functional genomics
1/14
Earn XP
Description and Tags
omics
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
15 Terms
DNA sequencing
Is used for: mapping the whole genome, identifying genomic variation, genetic diseases, microbiomes (composition of microbiome in the gut: take a sample and determine genome of species), genome editing.
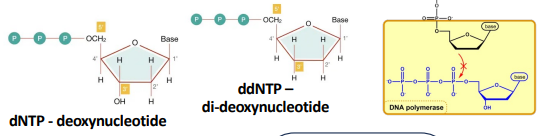
DNA sequencing - Sanger method
A DNA template (DNA to be read) is put into solution with a dNTP reaction mix which contains a small amount of fluorescently labeled ddNTPs. The template is copied by DNA polymerase, thus forming a chain of dNTPs. When a ddNTP is attached, the chain elongation stops. ddNTPs are implemented randomly, thus forming many chains of the same DNA but at different lengths. The chains are then put through a gel or capillary and separated by electrophores, which causes smaller chains (light weights) to travel faster than larger, and thus separating them based on chain length. A machine can then recognize the fluorescently labelled ddNTPs, and because of the different lengths of the chain, the sequence can be read out.
dNTP - high concentration
ddNTP - low concentration . ddNTP has a H where dNTP has an OH. dNTP uses this OH to elongate the chain, but this cannot happen with ddNTP due to the absence of OH.
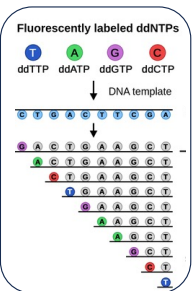
1st generation DNA sequencing
Is the Sanger method + using capillary gel electrophoreses. The capillary will separate based on length, smaller chains will go out first. At the end of the capillary a laser beam will activate the fluorescently labelled ddNTP and a photomultiplier will read out what base it is.
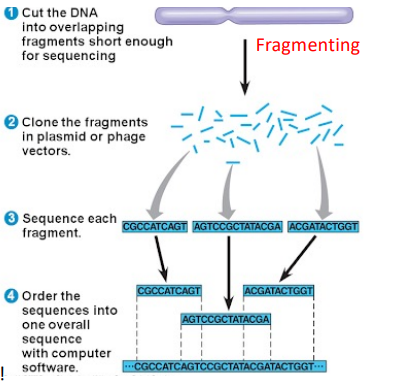
Whole genome sequencing (shot gun sequencing)
The DNA template is fragmented using sonication resulting in multiple small DNA fragments that overlap each other in sequence. The fragments are cloned in a plasmid or phage vectors resulting in a huge amount of tiny DNA fragments. Then Sanger sequencing is used to read out the DNA sequence of each fragment, then they are ordered into one overall sequence with computer software.
Next-generation sequencing (NGS)
Step 1: Genomic DNA is fragmented by sonication or by enzymes and put into a solution that contains adapters. These adapters will adhere to the ends of the fragments (both ends). Then the doubled strands are separated to result in single strands. The fragments with adapters attached are then put attached onto a flow cell. On the flow cell are oligonucleotides that match the sequence of the adapters, and thus they can attach to the adapters. The reverse strand is made so that it is directly attached to the oligonucleotide, attached to the flow cell. The forward strands (original templates) are washed away. The fragments attached to the flow cell are then amplified multiple times by PCR, causing clusters of identical fragments (every cluster has its own sequence) this is done so that the fluorescent signal will be strong. First the unattached end of the reverse strand is annealed to a second oligo on the flow cell. Then the forward chain is made from replicating the reverse strand, the forward strand is attached directly to the oligo. Then the doubled strands are melted to result in two single strands. This happens multiple times, resulting in clusters. The reverse strands get cut, leaving the forward strands for sequencing.
Step 2: a sequencing primer binds to the forward strands at a specific location. Fluorescently labelled dNTPs are added along with DNA polymerase. The dNTPs contain a temporary terminator that can be washed away. Because of this only one dNTP can be added at a time. Then a camera will detect the fluorescence of a cluster and determine the base at that position. Then everything is washed away, including the fluorescent label and terminator of the dNTP. Another solution of dNTPs is added and the chain elongates by 1 dNTP, and the base is determined again.
Step 3: The read outs are pass through a filter to get demultiplexed, using the attached indexes (in the adapters) to identify and sort reads from each sample. Then the reads get mapped to the reference genome
Billions of strands can be sequenced at once which makes it way faster compared to Sanger sequencing that can only sequence one fragment at a time.
A read out can be done by adding fluorescently labelled dNTPs. Then every clusters has a certain color depending on the base at that position of the template chain. Thus by imaging after adding dNTPs, the base at that position can be determined. Since dNTPs allow for the elongation after adding them, the chain will elongate with another fluorescently labelled dNTP and after imaging, the base can be determined again.
RNA can also be read out with NGS, but it first needs to be turned into DNA before it can get sequenced.
For NGS, the genome sequence is needed since you need to reference the read out strands to this sequence. Thus it is useful not for discovering the genome of something, but to compare the genome to an already known genome, for example when testing for genetic diseases or mutations.
Nanopore DNA sequencing
A nanopore is a pore into a membrane. In this pore a DNA/RNA strand can pass. An electric field (voltage) is applied over the pore. When the strand enters, this voltage changes slightly, and uniquely for each base. This difference in voltage is then measured and the sequence is determined. Methylation can also be seen, because methylation of a nucleotide changes the voltage difference.
The advantages are that you can work on native DNA or RNA, no amplification or fragmentation is required. It can also identify base modifications in he nucleotides. It is really fast and it has no length limitations.
The disadvantage is that it has a slightly higher per-base error-rate than NGS.