Lecture 23 - Genetic Drift
1/16
Earn XP
Description and Tags
included on exam 4
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
17 Terms
random factors in evolution
mass extinction
mutation
genetic drift
mass extinction
a random factor in evolution
loss of many species resulting from random occurrences (e.g. asteroids, volcanoes)
mutation
a random factor in evolution
a random process that gives rise to new alleles each generation at a relatively constant rate
adds allelic diversity; affects allele frequencies at a given locus
genetic drift
a random factor in evolution
change in allele frequencies due to sampling from a limited population
reduces allelic diversity; affects allele frequencies at all loci in the genome
2 main examples: founder effect, bottleneck
note: there is a very fine line between genetic drift and natural selection (does this trait directly influence the fitness of an individual, or not?)
reasons genetic drift happens
1) populations are NOT infinitely large
2) sampling error randomly changes allele frequencies if size is not infinite
by chance, some individuals/genes may leave more copies in the next generation → changes allele frequencies
predicability of genetic drift
genetic drift is random → cannot predict exactly what/when will happen
but we CAN predict probability of different results happening
genetic drift in populations
effect is faster and more dramatic in small population
each population follows a unique path (because drift is random)
with enough time, drift can cause substantial changes in allele frequencies (even in large populations)
in absence of other evolutionary forces, drift causes eventual fixation of some alleles and loss of others
as alleles drift to fixation, heterozygote frequency declines in the population
probability of fixation of an allele
X(1/2N) = X/2N
N = population size
(2N = number of alleles at locus A)
X = initial number of copies of A1
equation: genetic drift of heterozygotes in the next generation
Hg+1 = Hg(1 - 1/2N)
Hg+1 = expected heterozygote frequency in the next generation
Hg = observed heterozygosity in the current generation
for H values, use frequencies
for N, use actual sample size
equation: genetic drift of heterozygotes in any generation
Ht = H0(1 - 1/2N)t
t = time (number of generations)
0 = initial generation
effective population size (Ne)
the portion of the population that actually mates
always smaller than cencus size
fewer individuals actually contribute to the next generation’s gene than all the individuals that could
assume that Ne = N for this class, but know this is not often the case in real life
why effective population size is smaller than census size
1) variance in progeny production (breeding success): extreme in polyandry and polygyny
2) skewed sex ratio
3) overlapping generations → inbreeding
4) fluctuations in N (small N has huge impact: genetic bottleneck)
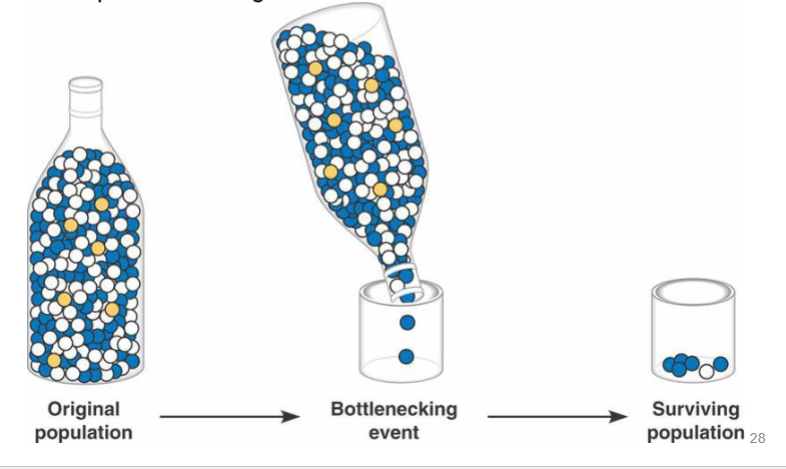
bottleneck effect
if population size is severely reduced, then only a small sample of alleles is left
even if these alleles are not under selection, allele frequency is change among survivors
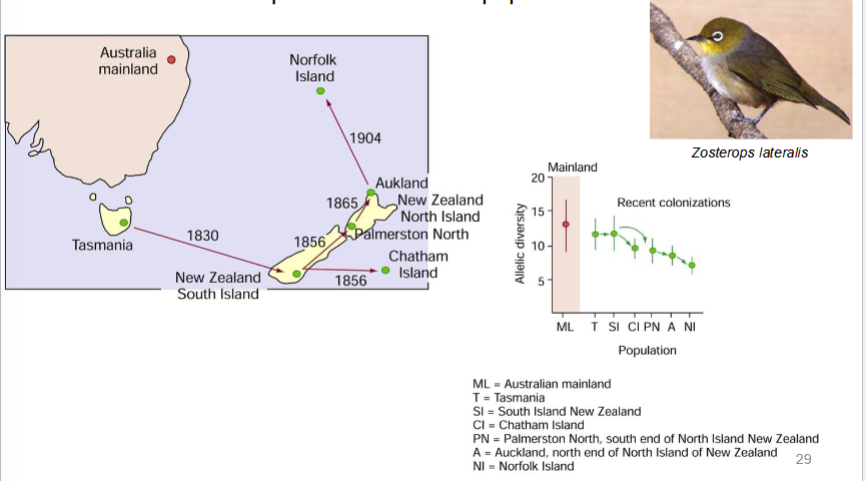
founder effect
if a population is founded by a small number of individuals, chance alone will cause different allele frequencies in the new population
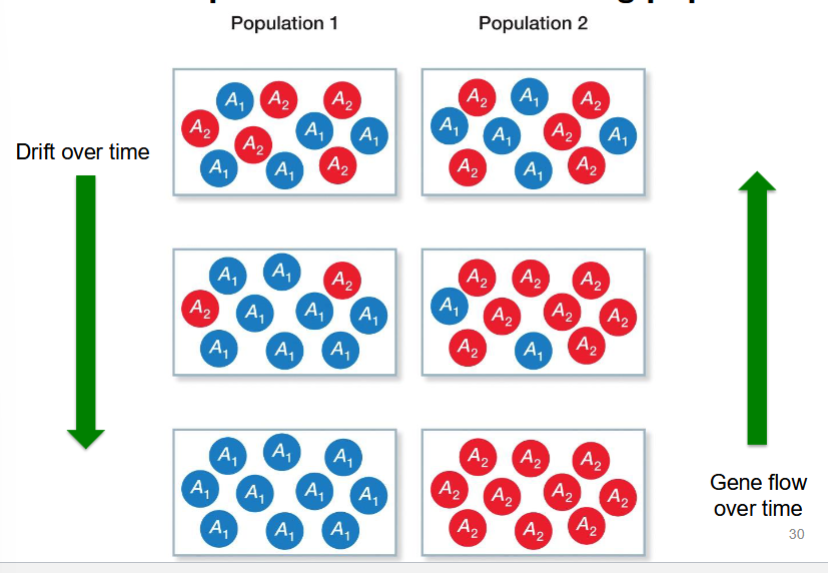
relation between gene flow and genetic drift
they have opposite effects on allele frequencies within and among populations
drift increases fixation, decreases variation
gene flow decreases fixation, increases variation
FST considering gene flow AND drift
FST = 1 / 4Nem+1
FST = fixation index
Ne = effective population size
m = migration rate
note: can be rearranged to find migration rate
equation: calculating m from FST
Nem = [(1/FST) - 1] / 4
even a little bit of gene flow can prevent genetic drift
![<p>N<sub>e</sub>m = [(1/F<sub>ST</sub>) - 1] / 4</p><p>even a little bit of gene flow can prevent genetic drift</p>](https://knowt-user-attachments.s3.amazonaws.com/7acdd6bf-9b84-4d2c-a0e1-a3254cbec9bb.png)