Microbial phylogeny + taxonomy
1/41
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
42 Terms
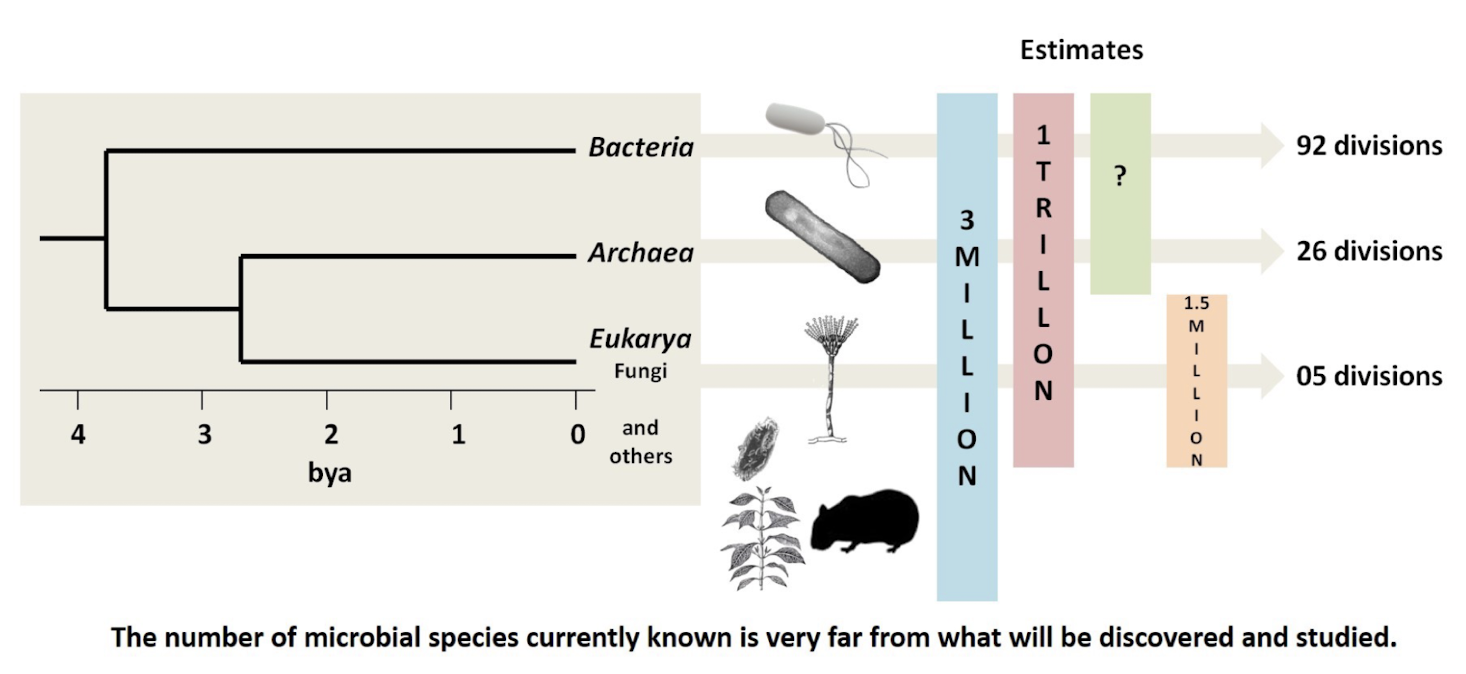
What is the estimated number of microbial species, and why is it uncertain?
~3 million species of organisms are formally described, but microorganisms may number up to 1 trillion species
Uncertainty arises because the vast majority are uncultivable, reside in unexplored habitats, or are invisible without molecular tools.
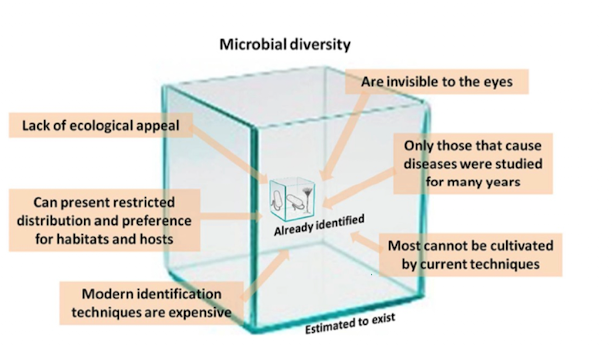
Why is the “unknown microbial world” so large?
Microbes can be:
Invisible to the eye
Restricted to extreme niches
Unculturable
Lacking clinical/ecological appeal
Detectable only with advanced molecular techniques
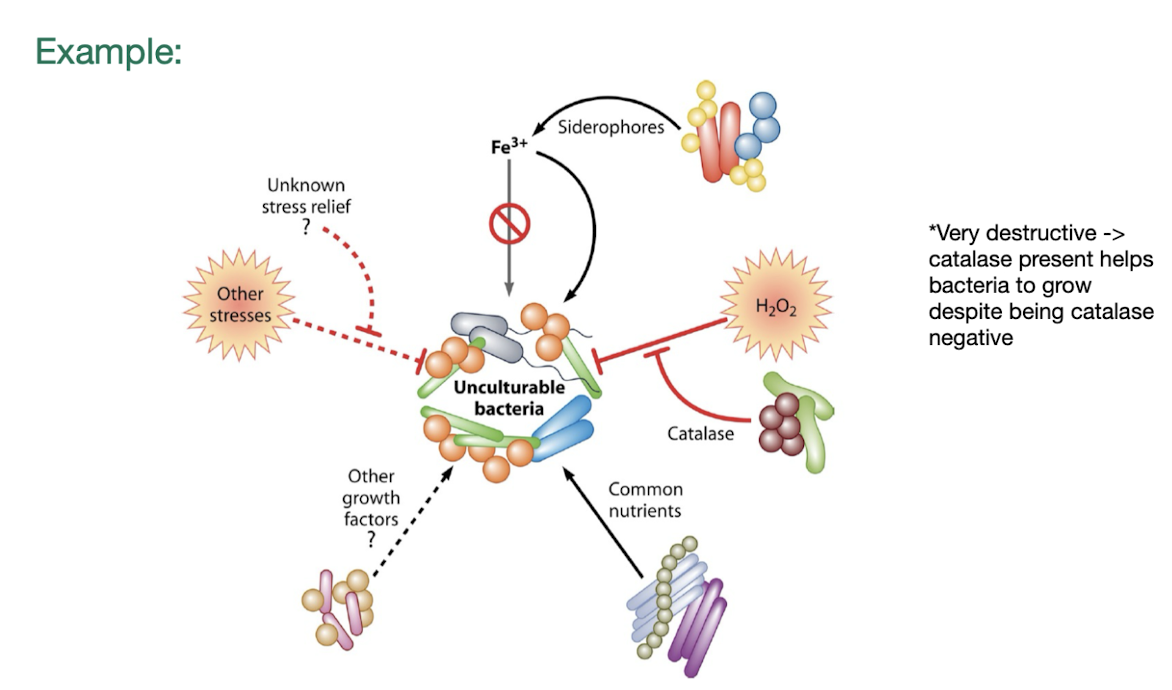
What is meant by uncultivable microbes?
Refer to microbial species that cannot be grown or maintained in laboratory settings due to specific growth requirements, environmental conditions, or unknown factors.
These microbes often exist in natural habitats, making them difficult to study using traditional culture techniques.
How do detection methods influence known microbial diversity?
Traditional culture-based methods detect only organisms that grow under lab conditions.
Growth depends on medium composition, incubation temperature, oxygen tolerance, and microbial interactions
Thus, species requiring specific nutrients or partners remain undetected.
How does co-culturing reveal unculturable microbes?
Some bacteria depend on helper strains that supply nutrients (amino acids, vitamins, carbon sources) or detoxify the environment (e.g., catalase removing ROS).
Co-culturing can therefore make previously “uncultivable” bacteria grow
How do habitats shape microbial diversity?
Heterogeneous/distributed habitats → occupied by generalist species
Homogeneous/stable habitats → occupied by specialist species and are influenced by natural selection and adaptation to specific environmental conditions based on biotic/ abiotic factors
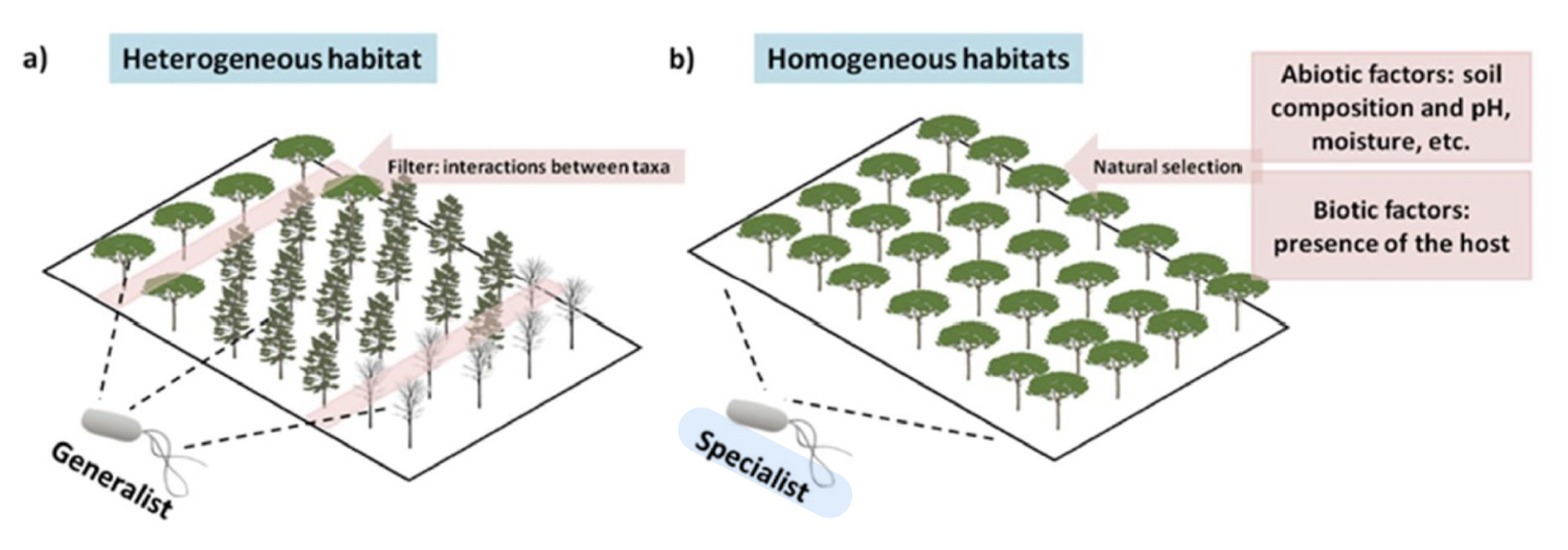
What are generalist species?
Organisms that can thrive in a wide variety of environmental conditions and habitats e.g. Bacillus species
Possess a broad range of resources they can utilise, allowing them to be flexible in their survival and reproduction.
Evolution of these species is influenced based on interaction b/w various taxa
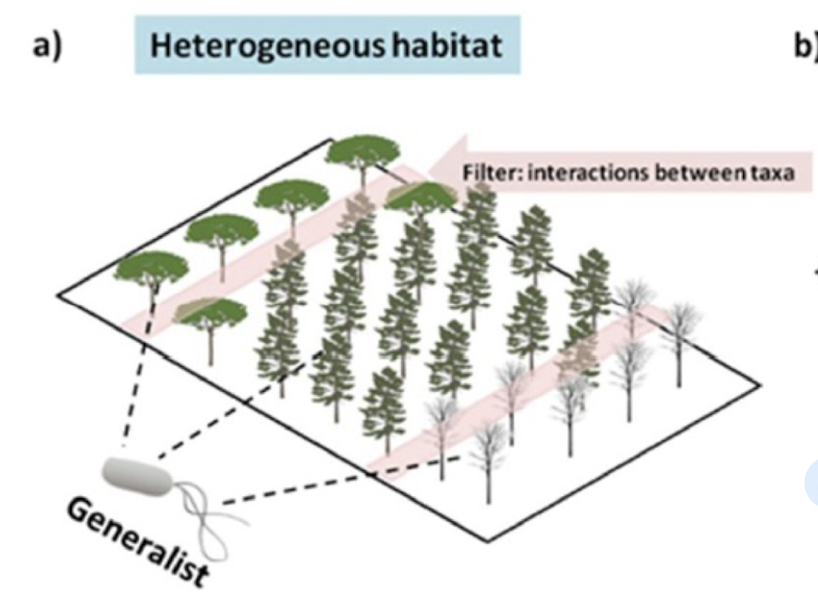
Microbial evolution occurs through ______
Mutations:
Mutations are inherited in offspring and maintained if beneficial (natural selection)
Leads to biodiversity
Thus, no. of mutations can indicate the amount of evolutionary change in a population over time.
Evolutionary chronometer
genes and proteins which can be used to measure evolutionary distance between species based on their genetic differences.
What Makes a Good Evolutionary Chronometer?
Gene must have:
Universal distribution
Functional homology
Conserved + variable regions (sequence homology)
Sequence changes proportional to evolutionary time
Examples: ATPase, RecA, 16S/18S rRNA.
What is SSU sequencing?
SSU sequencing refers to small subunit ribosomal RNA sequencing, a method used to analyse genetic material from microorganisms for phylogenetic studies.
Why is 16S rRNA widely used in microbial identification?
Universally present in bacteria/archaea
Contains conserved regions for primer binding and variable regions for discrimination
Large, stable, slow-evolving structure
Supported by extensive databases (RDP - ribosomal database project)
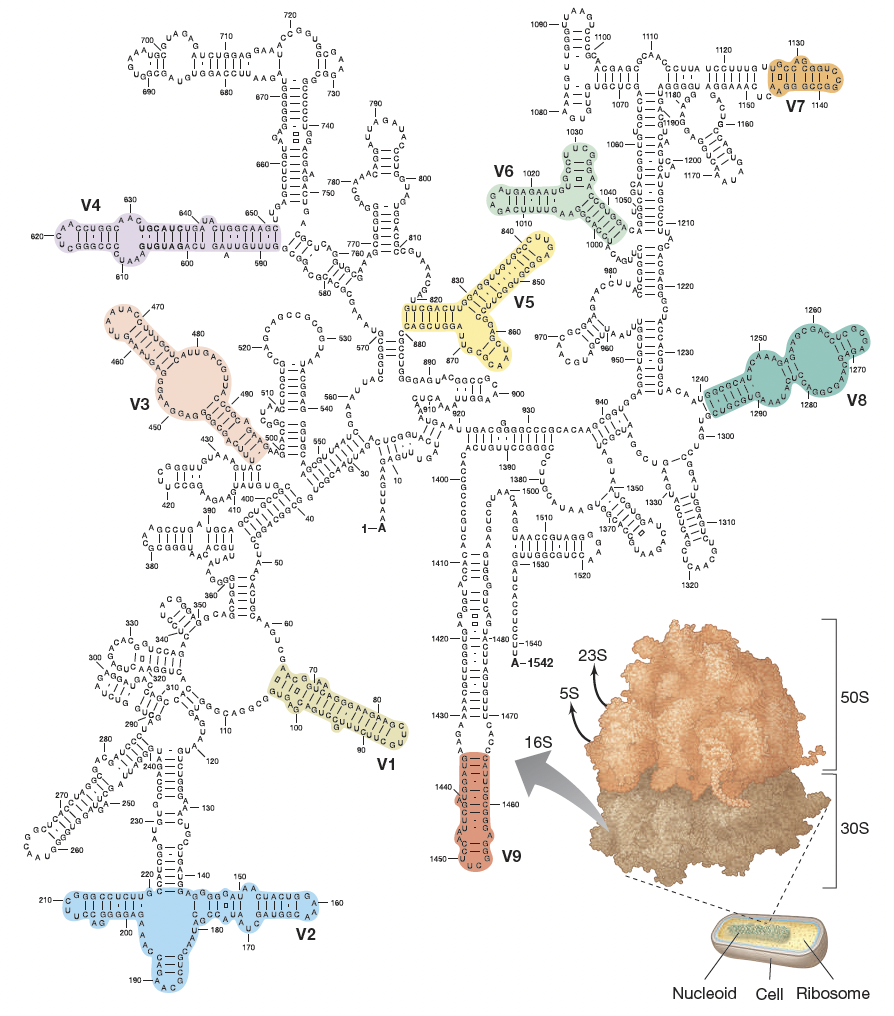
Limitations of 16S rRNA?
Cannot resolve closely related species (low sequence divergence)
Some organisms share nearly identical 16S but have different genomes and phenotypes
e.g. E. coli and Shigella dysenteriae can have very few (e.g., 3 bp) differences in their 16S rRNA but are clinically distinct pathogens
Unhelpful for strain-level epidemiology
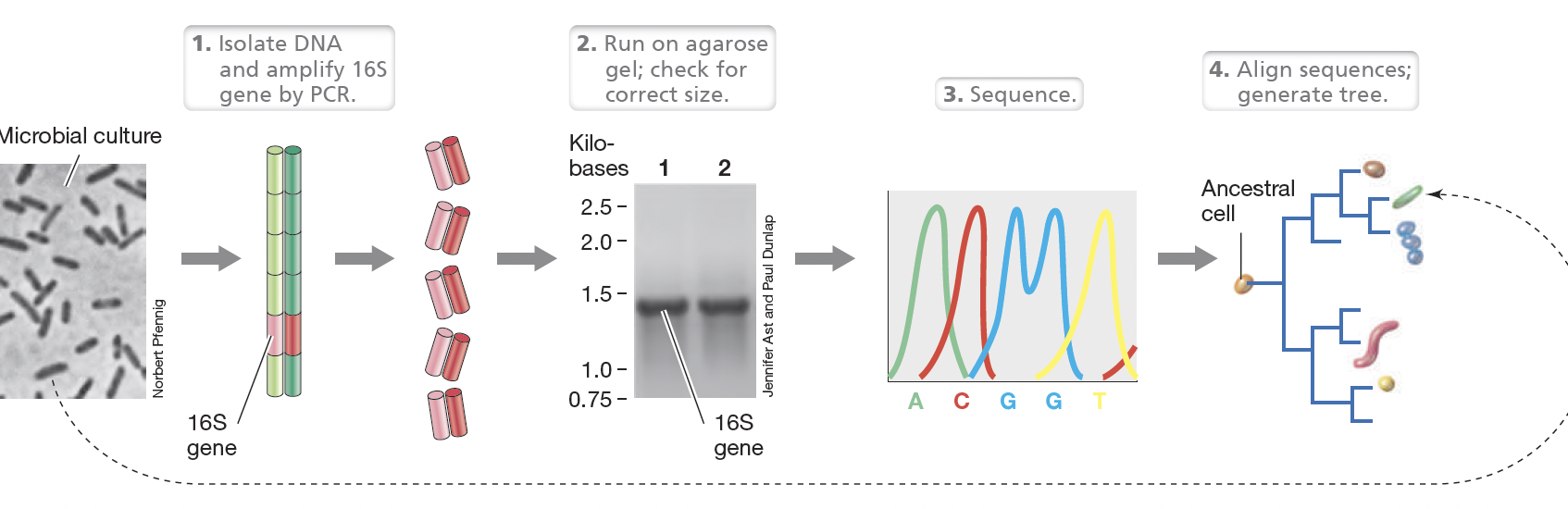
PCR amplification of the 16S rRNA gene from an unknown microbial culture workflow:
Isolate Pure culture → extract DNA
PCR amplify 16S using conserved primers, run PCR product on agarose gel
Sequence
Align sequences → calculate evolutionary distances then calculate the corrected evolutionary distance and construct
the tree.
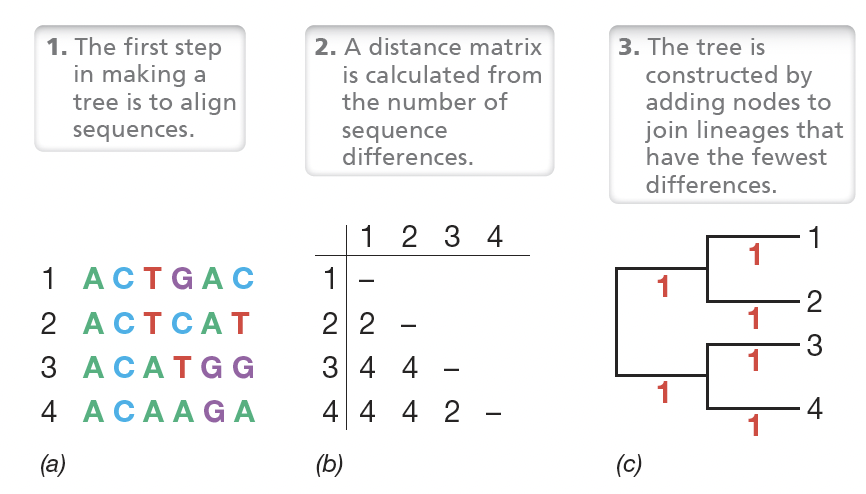
Build phylogenetic tree using models (parsimony, etc.)
What is the parsimony method in constructing phylogenetic trees?
Assumes the minimal amount of changes to diverge two sequences actually occurred during evolution.
• Other methods assume some bases more likely to change than others.
• Thus all trees are approximations of the true phylogeny of the group
How can PCR primers be designed for 16S sequencing?
Primers can be designed that target distinct species (or all species) within a genus, all genera within a phylum, or all phyla within a domain,
“Universal” PCR primers that will amplify the SSU rRNA gene from any organism
What is “corrected evolutionary distance”?
A statistical adjustment that accounts for the fact that some base changes occur more often than others.
Used to build more accurate phylogenetic trees
What are rRNA signature sequences?
Specific oligonucleotide motifs in 16S/18S rRNA that differentiate domains or genera (e.g., sequences unique to Archaea vs Bacteria)
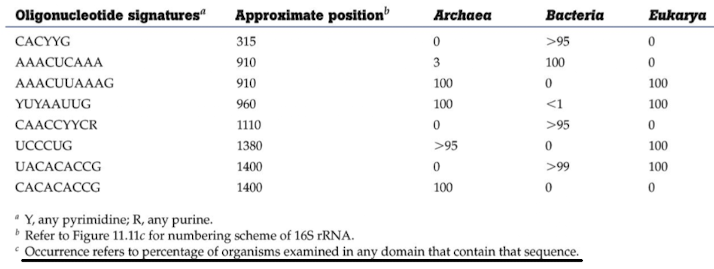
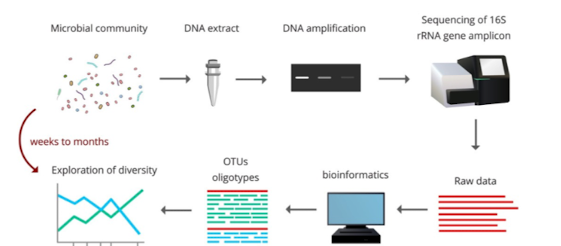
How does PCR-based community analysis expand known diversity?
Amplifying 16S genes directly from environmental DNA allows identification of organisms without cultivation, revealing novel taxa
What is FISH?
Fluorescence in situ hybridization (FISH) is a technique used to detect and localize the presence or absence of specific DNA sequences in microbial cells.
Uses fluorescent probes that bind to complementary DNA sequences, enabling visualization of specific microorganisms within their natural environments.
Role of FISH in microbial systematics?
Labeled nucleic acid probes hybridize to complementary rRNA inside intact cells, enabling visualisation and identification in situ. Useful in clinical diagnostics and ecology
Why use phenotypic characteristics if 16S is available?
Phenotypes reflect functional capabilities (metabolism, cell structure)
Some organisms with similar 16S differ drastically in phenotype
Essential for classification, diagnostics, and understanding domain-level traits
Give major distinguishing traits among the 3 major domains of microbial life:
Bacteria: Peptidoglycan cell walls; ester-linked lipids; 70S ribosomes
Archaea: No peptidoglycan; ether-linked lipids; unique metabolic pathways; extremophiles
Eukarya: Membrane-bound organelles; 80S ribosomes; diverse cell structures
Give examples of traits unique to specific domains.
Archaea: methanogenesis
Bacteria: nitrogen fixation, photosynthesis (various modes)
Eukarya: complex multicellularity
What is the binomial system of nomenclature?
• Genus name followed by species name
• Italicized
• Capital letter for first letter of Genus name.
– Bacillus subtilis or B. subtilis
• Only genus name can be abbreviated
• A genus name can be used alone but never a species name
developed by Carl Linnaeus
What is taxonomy & what does bacterial taxonomy rely on?
Taxonomy is the science of classification of living organisms into distinct groups
Bacterial taxonomy relies on characteristics such as morphology, genetic analysis, and biochemical properties.
What does DNA–DNA hybridization measure?
Overall genomic similarity between two organisms.
>70% hybridization traditionally indicates same species.
Useful for distinguishing closely related species
How does ribotyping differ from 16S sequencing?
Ribotyping uses restriction enzyme digestion to create a unique DNA banding pattern (“molecular fingerprint”).
Extremely specific; useful for strain discrimination i.e. in Clinical labs, food microbiology (detecting pathogens like E. coli), and environmental monitoring.
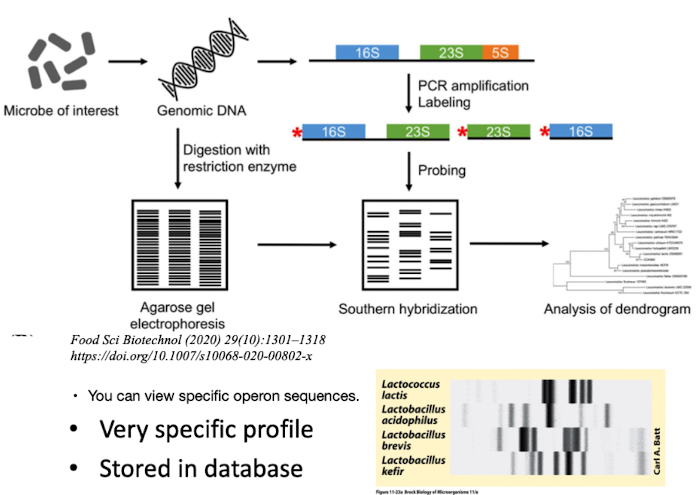
How is ribotyping performed?
Restriction Digestion: DNA is cut with restriction enzymes, creating fragments of the rRNA gene regions (16S, 23S, 5S).
Gel Electrophoresis: These fragments are separated by size.
Hybridization: Labeled rRNA probes bind to specific fragments.
Visualization: A unique banding pattern (riboprint) is produced, representing the organism's genetic makeup.
MLST (Multilocus Sequence Typing)
Used for characterizing bacterial species using DNA sequences from multiple housekeeping genes assigning a unique "sequence type" (ST) to each isolate for standardised tracking in public databases
Allows for the differentiation of closely related strains and provides insights into phylogenetic relationships.
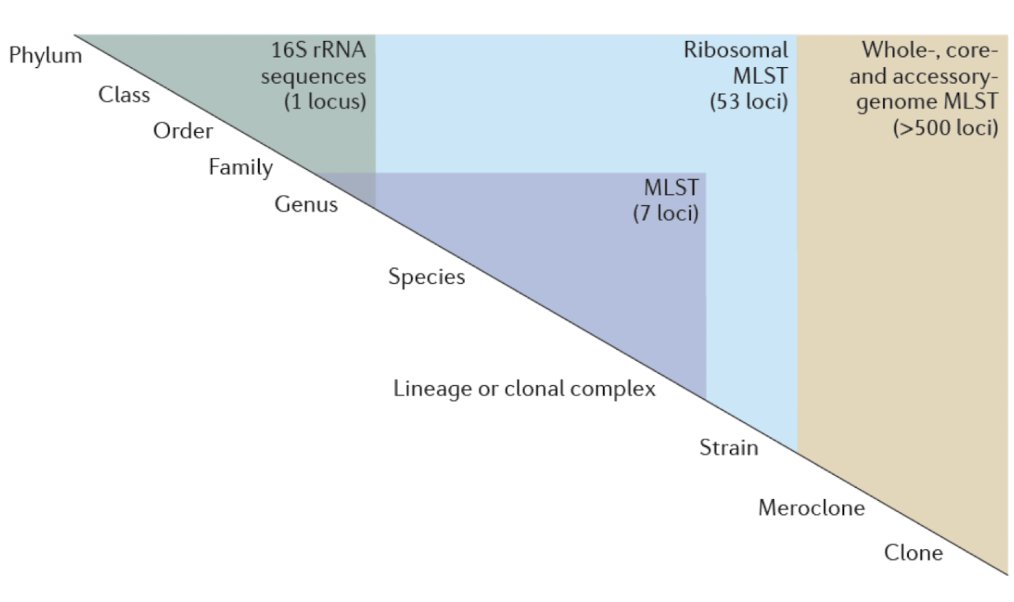
How is MLST performed?
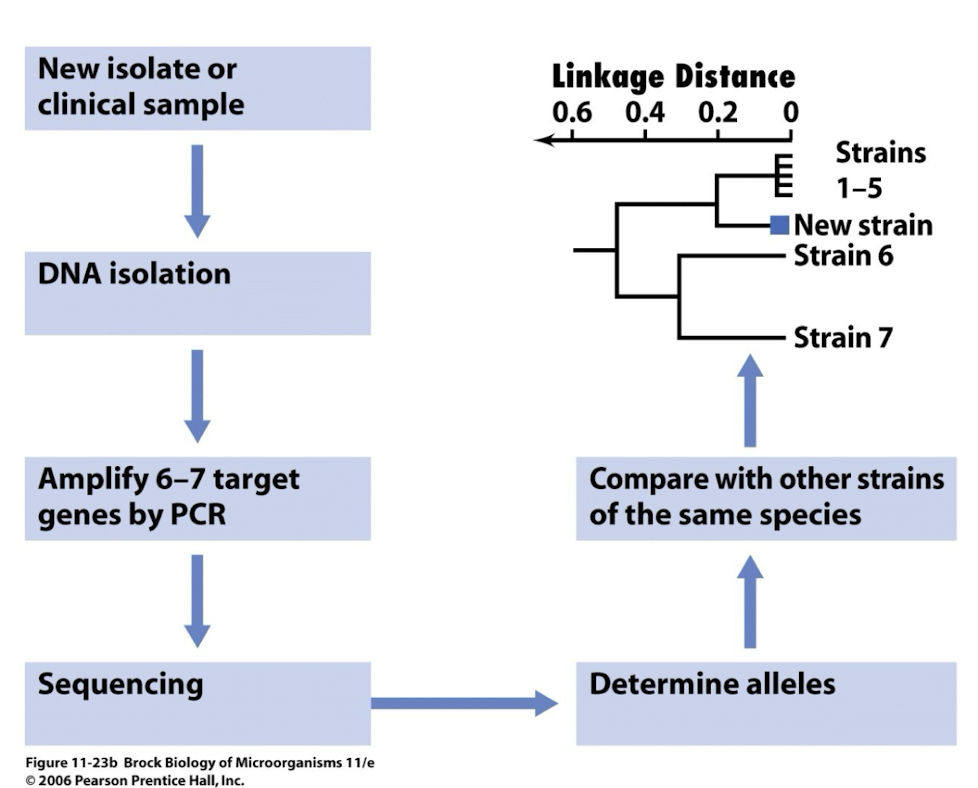
• After DNA is isolated, generally 7 different “housekeeping genes” are amplified by PCR
• Each gene can have up to 30 different alleles
• Up to 450bp from each gene amplified and sequenced
• Each allele is then compared to known alleles in database
• Linkage difference can be between 0 (identical) and 1 (very different)
Why use MLST?
Analyses ~7 housekeeping genes, allowing high-resolution strain identification:
Epidemiology applications
Distinguishing pathogenic vs commensal strains e.g. different E. coli pathotypes
How does FAME identify bacteria?
FAME (Fatty Acid Methyl Ester) analysis identifies bacteria by profiling their fatty acid composition, helping to differentiate species based on unique patterns.
Requires pure culture; however culture conditions can influence the FA profile for bacteria
Useful when DNA methods fail
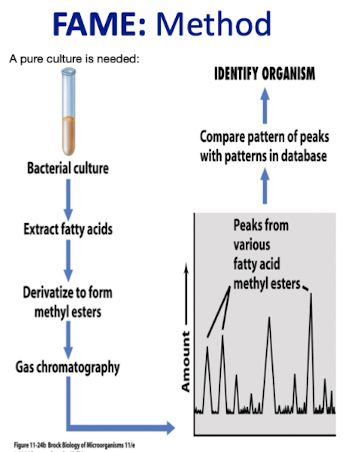
What variations in FA are looked at when analysing bacteria using FAME?
Variations in :
– Chain Length
– Saturated v Unsaturated groups
– Rings
– Branched chains
– Hydroxy groups
How are microbial species defined?
Groups of strains sharing many properties; compared to type strains in ATCC/NCTC collections.
>3% difference in 16S often indicates a new species
What are strains?
A population of microbes derived from a single individual or pure colony
Types of microbial strain differences?
Biovars: biochemical traits
Morphovars: morphology
Serovars: antigenic differences
Reflect genomic plasticity
Speciation
The process by which new distinct species arise through genetic divergence.
This often involves populations becoming isolated, leading to variations that define the new species.
Mechanisms of microbial speciation?
Adaptive mutation to new ecosystems
Lateral gene transfer (plasmids, phage DNA)
Rapid selection under environmental pressure
Why combine multiple methods for taxonomy?
Some microbes have nearly identical 16S sequences but very different genomes + phenotypes.
Using genomic, phenotypic, and ecological data provides reliable classification
What is the “two domain” hypothesis?
Evidence suggests Eukaryotes evolved from within Archaea, implying life may truly consist of Bacteria + Archaea only
What is Bergey’s Manual of Systematics of Archaea and Bacteria and why is it important?
Most widely accepted classification system by microbiologists for defining and organising prokaryotic organisms:
provides a comprehensive framework for identifying, classifying, and describing both Archaea and Bacteria, facilitating research and communication in microbiology.