Biological Molecules
1/69
Earn XP
Description and Tags
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
70 Terms
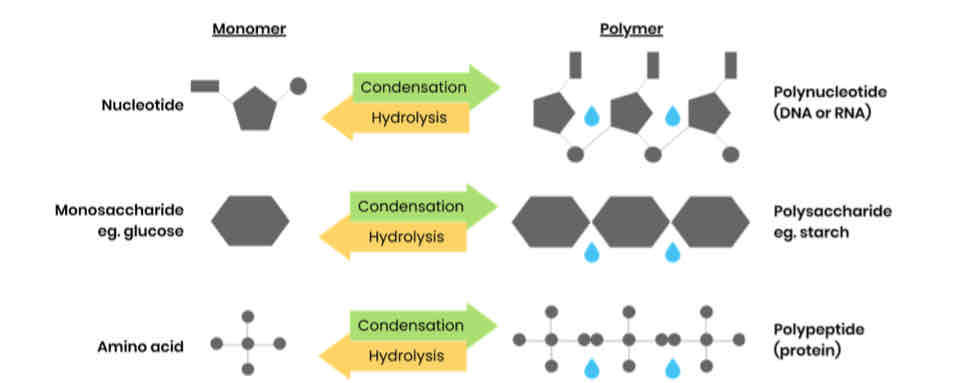
What are monomers and polymers?
● Monomers - smaller / repeating molecules from which larger molecules / polymers are made
● Polymer - molecule made up of many identical / similar molecules / monomers
What happens in condensation and hydrolysis reactions?

Give examples of polymers and the monomers from which they’re made
What are monosaccharides? Give 3 common examples
● Monomers from which larger carbohydrates are made
● Glucose, fructose, galactose
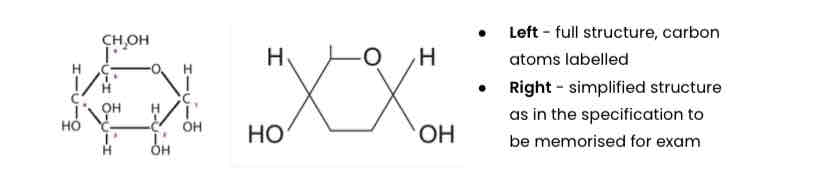
Describe the structure of α-glucose
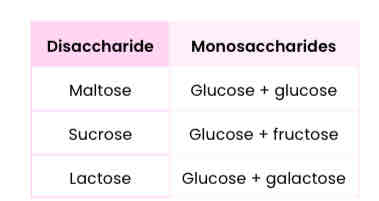
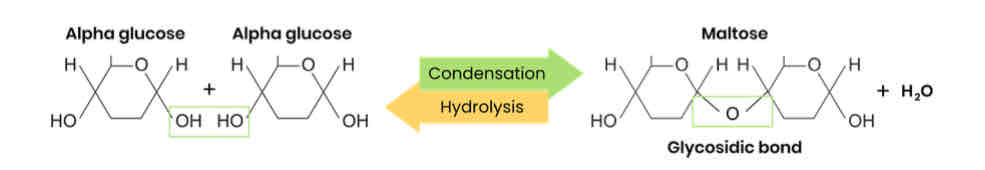
What are disaccharides and how are they formed?
● Two monosaccharides joined together with a glycosidic bond
● Formed by a condensation reaction, releasing a water molecule
List 3 common disaccharides & monosaccharides from which they’re made
Draw a diagram to show how two monosaccharides are joined together
What are polysaccharides and how are they formed?
● Many monosaccharides joined together with glycosidic bonds
● Formed by many condensation reactions, releasing water molecules
Describe the basic function and structure of starch and glycogen
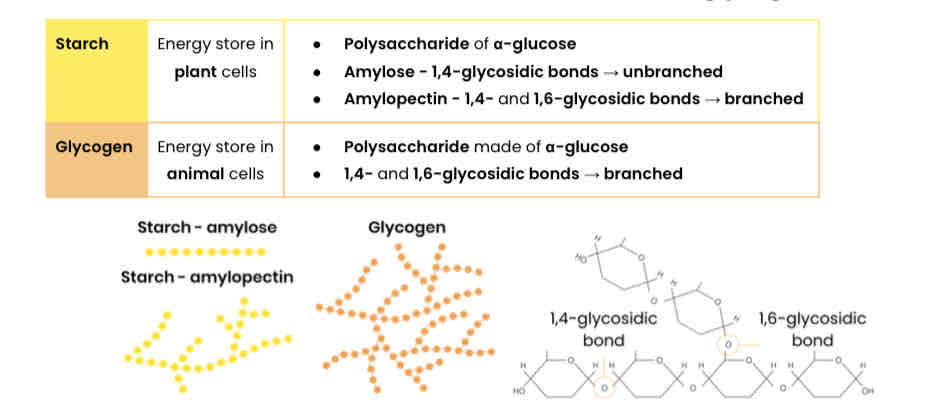
Explain how the structures of starch and glycogen relate to their functions
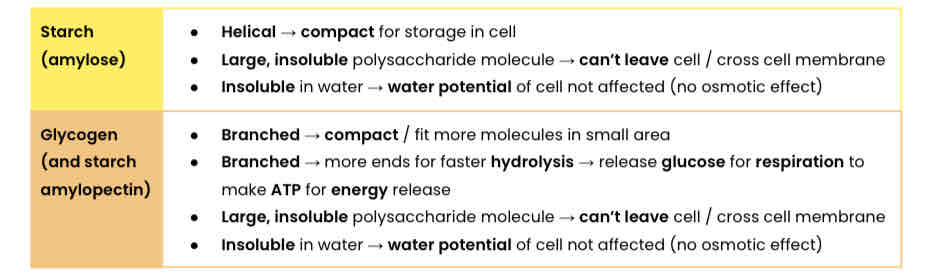
Describe the basic function and structure of cellulose

Explain how the structure of cellulose relates to its function
● Every other β-glucose molecule is inverted in a long, straight, unbranched chain
● Many hydrogen bonds link parallel strands (crosslinks) to form microfibrils (strong fibres)
● Hydrogen bonds are strong in high numbers
● So provides strength to plant cell walls
Describe the test for reducing sugars
Reducing sugars = monosaccharides, maltose, lactose
1. Add Benedict’s solution (blue) to sample
2. Heat in a boiling water bath
3. Positive result = green / yellow / orange / red precipitate
Describe the test for non-reducing sugars
Non-reducing sugars = sucrose
1. Do Benedict’s test and stays blue / negative
2. Heat in a boiling water bath with acid (to hydrolyse into reducing sugars)
3. Neutralise with alkali (eg. sodium bicarbonate)
4. Heat in a boiling water bath with Benedict’s solution
5. Positive result = green / yellow / orange / red precipitate
Suggest a method to measure the quantity of sugar in a solution
● Carry out Benedict’s test as above, then filter and dry precipitate
● Find mass / weight
Suggest another method to measure the quantity of sugar in a solution
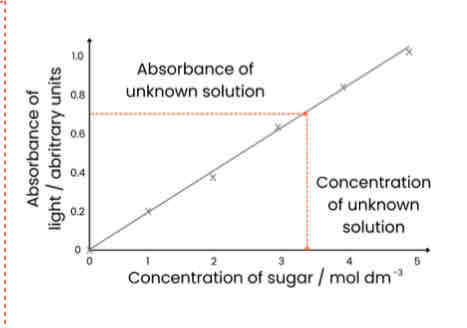
1. Make sugar solutions of known concentrations (eg. dilution series)
2. Heat each sample with Benedict’s for same time
3. Use colorimeter to measure absorbance (of light) of each known concentration
4. Plot calibration curve - concentration on x axis, absorbance on y axis and draw line of best fit
5. Repeat Benedict’s test with unknown sample and measure absorbance
6. Read off calibration curve to find concentration unknown sample
Describe the biochemical test for starch
1. Add iodine dissolved in potassium iodide (orange / brown) and shake / stir
2. Positive result = blue-black
Name two groups of lipid
Triglycerides and phospholipids
Describe the structure of a fatty acid (RCOOH)
● Variable R-group - hydrocarbon chain (saturated or unsaturated)
● -COOH = carboxyl group
Describe the difference between saturated and unsaturated fatty acids
● Saturated: no C=C double bonds in hydrocarbon chain; all carbons fully saturated with hydrogen
● Unsaturated: one or more C=C double bond in hydrocarbon chain (creating bend / kink)
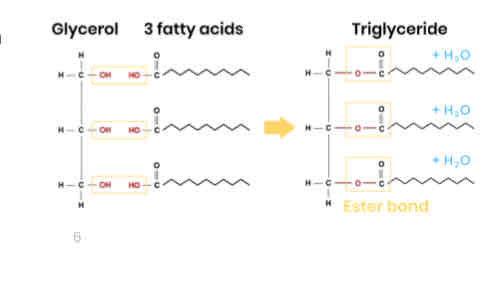
Describe how triglycerides form
● 1 glycerol molecule and 3 fatty acids
● Condensation reaction
● Removing 3 water molecules
● Forming 3 ester bonds
Explain how the properties of triglycerides are related to their structure
Function: energy storage
● High ratio of C-H bonds to carbon atoms
○ So used in respiration to release more energy
● Hydrophobic / non-polar fatty acids so insoluble in water (clump together as droplets)
○ So no effect on water potential of cell
Describe the difference between the structure of triglycerides and phospholipids
One of the fatty acids of a triglyceride is substituted by a phosphate-containing group
Describe how the properties of phospholipids relate to their structure
Function: form a bilayer in cell membrane, allowing diffusion of lipid-soluble (non-polar)/ small substances/ restricting movement of water-soluble (polar)/ larger substances
Phosphate heads are hydrophilic
○ Attracted to water
Fatty acid tails are hydrophobic
○ Repelled by water so point away
Describe the test for lipids
1. Add ethanol, shake (to dissolve lipids), then add water
2. Positive = milky white emulsion
Describe / draw the general structure of an amino acid
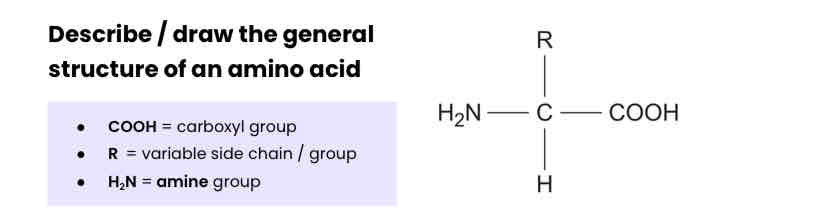
How many amino acids are common in all organisms? How do they vary?
The 20 amino acids that are common in all organisms differ only in their side group (R).
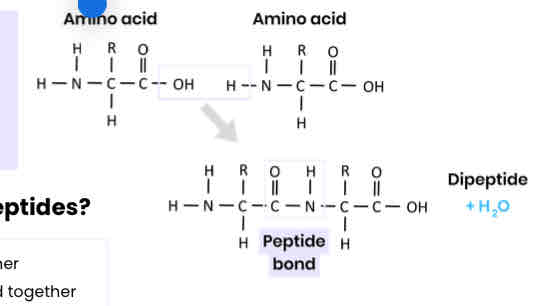
Describe how amino acids join together
● Condensation reaction
● Removing a water molecule
● Between carboxyl / COOH group of one and amine / NH2 group of another
● Forming a peptide bond
What are dipeptides and polypeptides?
A functional protein may contain one or more polypeptides.
● Dipeptide - 2 amino acids joined together
● Polypeptide - many amino acids joined together
Describe the primary structure of a protein
Sequence of amino acids in a polypeptide chain, joined by peptide bonds
Describe the secondary structure of a protein
● Folding (repeating patterns) of polypeptide chain eg. alpha helix / beta pleated sheets
● Due to hydrogen bonding between amino acids
● Between NH (group of one amino acid) and C=O (group)
Describe the tertiary structure of a protein
● 3D folding of polypeptide chain
● Due to interactions between amino acid R groups
(dependent on sequence of amino acids)
● Forming hydrogen bonds, ionic bonds and disulfide bridge
Describe the quaternary structure of a protein
● More than one polypeptide chain
● Formed by interactions between polypeptides
(hydrogen bonds, ionic bonds, disulfide bridges)
Describe the test for proteins
1. Add biuret reagent (sodium hydroxide + copper (II) sulphate)
2. Positive result = purple / lilac colour (negative stays blue) → indicates presence of peptide bonds
Proteins have a variety of functions within all living organisms. You need to be able to relate the structure of proteins to properties of proteins named throughout the specification eg. enzymes / antibodies.
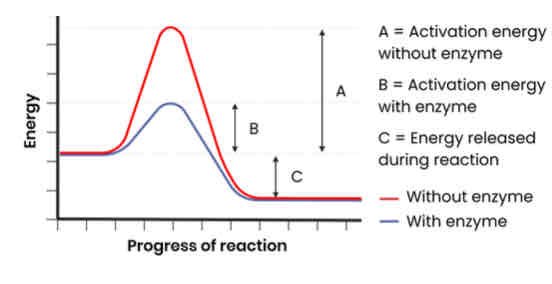
How do enzymes act as biological catalysts?
● Each enzyme lowers activation energy of reaction it catalyses
● To speed up rate of reaction
Enzymes catalyse a wide range of intracellular and extracellular reactions that determine structures and functions from cellular to whole-organism level.
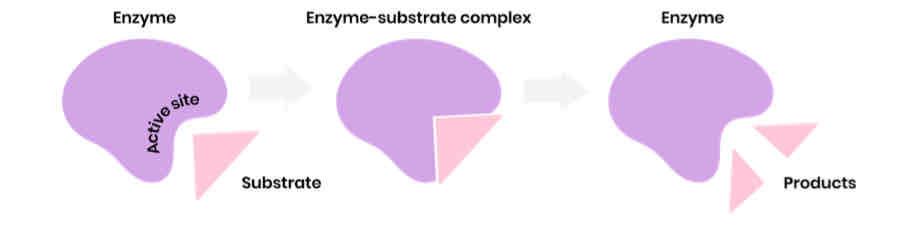
Describe the induced-fit model of enzyme action
1. Substrate binds to (not completely complementary) active site of enzyme
2. Causing active site to change shape (slightly) so it is complementary to substrate
3. So enzyme-substrate complex forms
4. Causing bonds in substrate to bend / distort, lowering activation energy
Describe how models of enzyme action have changed over time
● Initially lock and key model (now outdated)
○ Active site a fixed shape, complementary to one substrate
● Now induced-fit model
Explain the specificity of enzymes
● Specific tertiary structure determines shape of active site
○ Dependent on sequence of amino acids (primary structure)
● Active site is complementary to a specific substrate
● Only this substrate can bind to active site, inducing fit and forming an enzyme-substrate complex
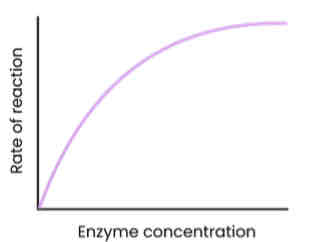
Describe and explain the effect of enzyme concentration on the rate of enzyme-controlled reactions
● As enzyme conc. increases, rate of reaction increases
○ Enzyme conc. = limiting factor (excess substrate)
○ More enzymes so more available active sites
○ So more enzyme-substrate (E-S) complexes form
● At a certain point, rate of reaction stops increasing / levels off
○ Substrate conc. = limiting factor (all substrates in use)
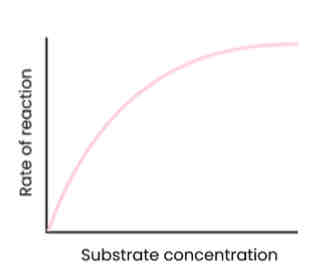
Describe and explain the effect of substrate concentration on the rate of enzyme-controlled reactions
● As substrate conc. increases, rate of reaction increases
○ Substrate conc. = limiting factor (too few enzyme molecules to occupy all active sites)
○ More E-S complexes form
● At a certain point, rate of reaction stops increasing / levels off
○ Enzyme conc. = limiting factor
○ As all active sites saturated / occupied (at a given time)
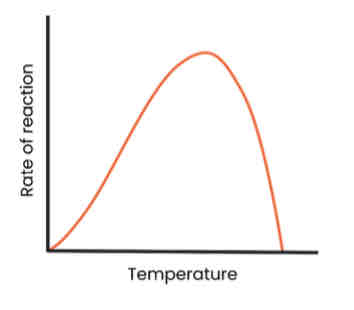
Describe and explain the effect of temperature on the rate of enzyme-controlled reactions
● As temp. increases up to optimum, rate of reaction increases
○ More kinetic energy
○ So more E-S complexes form
● As temp. increases above optimum, rate of reaction decreases
○ Enzymes denature - tertiary structure and active site change shape
○ As hydrogen / ionic bonds break
○ So active site no longer complementary
○ So fewer E-S complexes form
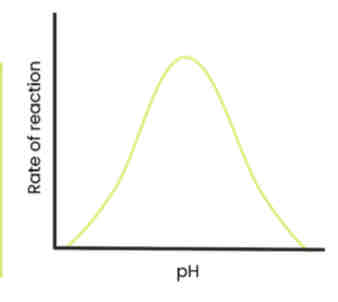
Describe and explain the effect of pH on the rate of enzyme-controlled reactions
● As pH increases / decreases above / below an optimum, rate of reaction decreases
○ Enzymes denature - tertiary structure and active site change shape
○ As hydrogen / ionic bonds break
○ So active site no longer complementary
○ So fewer E-S complexes form
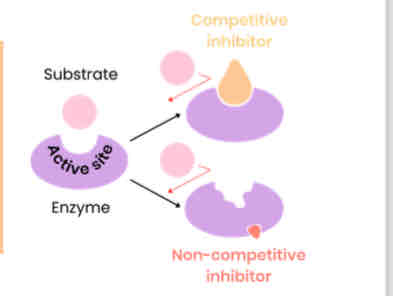
Describe and explain the effect of concentration of competitive inhibitors on the rate of enzyme-controlled reactions
● As concentration of competitive inhibitor increases, rate of reaction decreases
○ Similar shape to substrate
○ Competes for / binds to / blocks active site
○ So substrates can’t bind and fewer E-S complexes form
● Increasing substrate conc. reduces effect of inhibitors (dependent on relative concentrations of substrate and inhibitor)
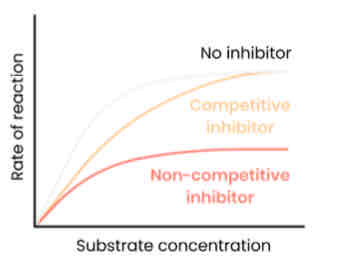
Describe and explain the effect of concentration of non-competitive inhibitors on the rate of enzyme-controlled reactions
● As concentration of non-competitive inhibitor increases, rate of reaction decreases
○ Binds to site other than the active site (allosteric site)
○ Changes enzyme tertiary structure / active site shape
○ So active site no longer complementary to substrate
○ So substrates can’t bind so fewer E-S complexes form
● Increasing substrate conc. has no effect on rate of reaction as change to active site is permanent
Describe the basic functions of DNA and RNA in all living cells
DNA—> Holds genetic information which codes for polypeptides (proteins)
RNA —>Transfers genetic information from DNA to ribosomes
Name the two types of molecule from which a ribosome is made
RNA and proteins
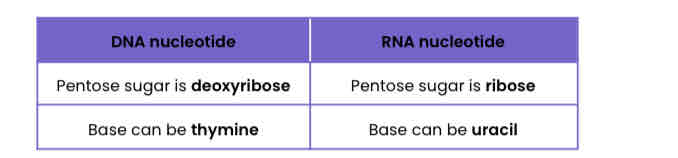
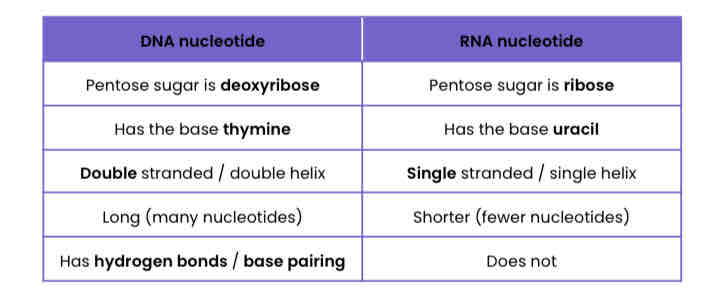
Describe the differences between a DNA nucleotide and an RNA nucleotide
Describe how nucleotides join together to form polynucleotides
● Condensation reactions, removing water molecules
● Between phosphate group of one nucleotide and deoxyribose/ribose of another
● Forming phosphodiester bonds
Why did many scientists initially doubt that DNA carried the genetic code?
The relative simplicity of DNA - chemically simple molecule with few components.
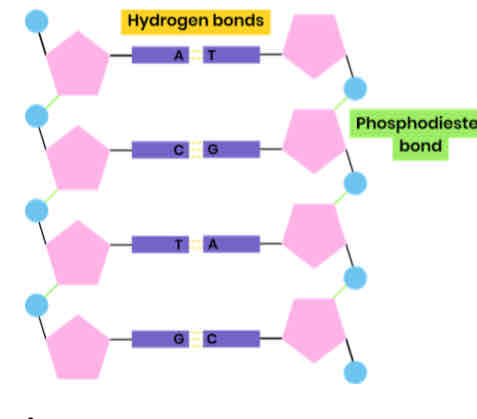
Describe the structure of DNA
● Polymer of nucleotides (polynucleotide)
● Each nucleotide formed from deoxyribose, a phosphate group and a nitrogen-containing organic base
● Phosphodiester bonds join adjacent nucleotides
● 2 polynucleotide chains held together by hydrogen bonds
● Between specific complementary base pairs - adenine / thymine and cytosine / guanine
● Double helix
Describe the structure of (messenger) RNA
● Polymer of nucleotides (polynucleotide)
● Each nucleotide formed from ribose, a phosphate group and a nitrogen-containing organic base
● Bases - uracil, adenine, cytosine, guanine
● Phosphodiester bonds join adjacent nucleotides
● Single helix
Compare and contrast the structure of DNA and (messenger) RNA
Suggest how the structure of DNA relates to its functions
● Two strands → both can act as templates for semi-conservative replication
● Hydrogen bonds between bases are weak → strands can be separated for replication
● Complementary base pairing → accurate replication
● Many hydrogen bonds between bases → stable / strong molecule
● Double helix with sugar phosphate backbone → protects bases / hydrogen bonds
● Long molecule → store lots of genetic information (that codes for polypeptides)
● Double helix (coiled) → compact (lots of information stored)
Suggest how you can use incomplete information about the frequency of bases on DNA strands to find the frequency of other bases
1. % of adenine in strand 1 = % of thymine in strand 2 (and vice versa)
2. % of guanine in strand 1 = % of cytosine in strand 2 (and vice versa)
Because of specific complementary base pairing between 2 strands
Why is semi-conservative replication important?
Ensures genetic continuity between generations of cells
Describe the process of semi-conservative DNA replication
1. DNA helicase breaks hydrogen bonds between complementary bases, unwinding the double helix
2. Both strands act as templates
3. Free DNA nucleotides attracted to exposed bases and join by specific complementary base pairing
4. Hydrogen bonds form between adenine-thymine and guanine-cytosine
5. DNA polymerase joins adjacent nucleotides on new strand by condensation reactions
6. Forming phosphodiester bonds
Semi-conservative meaning
each new DNA molecule consists of one original / template strand and one new strand
Use your knowledge of enzyme action to suggest why DNA polymerase moves in opposite directions along DNA strands
● DNA has antiparallel strands
● So shapes / arrangements of nucleotides on two ends are different
● DNA polymerase is an enzyme with a specific shaped active site
● So can only bind to substrate with complementary shape (phosphate end of developing strand)
Name the two scientists who proposed models of the chemical structure of DNA and of DNA replication
Watson and Crick
Describe the work of Meselson and Stahl in validating the Watson-Crick model of semi-conservative DNA replication
1. Bacteria grown in medium containing heavy nitrogen (15N) and nitrogen is incorporated into DNA bases
● DNA extracted & centrifuged → settles near bottom, as all DNA molecules contain 2 ‘heavy’ strands
2. Bacteria transferred to medium containing light nitrogen (14N) and allowed to divide once
● DNA extracted & centrifuged → settles in middle, as all DNA molecules contain 1 original ‘heavy’ and 1 new ‘light’ strand
3. Bacteria in light nitrogen (14N) allowed to divide again
● DNA extracted & centrifuged → half settles in middle, as
contains 1 original ‘heavy’ and 1 new ‘light’ strand; half settles near top, as contains 2 ‘light’ strands
Other models (eg. dispersive, conservative) not supported - bands would be in different places.
What is ATP?
Adenosine triphosphate
Describe the structure of ATP
● Ribose bound to a molecule of adenine (base) and 3 phosphate groups
● Nucleotide derivative (modified nucleotide)
Describe how ATP is broken down
● ATP (+ water) → ADP (adenosine diphosphate) + Pi (inorganic phosphate)
● Hydrolysis reaction, using a water molecule
● Catalysed by ATP hydrolase (enzyme)
Give two ways in which the hydrolysis of ATP is used in cells
● Coupled to energy requiring reactions within cells (releases / provides energy)
○ Eg. active transport, protein synthesis
● Inorganic phosphate released can be used to phosphorylate (add phosphate to) other compounds, making them more reactive
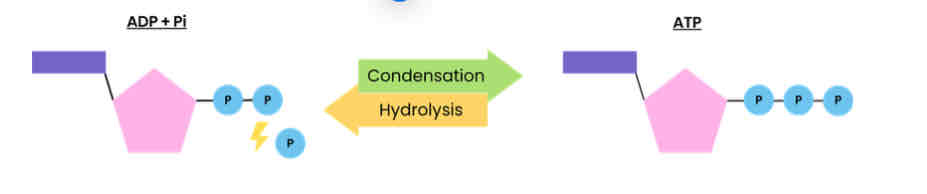
Describe how ATP is resynthesised in cells
● ADP + Pi → ATP (+ water)
● Condensation reaction, removing a water molecule
● Catalysed by ATP synthase (enzyme)
● During respiration and photosynthesis
Explain how hydrogen bonds occur between water molecules
● Water is polar molecule
● Slightly negatively charged oxygen atoms attract slightly positively charged hydrogen atoms of other water molecules
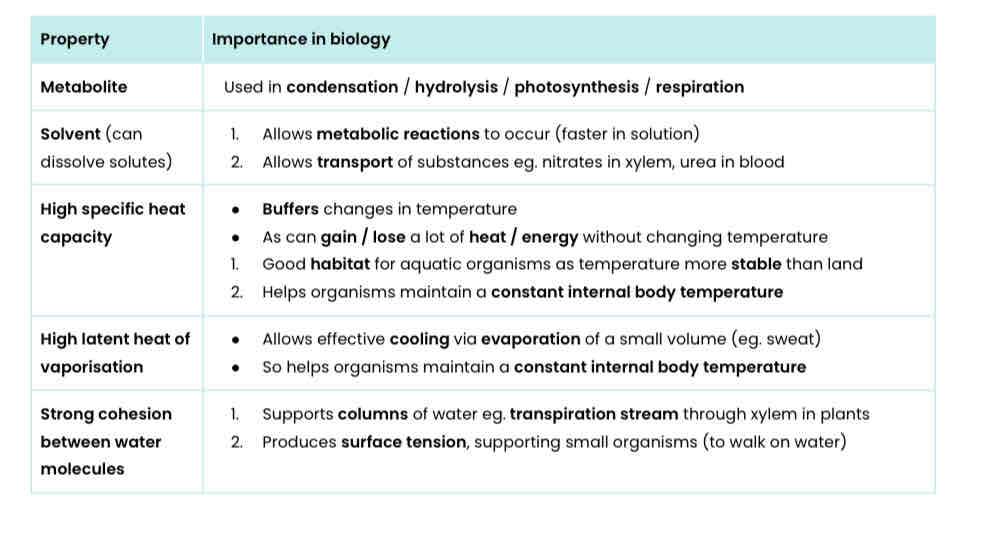
Explain 5 properties of water that are important in biology
Where are inorganic ions found in the body?
In solution in cytoplasm and body fluid, some in high concentrations and others in very low concentrations.
Describe the role of hydrogen, iron, sodium and phosphate ions
