Föreläsning 3: Production of heterologous proteins in bacteria and yeast
1/42
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
43 Terms
What is a heterologous protein
Protein that is produced by a host organism but originates from a different species
How do make a recombinant protein (general steps)
Gene encoding the protein of interest is isolated from an animal cell
It’s inserted into a cloning vector
The vector is introduced into a host organism
The host transcribes and translates the gene → protein
What are the key challenges producing a recombinant protein
Which cloning vector: must be compatible and efficient
How to get high protein yield
Which organism to express the protein in: depends on the complexity of the protein (post-translational modifications)
Recombinant host
Is inserted with the gene and produces the wanted protein.
Why is often E. coli used for production of recombinant proteins
Fast-growing
Easy to manipulate
High yields
Backed by decades of research
→ perfect for simple protein expression
Why use other hosts
For more complex proteins
Some proteins require post-translational modifications (like glycosylation) that bacteria can't perform
Choice of host depends on the type and complexity of the protein.
Why can’t we just put the gene into any plasmid?
A normal plasmid (eukaryotic) doesn’t have the right signals for E. coli to recognize and use the foreign gene. Instead, a special “expression vector” — a plasmid designed to make E. coli produce protein, is used. The expression vectors contain 3 important signals surrounding the gene:
P: promoter
R: ribosome binding site
T: terminator
Together these form the expression cassette where the foreign gene is inserted.
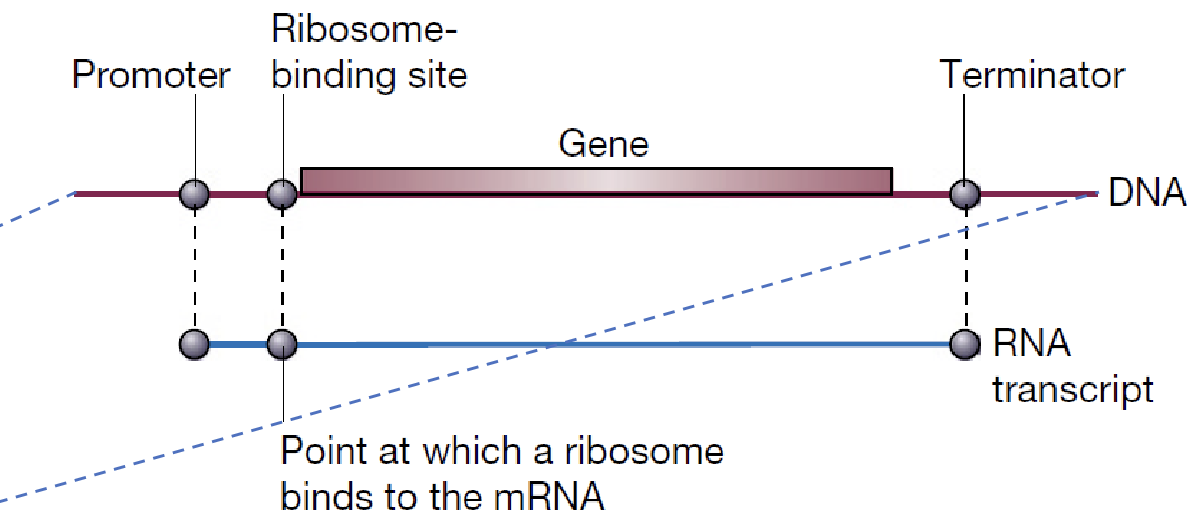
Promoter (P)
Tells the cell where to start making RNA and determines the rate of mRNA synthesis. More mRNA → more recombinant protein.
A strong promoter is needed to produce much protein.
Eukaryotic promoters (like the TATA box at -25) are not recognized by E. coli RNA polymerase, which instead needs bacterial promoter sequences like the -35 (TTGACA) and -10 (TATAAT) boxes.
→ To produce a eukaryotic (foreign) protein in E. coli, the gene must be placed under a bacterial promoter that E. coli can read and transcribe efficiently
Ribosome bindning site (R) (
Where ribosome attaches to mRNA
Terminator (T)
Stops DNA transcription (stem loop)
Why do we need regulation of promoters
Sometimes the protein can be toxic or use too much energy, so we don’t want it made all the time.
Describe regulatable promoters
Inducible: Protein is made only when you add a chemical like IPTG. Gene is switched on when chemical is added
Repressible: Protein is made unless a chemical turns it off.
Gene is switched off when chemical is added
Give some examples on promoters
lac: induced by IPTG
Trp: repressed by tryptophan
Tac (hybrid of lac & trp): induced by IPTG
λ: controlled by temp (turns on above 30°C)
T7: IPTG + T7 RNA polymeras
Why do some cassettes sometimes include fusion between E. coli foreign gene
Sometimes the foreign gene is hard to express by itself → fusion
The benefits are:
Avoiding weird secondary structure
Avoiding the gene from being degraded by the bacteria
Can act as signal peptide
Help for purification
There are 2 main roots that problems with producing heterologous proteins in E. coli can derive from. What are they?
Problems due to the sequence of the foreign gene
Problems Caused by the Host — E. coli Itself
There are three problems caused by the sequence of the foreign gene. Name them and their solutions
Foreign genes have introns:
Eukaryotic genes often contain introns (non-coding regions), but E. coli lacks the machinery to remove them.
As a result, introns stay in the mRNA, leading to incorrect or non-functional proteins.
Solution: Use cDNA (complementary DNA) made from spliced mRNA (which has no introns), allowing correct expression in E. coli
The foreign gene contains E. coli termination signals:
Not all of the gene is transcribed
Solution: Use in vitro mutagenesis to remove or alter the termination-like sequences without affecting the protein’s amino acid sequence. In vitro mutagenesis = introducing specific mutations into a DNA sequence in a test tube (outside of a living organism)
The foreign gene contains codon bias not ideal for E. coli
Different organisms prefer different codons for the same amino acid.
Solution use artificial gene synthesis to replace rare codons with those preferred by E. coli, optimizing translation without changing the protein.
There are three problems caused by the host cell. Name them and their solutions
Post-translational modifications are not done correctly (e.g glycosylation):
Some proteins require modifications like glycosylation after translation. E. coli cannot perform these eukaryotic-specific modifications.
Solution: use mutant E. coli strains engineered to perform limited modifications, though this is technically difficult.
Incorrect protein folding
Many eukaryotic proteins need help folding or forming disulfide bonds.
In E. coli, this can lead to misfolded proteins or insoluble aggregates (inclusion bodies).
Solution: Engineer E. coli to produce chaperone proteins that assist in correct folding.
Protein degradation:
E. coli may degrade foreign proteins using its own proteases
Solution: Use mutant strains of E. coli that lack certain proteases to prevent unwanted degradation.
What is the workflow for constructing production strain
Production strain = a specially engineered microorganism (like E. coli) that has been modified to produce a specific protein or product in large amounts.
Choosing a vector
Designing the expression vector
Constructing the plasmid
Transformation and selection
Producing the protein
Purification and analysis
Evaluate the result
Choosing a vector
Decide what type of vector to use to carry the gene:
Plasmid:
small circular DNA that replicate independently
Easy to manipulate
High copy number → gives more protein per cell
Episomes (integrative vectors):
Integrate into bacterial chromosome
lower copy number but more genetically stable
Useful when long-term, stable expression is more important than quantity
Designing the Expression Vector
The vector is built by inserting the foreign gene into a complete expression system. The vector must contain:
A strong promotor that works in (in E. coli: lac, T7)
A ribosome binding site so the protein can be translated
A terminator to end the transcription
A selectable marker so you can identify the cells that have taken up the plasmid (e.g antibiotic resistance gene)
An origin of replication (ori) so the plasmid can multiply inside the cell
Optional: fusion tags (like His-tag) to help with purification, or signal peptides for protein export
3. Constructing the Plasmid
Restriction enzymes cut the DNA
Ligation joins the gene and vector
Check that everything is correct, often using gel electrophoresis (size) or sequencing
Transformation and selection
You introduce the finished plasmid into the host. Not all cells will take up the expression vector → selection (e.g grow cells on a plate with antibiotics to exclude the unwanted cells):
Only the bacteria that have taken up the plasmid (and the resistance gene) will survive
These are the candidate production strain
5. Producing the Protein
Selected bacteria are grown in liquid culture under controlled conditions. You can:
Add IPTG or another inducer to trigger expression.
Monitor temperature, pH, and oxygen for optimal growth.
Purification and analysis
To isolate the protein, you:
Lyse the cells (break them open).
Use chromatography (often with fusion tags) to purify the protein.
Use analysis methods like SDS-PAGE or activity assays to check:
Is the protein pure?
Is it the right size?
Is it active?
7. Evaluate the Result
Once the protein is purified, you ask:
Is the purity good enough?
What is the yield (g protein/g substrate)?
How productive is the strain (g/Lh)?
Is the process stable (does it work the same every time)?
What is the advantage of using yeast and filamentous fungi for production of heterologous proteins
Yeasts are eukaryotes so closer to animals cells → can process and fold animal proteins more efficiently, especially when the recombinant protein originates from animals
You still need expression vectors to insert and express foreign genes.
What is the advantage of using Saccharomyces cerevisiae for production of heterologous proteins
A commonly used yeast species in biotechnology and brewing.
It contains a natural plasmid called the 2 µm plasmid, which is:
Good size: 6.3 kb
good copy number: 60 copies per cell.
What is the disadvantage of the 2 µm plasmid
Has weak selectable markers (less efficient for selecting transformed cells)
Solution:
A special strain of S. cerevisiae that lacks the LEU2 gene cannot make leucine and needs it supplied in the medium.
LEU2 gene can act as selectable marker - organisms with the gene can survive without leucine
If you insert the correct LEU2 gene into the plasmid, only transformed cells will survive on leucine-free medium.
So, LEU2 acts as a selectable marker: only cells that took up the plasmid can grow.
What is a shuttle vector
Plasmids that can function (replicate) in two different organisms
Describe yeast episomal plasmid (YEps)
Plasmids that derive from the 2 µm plasmid are called YEps
They are shuttle vectors: can function in both E. coli and S. cerevisiae
Usually for shuttle vectors, you build and amplify the plasmid in E. coli (easy to grow), then transfer it into yeast for protein production
Selective markers in shuttle vectors: ampR, LEU2, tetR.
How do you build and amplify the plasmid in E. coli and then transfer it to S. cerevisiae
Du klonar eller bygger plasmiden i E. coli.
ampR används här för att välja ut bakterier som har tagit upp plasmiden (de överlever på ampicillinplattor).
Du får många kopior av plasmiden genom att odla E. coli → enkel och billig DNA-produktion.
Andra steget: in i jäst
Sedan renar du plasmid-DNA:t från E. coli.
Det renade plasmid-DNA:t förs sedan in i jästceller (t.ex. med kemisk behandling eller elektroporering).
LEU2 används nu för att välja ut jästceller som tagit upp plasmiden – de överlever på leucin-fritt medium.
Alltså: pro
Name different yeast vectors
YIps (Yeast Integrative plasmids)
YRps (Yeast replicative plasmids)
YCps (Yeast centromeric plasmids)
Describe YIps
Must integrate into the yeast genome
Very stable, but only one copy per cell
Describe YRps
Have yeast origin of replication
More copies per cell but less stable
Describe YCps
Include a centromere sequence
Stable but one copy per cell
What is yeast artificial chromosome (YAC)
The YAC is the largest cloning vector used in yeast (can carry up to 1000 kb of DNA).
It includes key parts of a chromosome: telomeres, centromere, and origin of replication.
Useful for cloning very large pieces of DNA, like entire genes or genomic fragments.
What is the problem of using yeast instead of E.coli
The proteins produced by yeast are hyperglycosylated - get too many sugar chains added. This alters the protein structure and can reduce its function in humans.
What is the advantages of using S. cerevisiae
High yield
Safe
Well studied
Name other useful yeast and filamentous fungi
Pichia pastoris: can produce very high amounts of protein (up to 30% of total cell protein)
Aspergillus niger: Secretes proteins directly into the medium, which makes purification easier.
Describe using insect cells
These cells (e.g., from Drosophila) are widely used because they have strong expression systems. This leads to high yields of recombinant protein, especially when using baculoviruses as vectors. Baculoviruses are viruses that naturally infect insect cells and are modified to carry the gene of interest.
Describe using mammalian cells
These provide the most accurate environment for producing human or other animal proteins. They perform complex post-translational modifications like glycosylation exactly as they would happen in the body. However, mammalian systems are expensive and slow, and they often result in lower protein yields.
Describe the genetic tools in mammalian systems
Cloning vectors are not necessary but can boost the yield especially if strong promotor
Viruses like SV40 can serve as cloning vectors.
Microinjection: directly injecting the DNA in the nucleus of a cell, often in a fertilized egg to create transgenic animals
Pharming = protein production in live animals
A common approach is to insert the gene into the milk-producing genes (e.g., using the β-lactoglobulin promoter).
This way, the protein is secreted in the milk and can be collected and purified easily.
What are the advantages and disadvantages of the different host systems