cause normal proteins to misfold into amyloids
This leads to TSEs (transmissible spongiform encephalopathies)
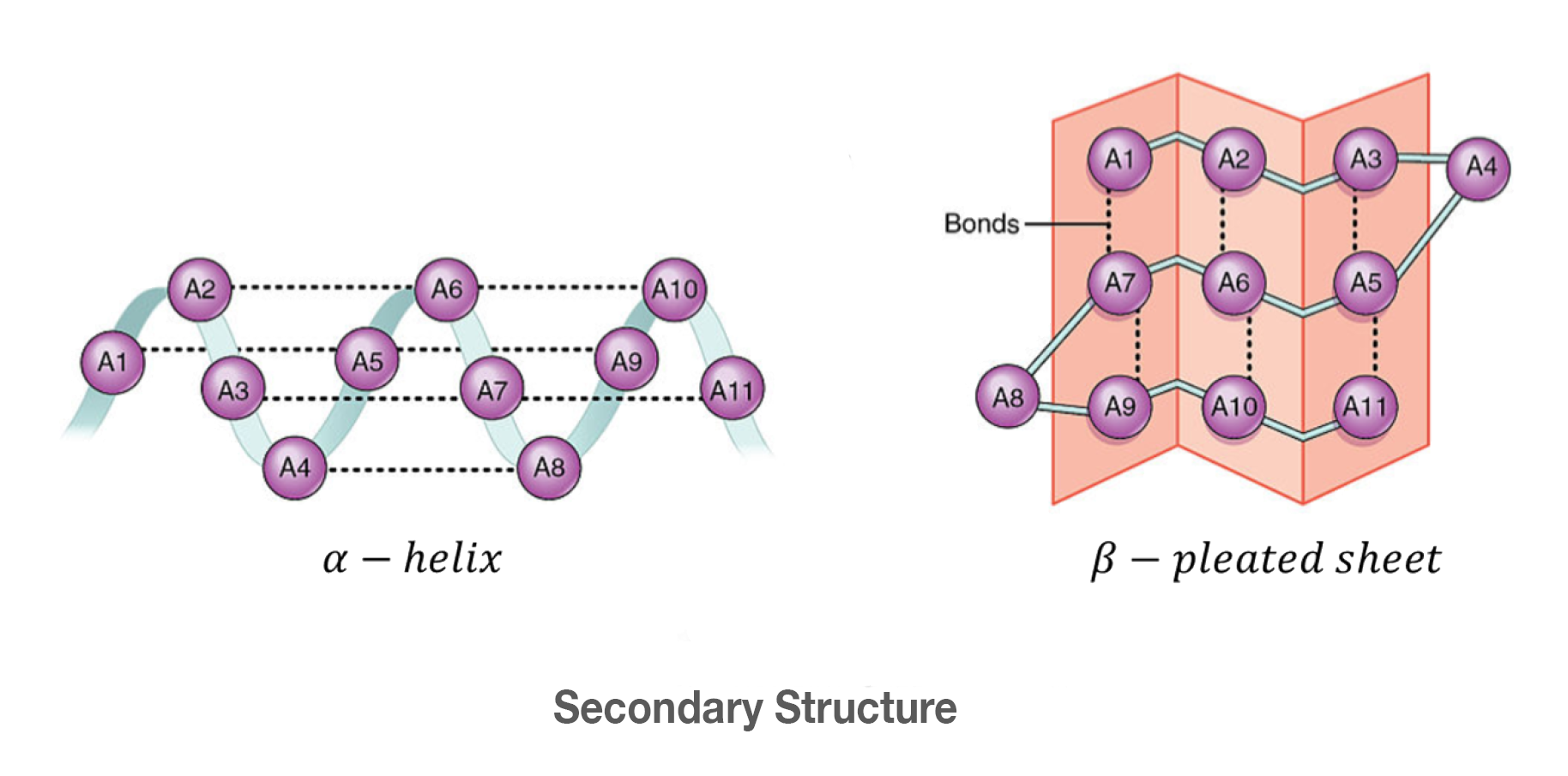
secondary structure
Formed by hydrogen bonds in the backbone of the polypeptide
α-Helices
Spiral-shaped
Often:
Found in cell membranes (hydrophobic amino acids face outward)
Used for protein–protein interactions
β-Sheets
Flat, rigid structures
Can be:
Parallel
Antiparallel
Functions:
Form protein cores
Can create β-barrels (large pores)
domains
Functional regions in the tertiary structure of proteins composed of a specific combination of secondary structures
Same domains can appear in different proteins
Most proteins have multiple domains
tertiary structure
Overall 3D shape of one polypeptide
Formed by interactions between R groups (side chains) through noncovalent bonds
Creates protein domains
proteins organized into families based upon homologous domains + functions
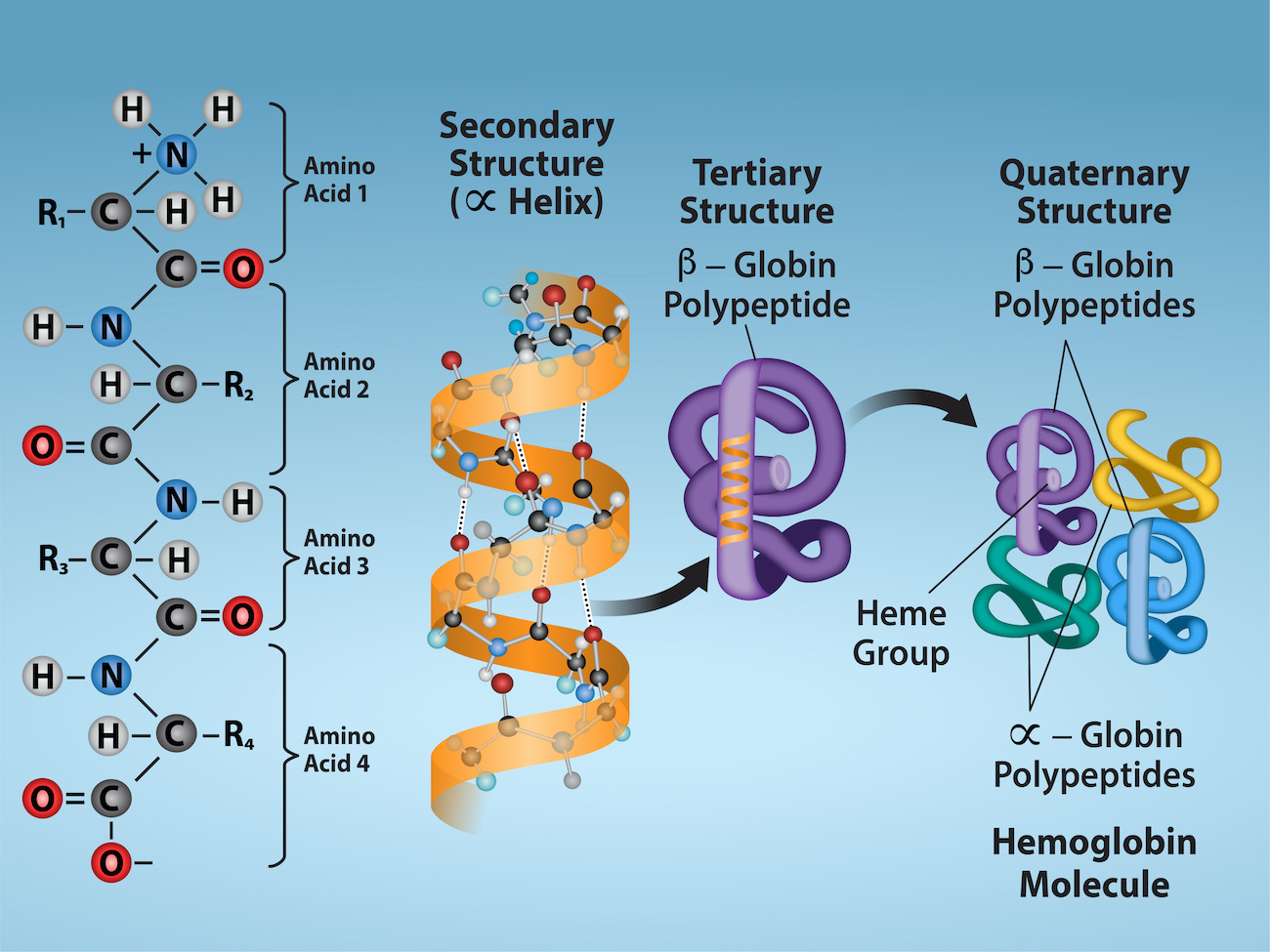
quaternary structure
Proteins made of multiple polypeptide chains (subunits)
Subunits can be:
Identical
Different
Examples:
Collagen → triple helix
Elastin → cross-linked polypeptides
Stabilized by:
Non-covalent interactions
Disulfide bonds (between cysteine residues)
how are disulfide bonds formed?
formed btwn cystine residues
cross links polypeptides
How Proteins Work
Binding
All proteins bind other molecules (ligands)
Binding uses non-covalent interactions
Shape + chemistry must match
Enzymes:
Bind substrates
Lower activation energy
Speed up chemical reactions
They are not consumed in the reaction
Enzymes do not require an input of energy from ATP for activation (most of the time)
chemically transform the ligands they bind to!
protein regulation
Proteins are regulated so cells can respond to their environment.
allosteric regulation
feedback regulation
phosphorylation
multiple binding sites
G-proteins
allosteric regulation
binding of an effector molecule to a specific site distinct from the active site, inducing a conformational change
regulation by binding outside of catalytic site
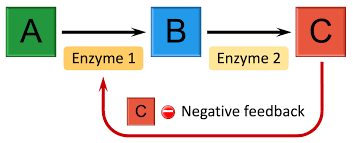
feedback inhibition
product shuts off pathway, stops rxn from continuing to occur
positive feedback
The end product enhances the production of more products.
feed-foward regulation
early signal prepares later steps
proteins have multiple binding sites
Catalytic sites
Regulatory sites
This lets them integrate multiple signals from environment
bind noncovalently
phosphorylation
a type of covalent protein regulation
a phosphate group is added to serine, threonine, or tyrosine amino acid residues, acting as a molecular switch to activate or deactivate proteins
also effects:
- Function
Localization
Degradation
can cause conformational change
creates docking sites for other proteins to bind, hence promoting assembly into larger complexes
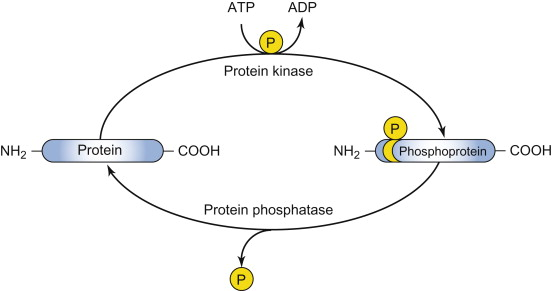
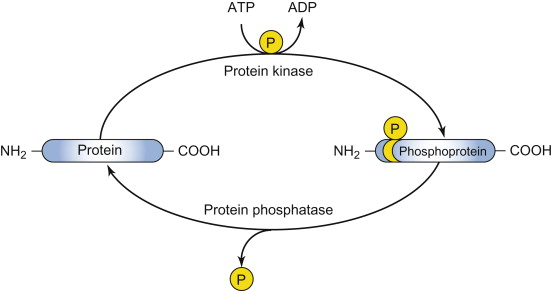
protein kinase
Enzyme that catalyzes the transfer/addition of a phosphate group from ATP to a specific amino acid side chain on a target protein.
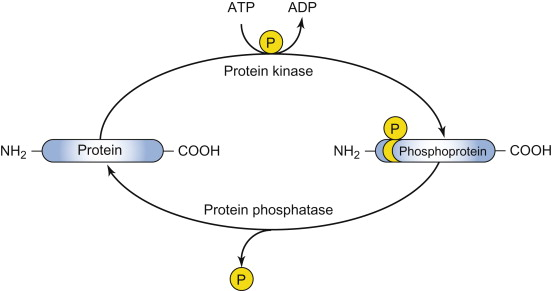
protein phosphatase
Enzyme that catalyzes the removal of a phosphate group from a protein, often with high specificity for the phosphorylated site.
G-proteins
Intracellular signaling protein whose activity is determined by its association with either GTP or GDP. Includes both trimeric G proteins and monomeric GTPases, such as Ras.
Regulated by:
Binding GTP (active)
Hydrolyzing GTP → GDP (inactive)
Controlled by:
GAPs → speed up GTP hydrolysis (turn OFF)
GEFs → replace GDP with GTP (turn ON)
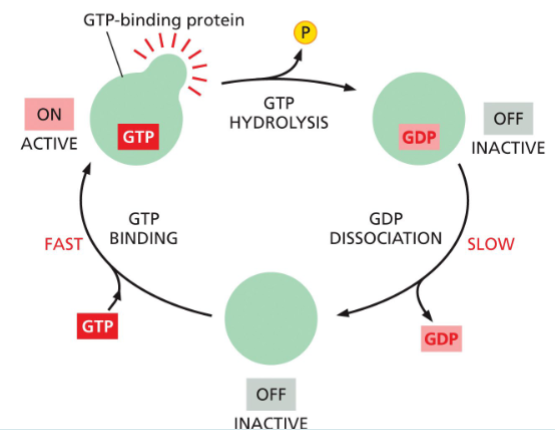
g-proteins are active when?
when GTP is bound
they can hydrolyze this GTP to GDP—which releases a phosphate and flips the protein to an inactive conformation
the active conformation is regained by dissociation of the GDP, followed by the binding of a fresh molecule of GTP
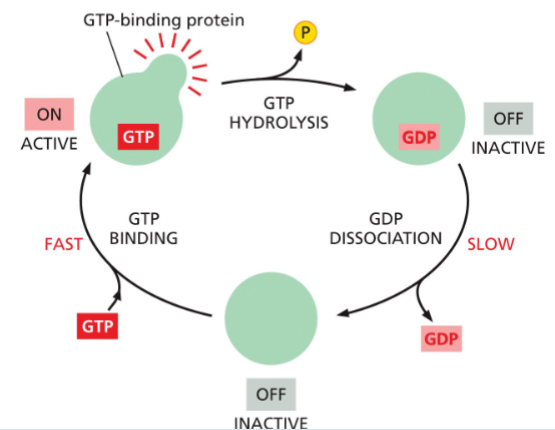
g-proteins are inactive when?
inactive when they’re bound to GDP
A GEF helps the G-protein release GDP and bind GTP → turns it ON
The G-protein’s own GTPase activity (often sped up by GAPs) hydrolyzes GTP to GDP → turns it OFF
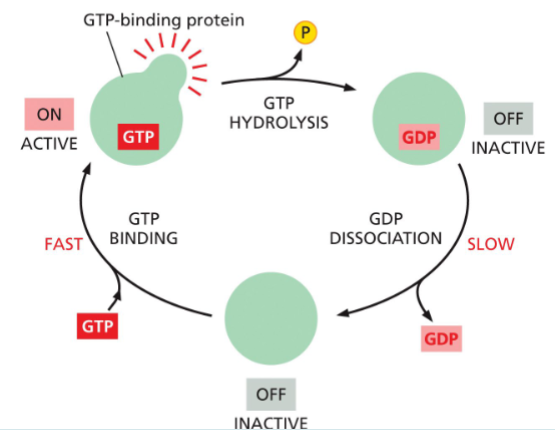
GEF
guanine nucleotide exchange factors
swaps GDP for GTP → turns protein ON
GAP
GTPase activating proteins
Speeds up turning the G-protein OFF
How:
Makes the G-protein hydrolyze GTP faster
speeds up the GTPase → turns protein OFF faster
GTPase
breaks GTP into GDP → turns protein OFF
An ability the G-protein already has
motor proteins
Protein such as myosin or kinesin that uses energy derived from the hydrolysis of a tightly bound ATP molecule to propel itself along a protein filament or other polymeric molecule.
Use ATP hydrolysis
Produce directed movement in cells
motor proteins are also ATPases. A great deal of free energy is released when ATP is hydrolyzed, making it very unlikely that the protein will undergo the reverse shape change needed to move backward
protein degradation
Proteins tagged with polyubiquitin
Sent to the proteasome for destruction
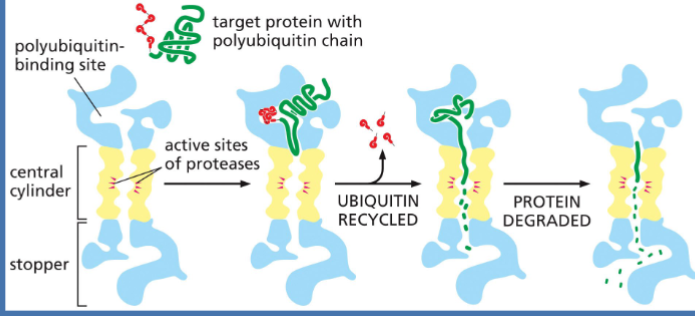
polyubiquitin
targets proteins for degradation
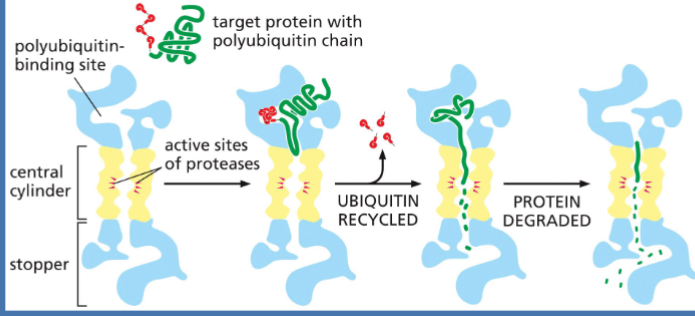
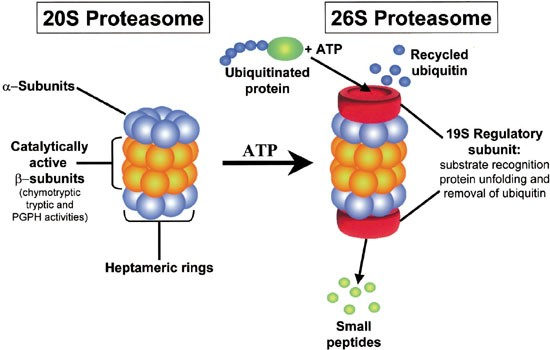
proteasome
where proteins r sent after being tagged w/ ubiquitin to be degraded
a massive, multi-subunit protease complex
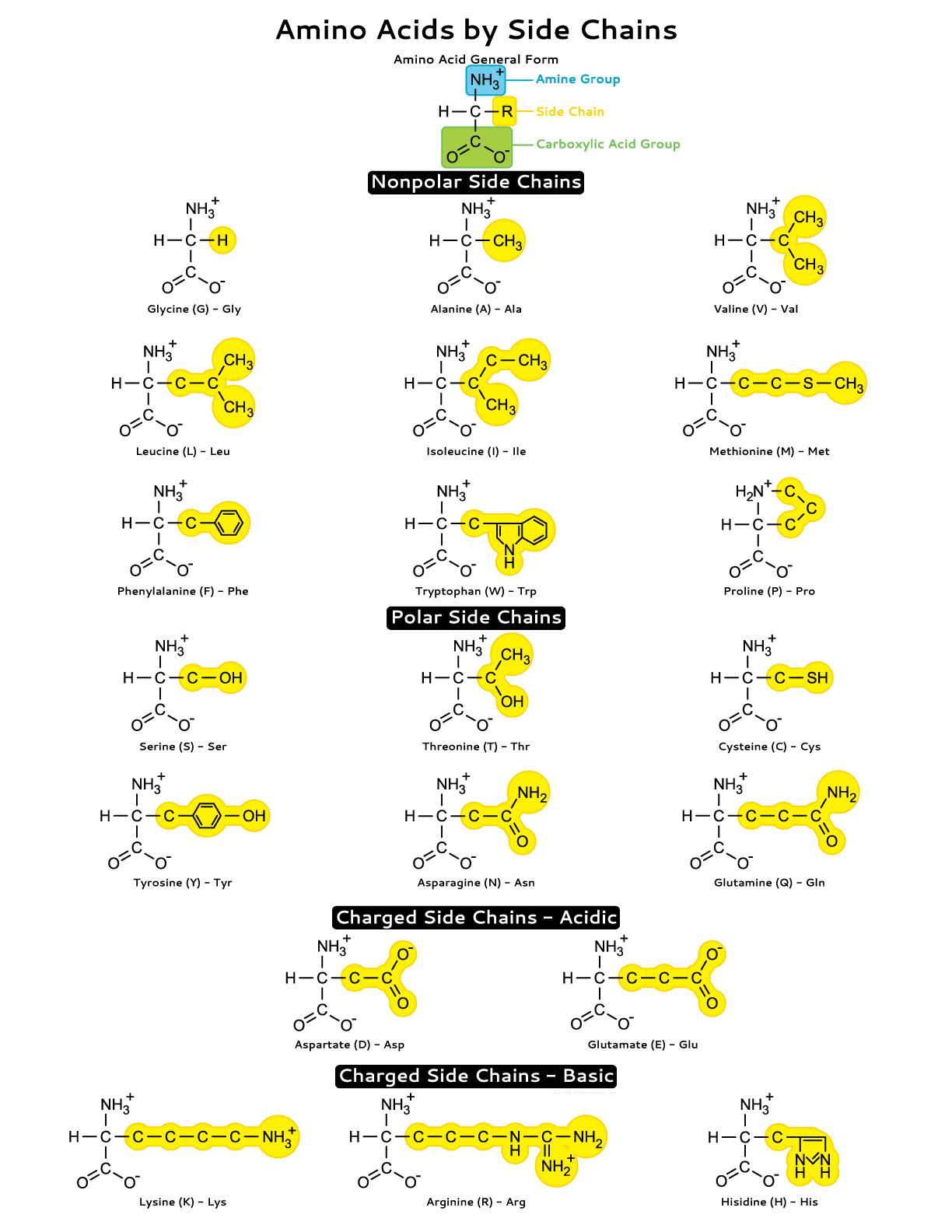
side chains
Portion of an amino acid not involved in forming peptide bonds; its chemical identity gives each amino acid unique properties.
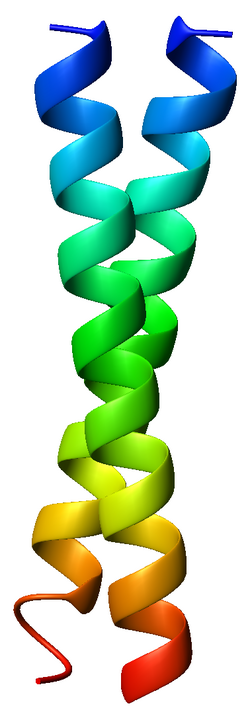
coiled coil
Stable, rodlike protein structure formed when two or more α helices twist repeatedly around each other.
hydrophobic side chains facing inward
amyloid structures
misfolded proteins can cause this which leads to disease
prions are considered “infectious” because the amyloid form of the protein can convert properly folded molecules of the protein into the abnormal, disease-causing conformation
covalent cross-linkages
To help maintain their structures, the polypeptide chains in such proteins are often stabilized by covalent cross-linkages. These linkages can either tie together two amino acids in the same polypeptide chain or join together many polypeptide chains in a large protein complex
most common type is disulfide bond btwn 2 cysteines
ATPases
a crucial group of enzymes that catalyze the hydrolysis of adenosine triphosphate (ATP) into adenosine diphosphate (ADP) and inorganic phosphate, releasing energy to power essential cellular processes
How are the 4 levels of protein structure defined?
Primary: Amino acid sequence held together by peptide bonds.
Secondary: Local folding (α-helices, β-sheets) stabilized by hydrogen bonds.
Tertiary: Overall 3D shape of one polypeptide due to side-chain interactions.
Quaternary: Association of multiple polypeptide chains into one functional protein.
What are the roles of covalent vs noncovalent interactions in protein formation?
Covalent bonds (peptide bonds, disulfide bridges) provide strong, permanent structure.
Noncovalent interactions (hydrogen bonds, ionic bonds, hydrophobic interactions) drive folding and stabilize shape.
What is the causative agent of a TSE and how does this disease arise?
Prions (misfolded proteins).
They cause disease by inducing normal proteins to misfold, leading to brain damage.
By what types of processes do proteins mature?
Folding into correct shape.
Post-translational modifications (cleavage, phosphorylation, glycosylation).
Assembly with other subunits if needed.
What are the types of mechanisms by which protein activity can be regulated?
Allosteric regulation (binding of regulators).
Covalent modification (e.g., phosphorylation).
Protein degradation or activation by cleavage.
Changes in localization within the cell.
hydrolase
The splitting of a compound into fragments by the addition of water,
General term for enzymes that catalyze a hydrolytic cleavage reaction
nuclease
Breaks down nucleic acids by hydrolyzing covalent phosphodiester bonds between nucleotides
protease
Breaks down proteins by hydrolyzing peptide bonds between amino acids
isomerase
Catalyzes the rearrangement of bonds within a single molecule
rearrangement into a diff isomer
polymerase
Catalyzes polymerization reactions such as the synthesis of DNA and RNA
oxidoreductase
General name for enzymes that catalyze reactions in which one molecule is oxidized while the other is reduced. Enzymes of this type are often called oxidases, reductases, or dehydrogenases