Biochemistry 3400- Midterm 1
1/160
Earn XP
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
161 Terms
How do living organisms store and propagate genetic information?
Storage = DNA/RNA sequences (the “genetic code”).
Propagation = replication + inheritance + expression through transcription and translation.
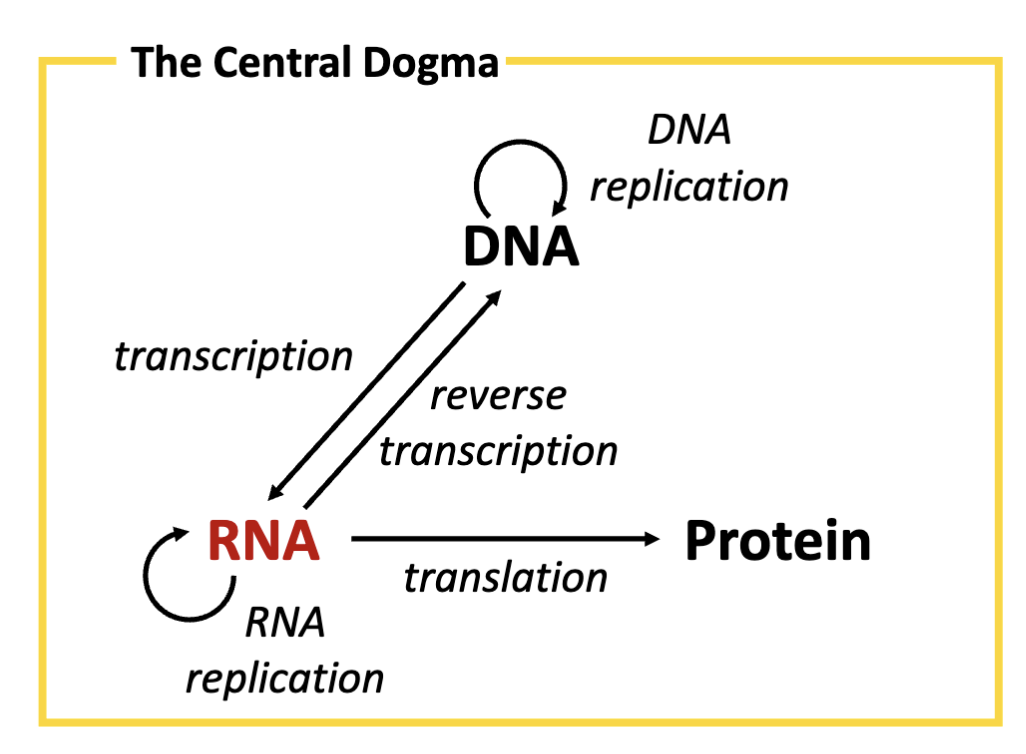
What is the central dogma?
storage of flow of information from DNA →RNA
Who created the central dogma?
Francis Crick in 1958 (once the structure of DNA was solved).
What is the role of DNA (in relation to the central dogma)?
Deoxyribonucleic acid (DNA) serves as the genetic instructions for genes. DNA is its own template, synthesized by the process of DNA replication
What is the role of RNA (in relation to the central dogma)?
Ribonucleic acid (RNA) is an intermediary molecule mediating the transfer of genetic instructions from DNA in the nucleus to the site of protein synthesis in the cell. RNA is created from a DNA template by
the process of transcription.
What is the role of proteins (in relation to the central dogma)?
Proteins mediate many biochemical functions (e.g, catalyses, regulation) critical to life. Proteins are synthesized by translation, converting the genetic instructions transcribed in RNA into protein.
What is the role of non-coding RNA?
Non-coding RNA are functional molecules. Ribosomal RNA (rRNA) is a component of ribosomes, the cellular machinery responsible for
protein translation. rRNA binds to mRNA and recruits transfer RNA (tRNA) delivering amino acids to the ribosome for protein translation
Is the central dogma linear?
When Crick proposed the central dogma it was thought to be linear but this is not the case
What is the role of coding RNA?
Messenger RNA (mRNA) is produced from template DNA by transcription. RNA can also be replicated by an RNA-dependent RNA polymerase
What experiments prove the central dogma?
Gregor Mendel- Heredity
Edmond Wilson + Nettie Stevens - independently identified sex linked chromosomes
Walter Sutton- proposes chromosome theory of inheritance
Thomas Hunt Morgan- Demonstrates chromosome theory of inheritance
Oswald Avery- demonstrates that DNA is the carrier of genetic information
Frederick Griffith- demonstrates that DNA is the carrier of genetic information
Martha Chase+ Alfred Hershey- conclusively demonstrate viruses infect host with DNA
What was Mendel’s experiment?
Experiment tracking the distribution of traits over successive generations of garden pea plant progeny.
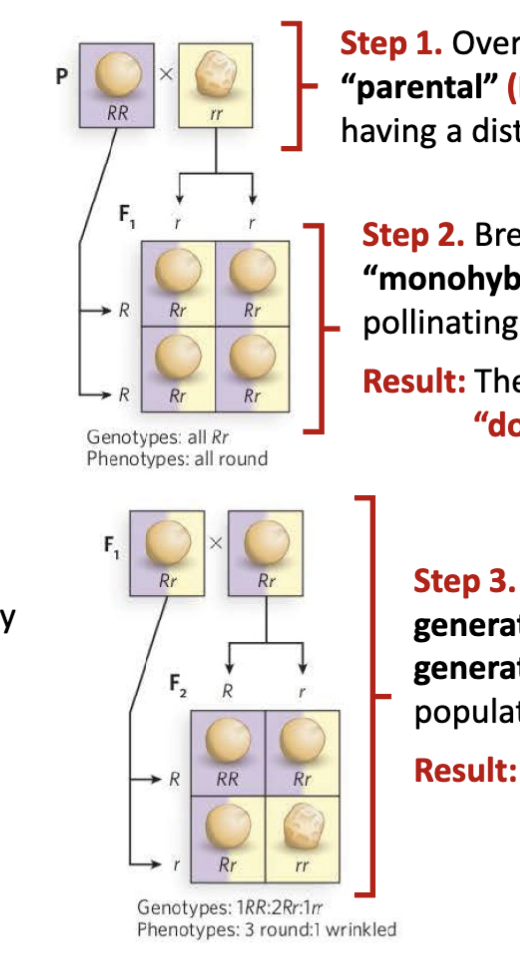
What were the steps of Mendel’s experiment?
Step 1. Over multiple generations, breed two “parental” (P) populations of pea plant, each having a distinct and homogeneous trait
Step 2. Breed a single generation of “monohybrid offspring” (F1) by cross- pollinating the two P populations
Result: The F1 generation has a single “dominant” trait
Step 3. Self-cross pollinate the F1 generation to produce a second
generation (F2). This produces a population with both traits.
Result: The F2 generation has a 3:1 ratio of dominant and recessive traits
Legend: R round r wrinkled RR and rr =P (parental generation)
What are Mendel’s laws?
First law: “Alleles of the same gene segregate independently
into gametes (the reproductive cell of the parent)”
Second law: “Different genes assort independently during
gamete formation”
What is the difference between a phenotype and a genotype?
phenotype physical trait vs genotype genome
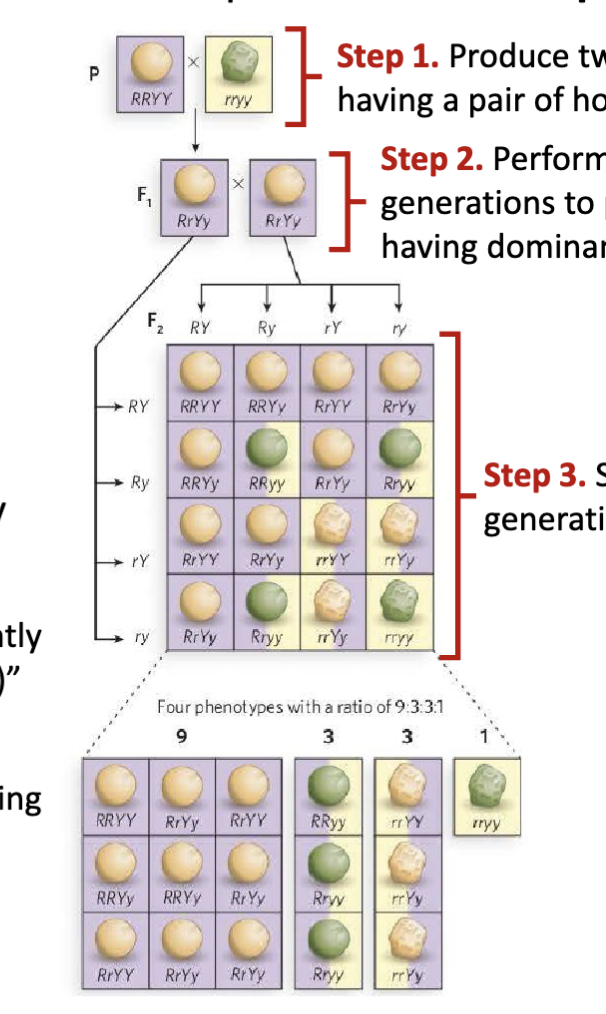
What are the features of Mendel’s dihybrid cross experiment?
Step 1: Produce two P generations each having a pair of homogeneous traits
Step 2: Perform dihybrid cross of P generations to produce F1 generation having dominant traits
Step 3: Self-cross pollinate the F1 generation to produce F2 generation
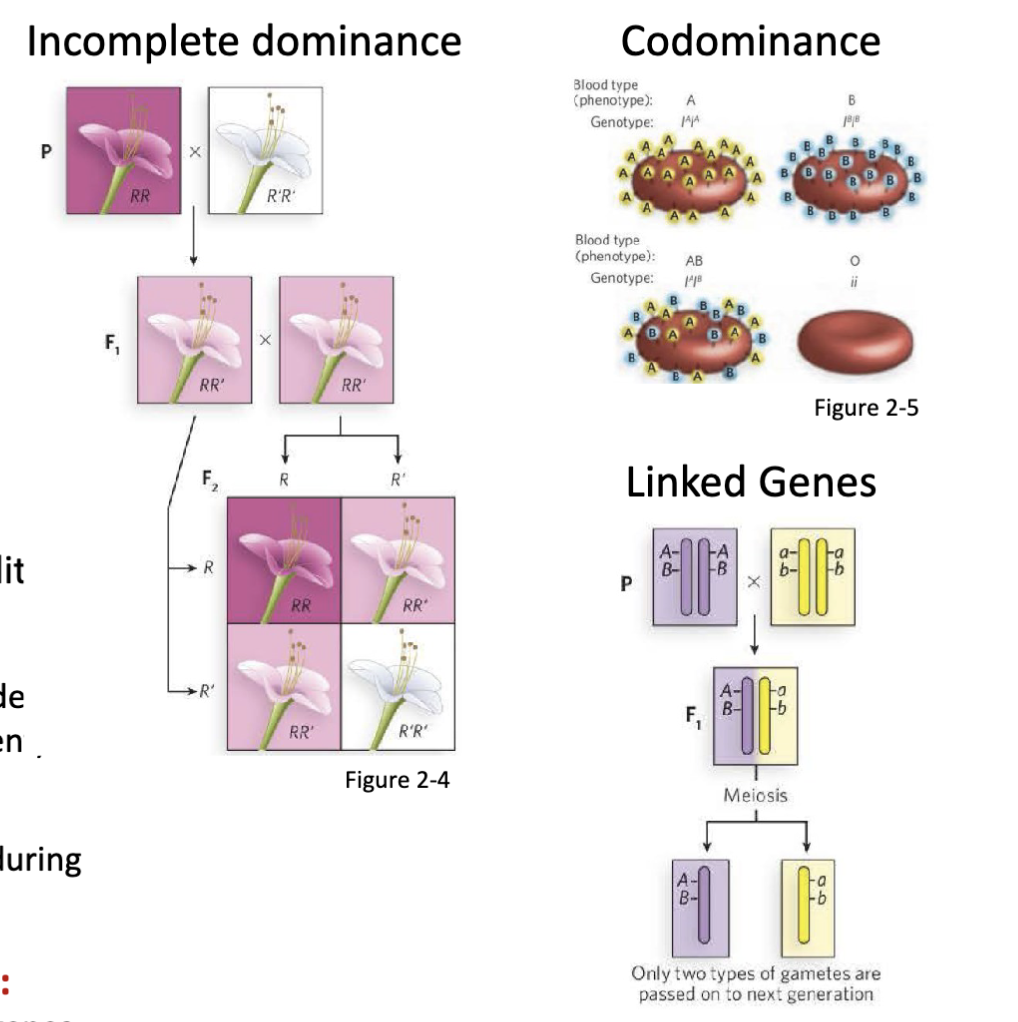
What are some examples of non-Mendelian inheritance?
Incomplete dominance, codominance, linked genes
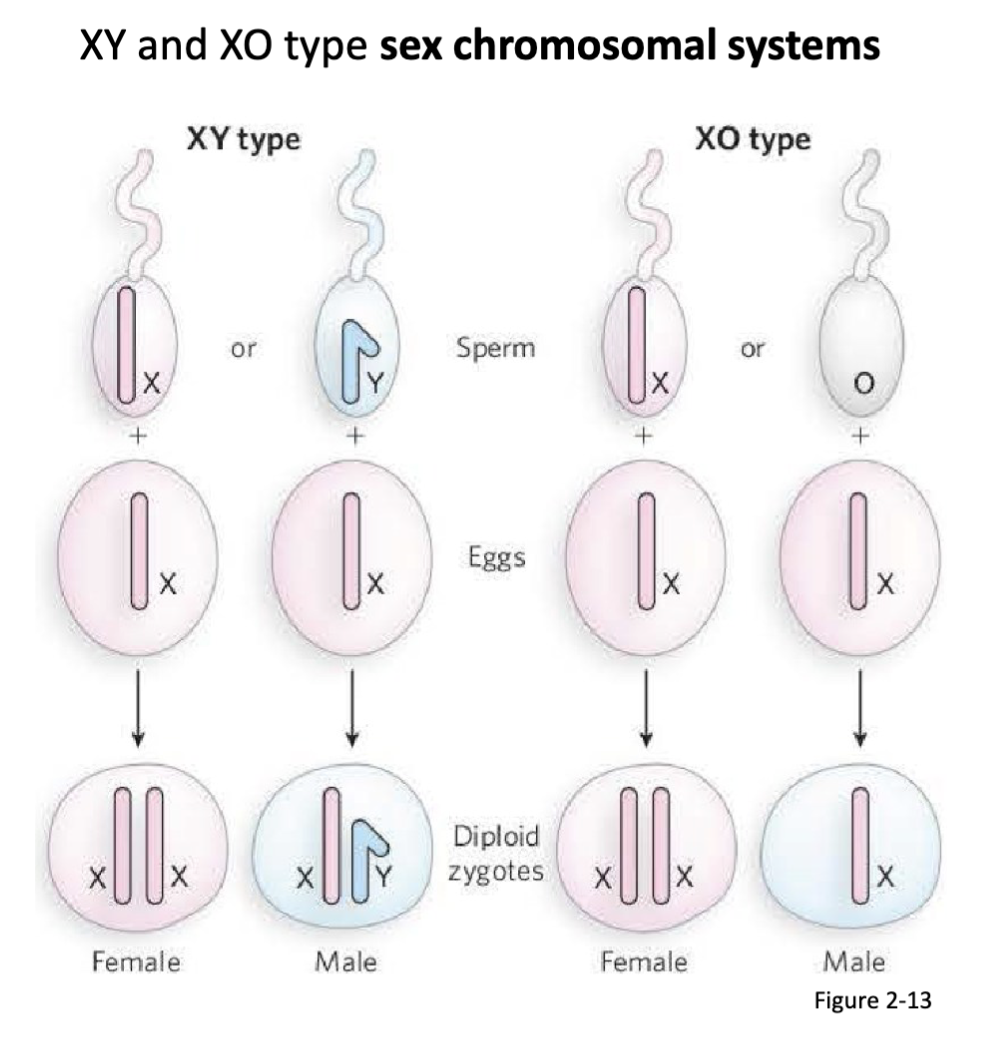
How was the chromosomal basis for biological sex determined?
Both Wilson and Stevens independently observe that male gametes posses an accessory (X) chromosome with: (1) a morphologically distinct partner, or (2) no counterpart chromosome.
conclude that these accessory chromosomes are the determinants of male and female sex
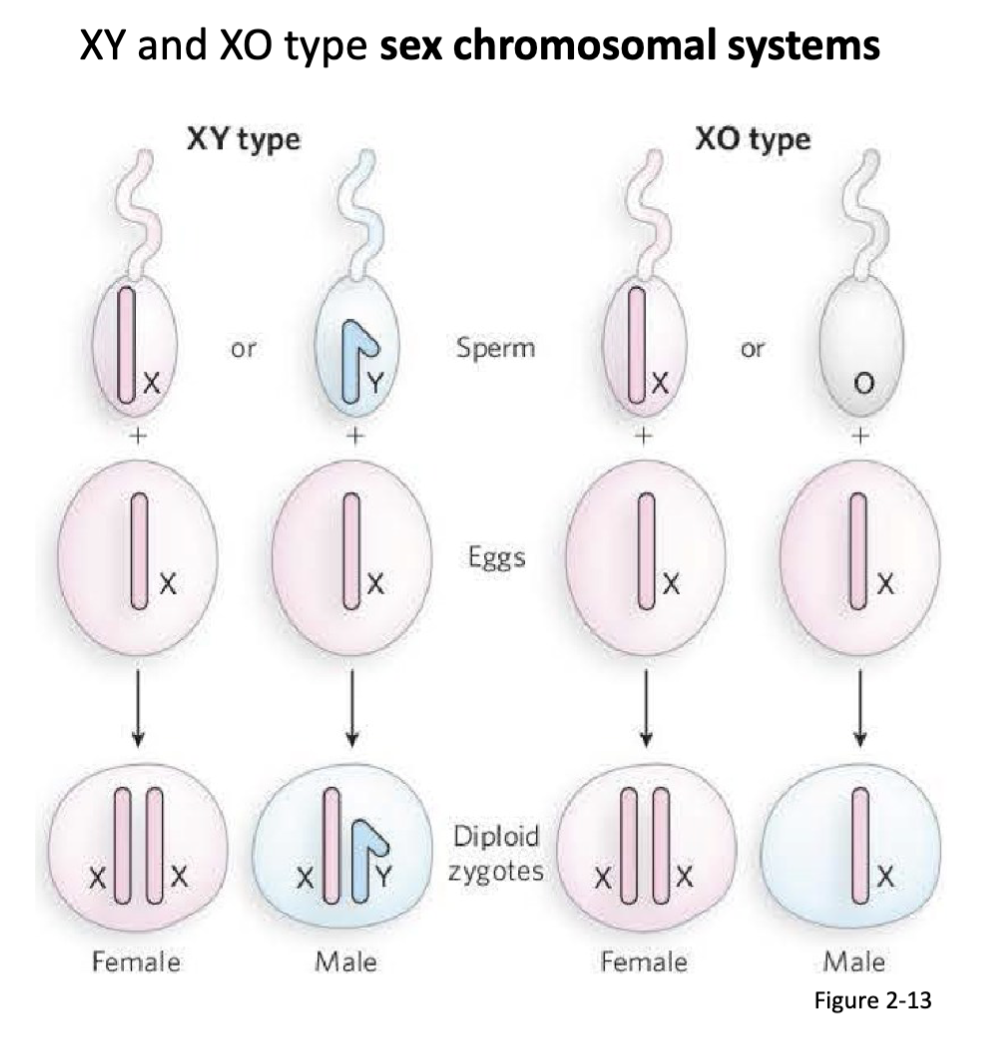
Who was involved in determining the chromosomal basis for biological sex?
Edmond Wilson and Nettie Steven
Who connected chromosomes to Mendel’s “particle” of heredity?
Walter Sutton proposed the theory
It was proven by Thomas Hunt Morgan
What was the proposed theory that connected Mendel’s theory of heredity to chromosomes?
Mendel showed that the population distribution of some traits can be predicted.
Mendel proposed that a pair of particles, referred to as “alleles”, mediate the sorting of these traits.
Sutton postulates that sex chromosomes could be Mendel’s alleles providing a molecular basis for heredity.
How was the connection between chromosomes and heredity proven?
Fruit flies have an eye colour trait:
Morgan studies fruit flies (Drosophilia melanogaster) notes that the predominant red-eyed phenotype (wild-type) is occasionally lost in males producing a rarer white-eyed phenotype (mutant).
Testing the chromosome theory of inheritance:
Morgan is familiar with both Mendel and Sutton’s work. Morgan hypothesizes that the eye colour allele is located
on the sex chromosomes and sets out to demonstrate
chromosome theory of inheritance using fruit flies
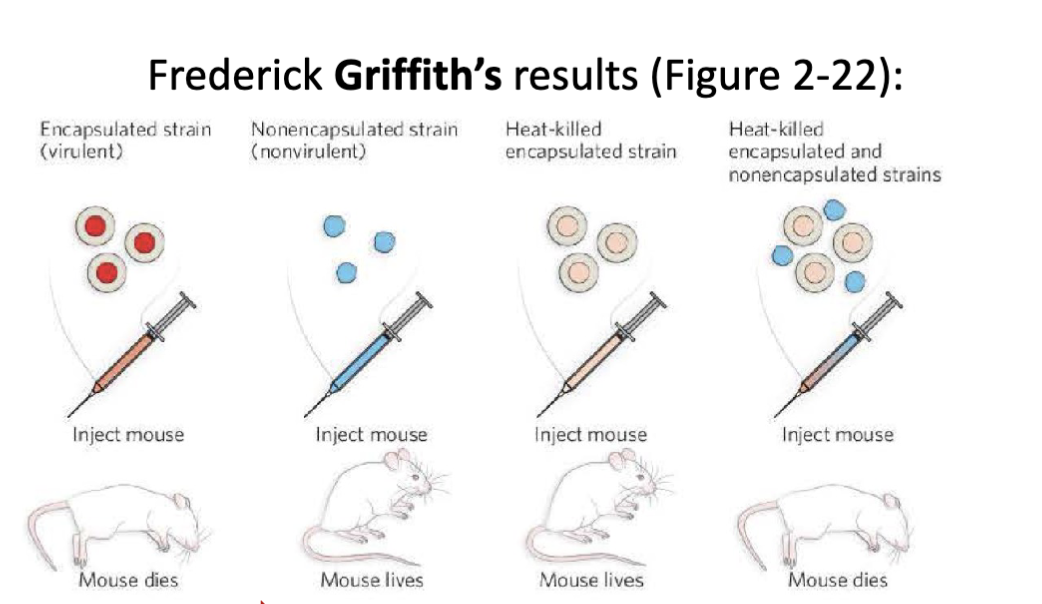
Once the molecular basis for heredity could be attributed to chromosomes what was the next experiment performed to answer the question: What is molecule (protein or DNA) is responsible for genetics?
Griffith’s experimental set up: There are two strains of pneumonia causing bacteria (Streptococcus pneumoniae). One strain is virulent (smooth phenotype) and the other strain is non-virulent (rough phenotype.
The non-virulent bacteria can be transformed by the molecular carrier of genetic information (e.g. virulence)
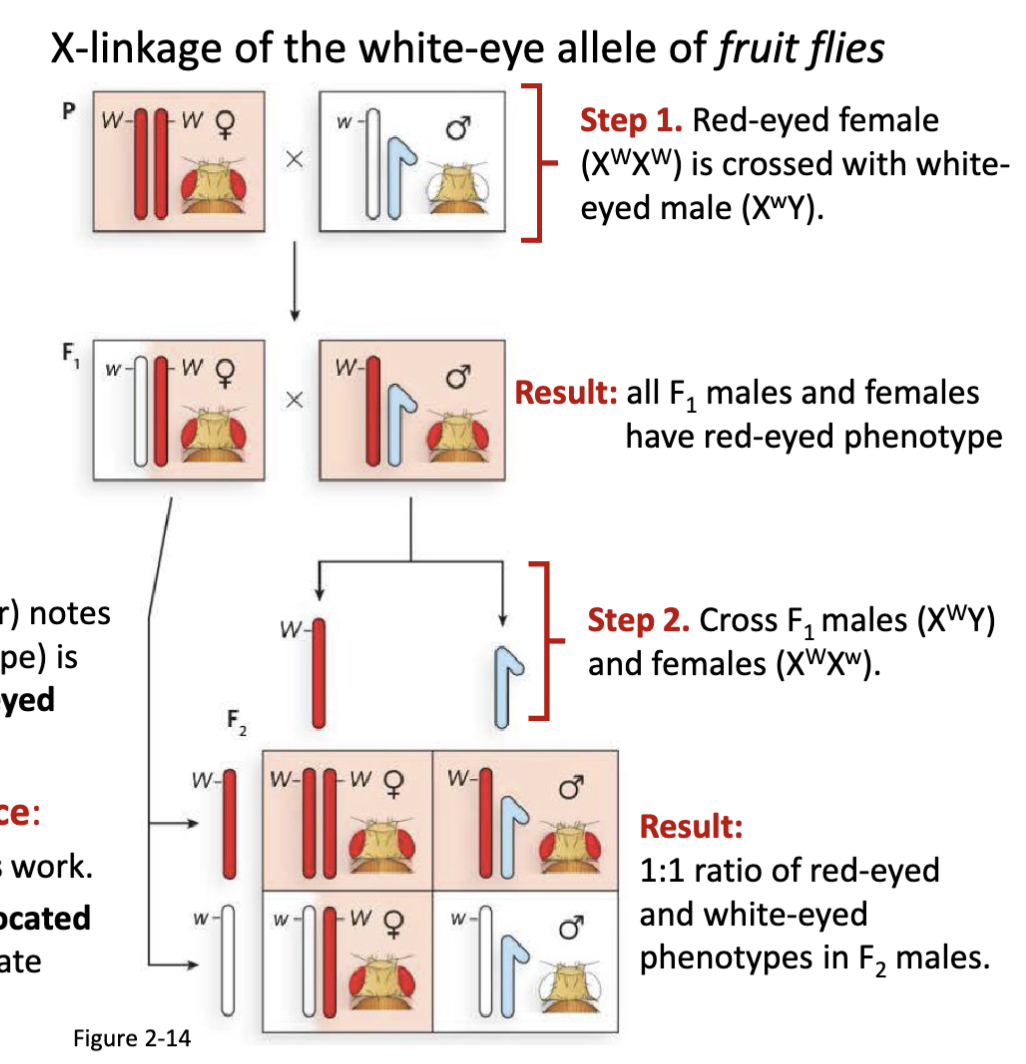
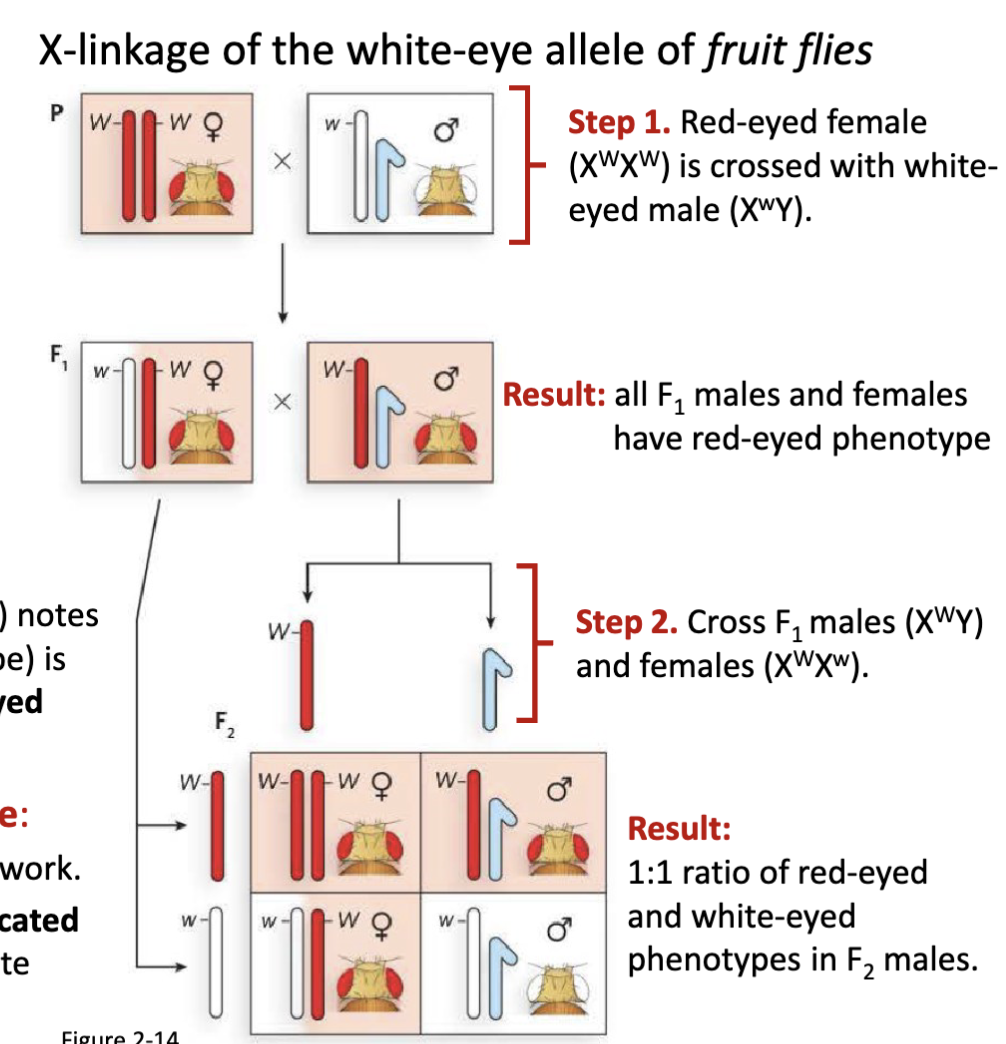
What were the details of Morgan’s fruit fly experiment?
Step 1. Red-eyed female (XWXW) is crossed with white-
eyed male (XwY)
Result: all F1 males and females have red-eyed phenotype
Step 2. Cross F1 males (XWY) and females (XWXw)
Result: 1:1 ratio of red-eyed and white-eyed phenotypes in F2 males
fruit flies were cheap to maintain
*W for red w for white
1:1 ratio in F2 is only applicable to the males not females
first proof for chromosome theory of inheritance
Was Griffith’s experiment enough to prove what molecule (protein or DNA) is responsible for genetics?
No it was not.
Frederick Griffith (1879-1941) demonstrates that a non- virulent strain of bacteria can be transformed into virulent (lethal) strain via an unknown molecular carrier of genetic information. Oswald Avery (1877-1955) demonstrates that the carrier molecule is DNA
Why was it unclear if proteins or DNA was the molecule responsible for genetics?
Chemical analysis of chromosomes shows that they contain both protein and deoxyribonucleic acid (DNA)
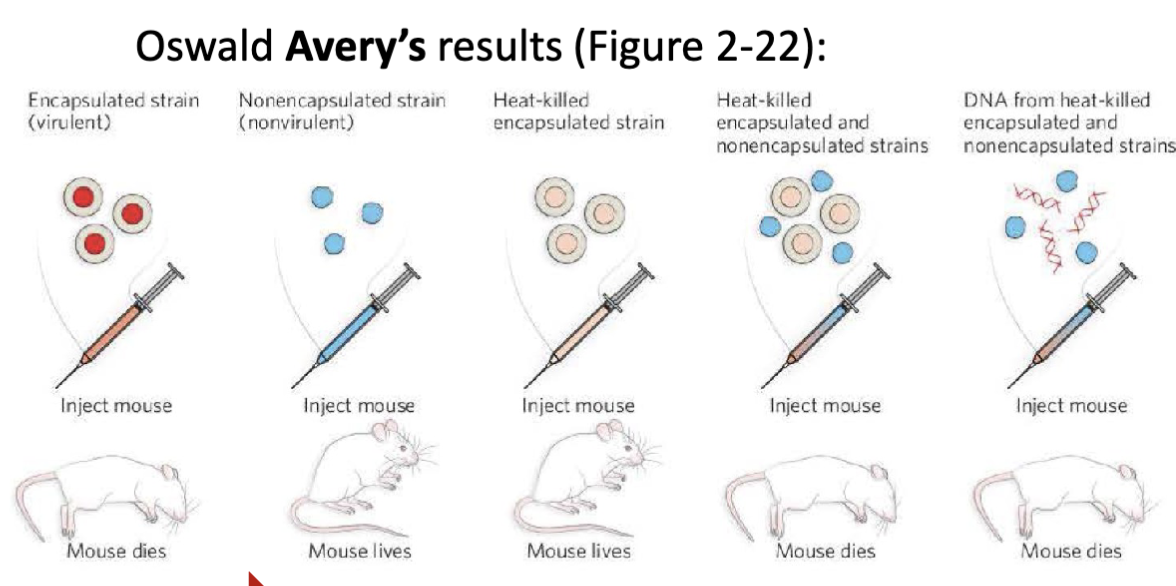
What was Oswald Avery’s experiment? What did this experiment prove?
Experimental setup:
DNA degrading enzymes
RNA degrading enzymes or
protein degrading enzymes
DNA is the molecular carrier of genetic information
The DNA from the heat-killed virulent strain entered the non-virulent strain and made it virulent.
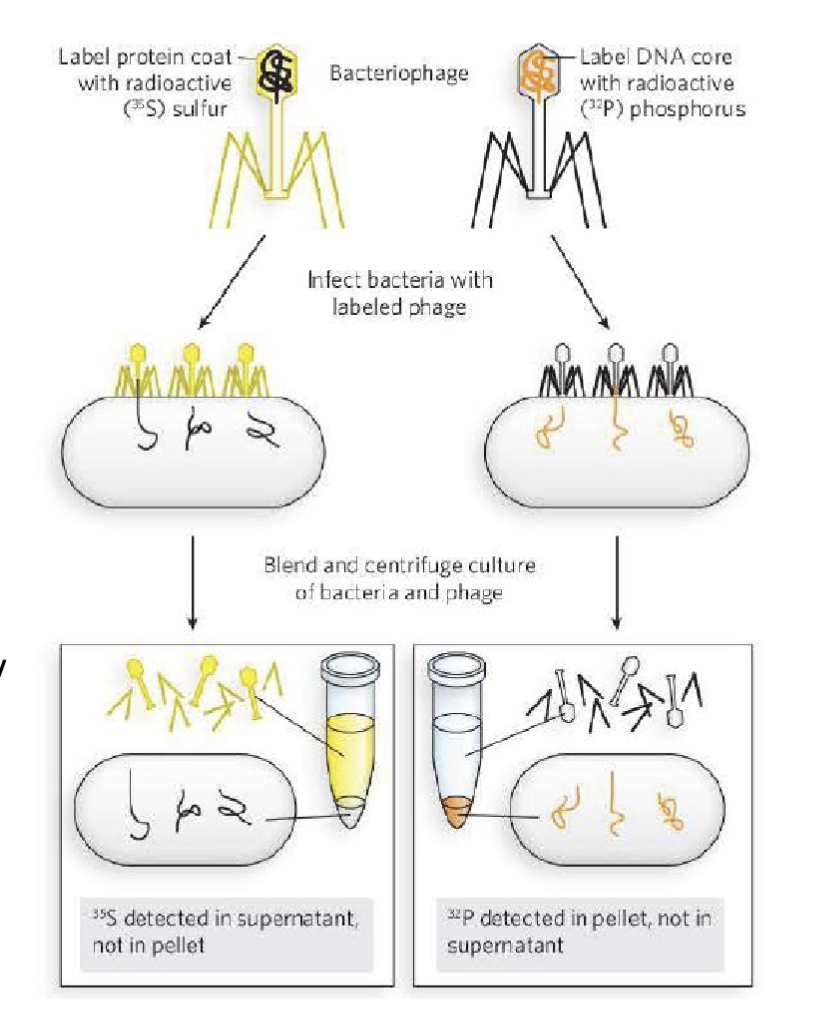
Who conclusively established that DNA is the carrier of genetic information? How was this done?
Martha Chase and Alfred Hershey
Conduct an experiment using T2 bacteriophage
By this time, it was understood that viruses propagate by infecting their host. Chase and Hershey prepared T2 bacteriophage, radiolabelled with either:
-32P isotope which is only incorporated in DNA
-35S isotope which is only incorporated in protein
sulfur is specific to the protein of the bacteriophage and the phosphorus is backbone of DNA
this proved that the DNA entered into the bacteria but the protein did not, therefore the DNA is the material that is transferred aka the genetic material
What is the process of transcription?
Transcription = copying DNA → RNA (mRNA), using RNA polymerase, with initiation, elongation, and termination steps.
What is the process of translation?
Translation = mRNA → protein, carried out by ribosomes, using tRNAs and the genetic code.
What is the difference between a dominant and a recessive trait?
Dominant = masks the effect of a recessive allele when both are present (heterozygous).
Recessive = hidden in heterozygous individuals, only visible in homozygous recessive.
What is the difference between a wild-type and mutant phenotype?
Wild type = baseline, “normal” phenotype in nature.
Mutant = altered phenotype caused by a DNA change.
Describe the rationale for Sutton’s hypothesis “alleles are located on chromosomes”
He hypothesizes that the chromosomes observed by Wilson and Stevens are Mendel’s “particles of genetic inheritance” (alleles).
Sutton’s rationale was that the parallel behavior of chromosomes and Mendel’s factors during meiosis strongly suggested that alleles reside on chromosomes.
Describe Morgan’s experiment demonstrating the chromosome theory of inheritance
The white-eye trait appeared only in males, and always linked with the X chromosome.
This pattern did not fit simple Mendelian ratios if the trait was autosomal.
Instead, it fit perfectly with sex-linked inheritance:
Males (XY) inherit their single X from their mother.
If that X carried the white-eye allele, the male would show the mutant phenotype (no second X to mask it).
Females (XX) needed two copies of the mutant allele to have white eyes, which was rare.
Morgan’s experiment with white-eyed Drosophila demonstrated that a specific trait (eye color) is inherited with the X chromosome, proving that genes reside on chromosomes and validating the chromosome theory of inheritance.
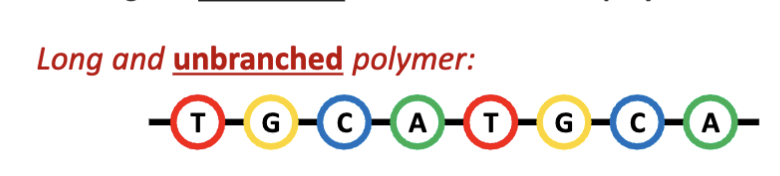
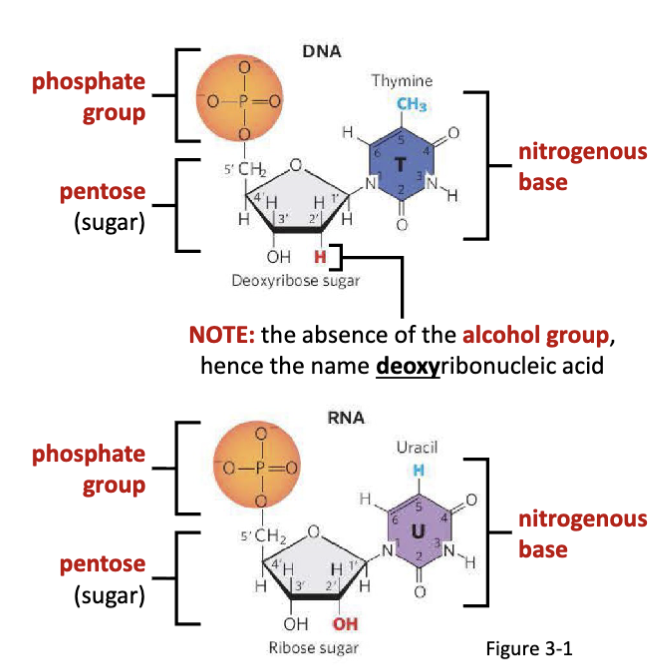
What are nucleic acids?
Deoxyribonucleic acid (DNA) and ribonucleic acid (RNA) are long and unbranched biomacromolecular polymers.
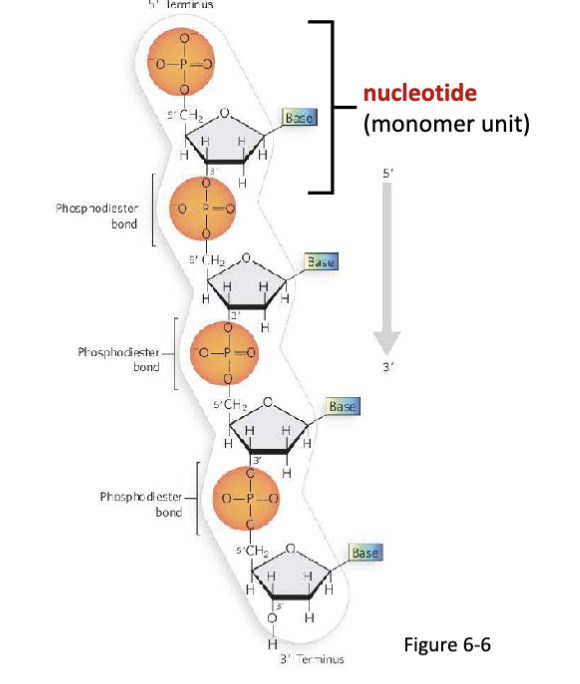
What are nucleotides?
Nucleotides are the monomer units of nucleic acids, joined by
phosphodiester bonds.
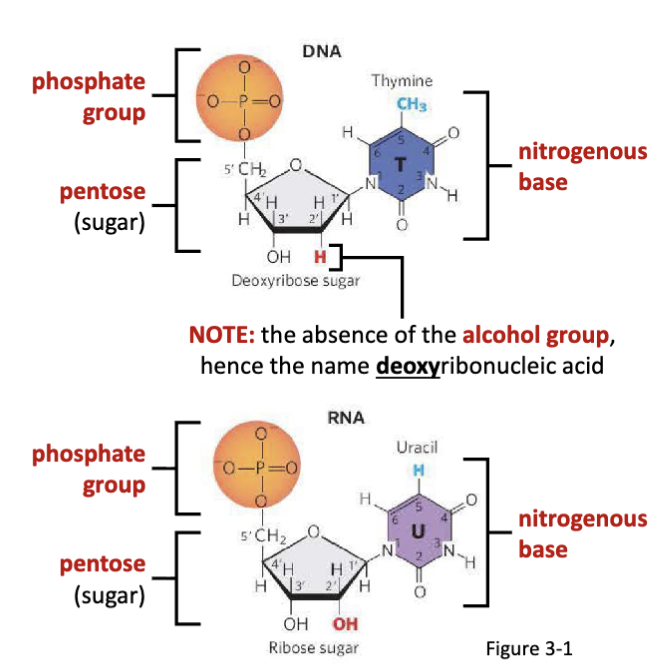
What is the structural difference between RNA and DNA?
the absence of the alcohol group= deoxyribonucleic acid
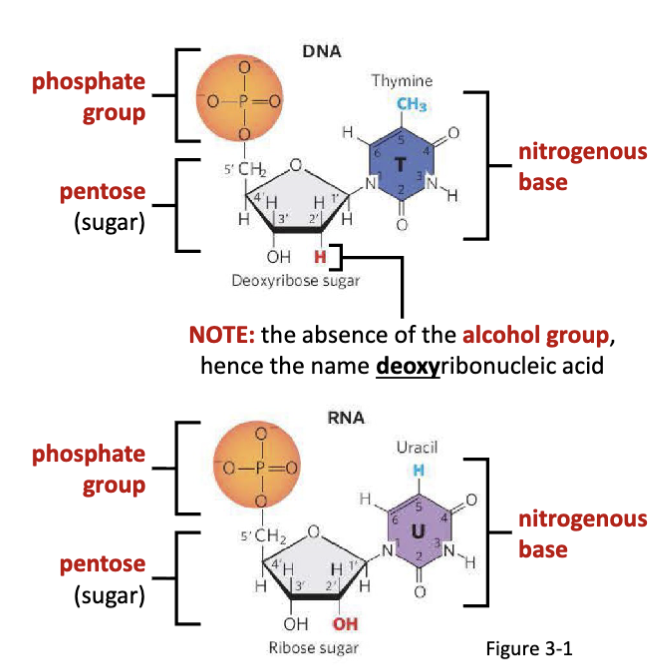
What are the components of nucleotides?
a nitrogenous base (varies by nucleotide identity)
a five-carbon “pentose” sugar
phosphate group(s)
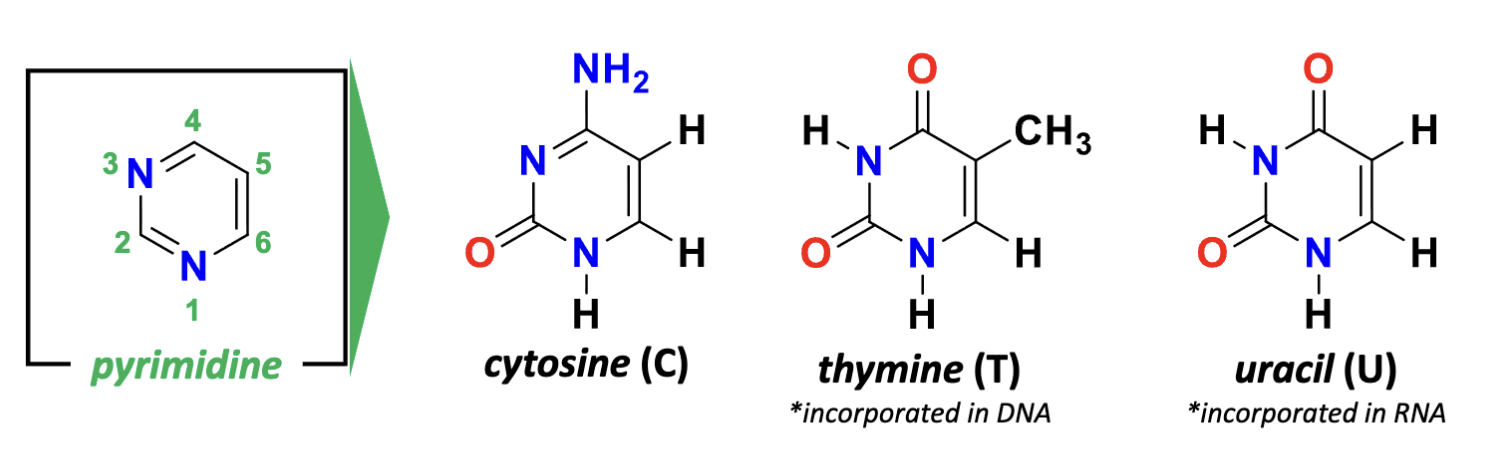
What are the two types of nitrogenous bases?
pyrimidines and purines
What are the features of pyrimidines?
(nitrogen at the bottom is 1) aromatic Hydrogens at 2,4,5,6
derivatives of pyrimidine= cytodine (4- amino 2-keto), thymine (4-keto, 2-keto, 5-methyl), uracil (looses the methyl group of thymine)
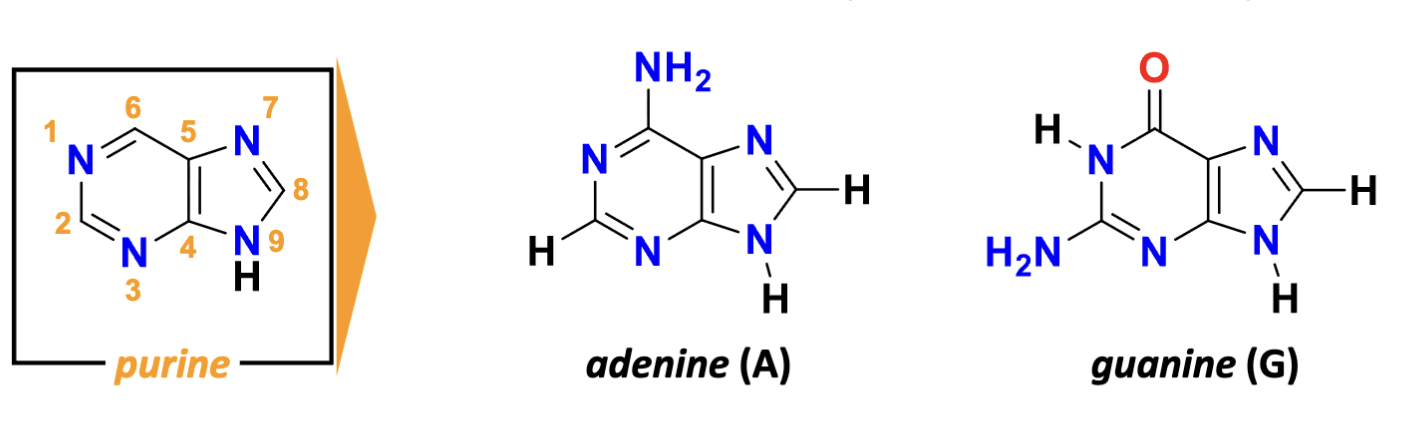
What are the features of purines?
purine:
adenosine (6-amino), guanine (2-amino group, 6-keto group)
What are the features of the sugar in RNA or DNA?
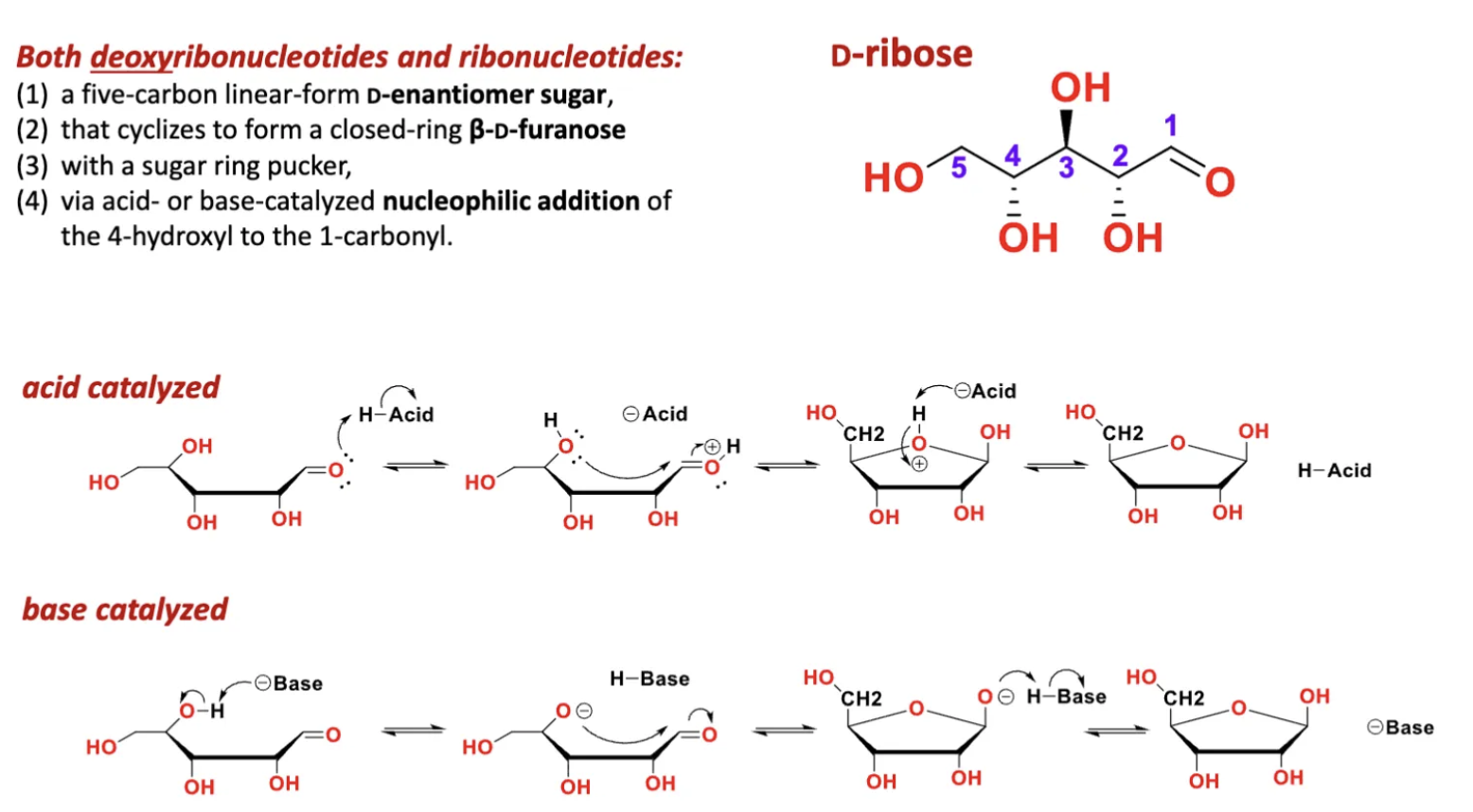
(1) a five-carbon linear-form D-enantiomer sugar
(2) that cyclizes to form a closed-ring β-D-furanose
(3) with a sugar ring pucker
(4) via acid- or base-catalyzed nucleophilic addition of the 4-hydroxyl to the 1 carbonyl
D-ribose: R stereochemistry for all the carbons
L ribose: less common, S configuration
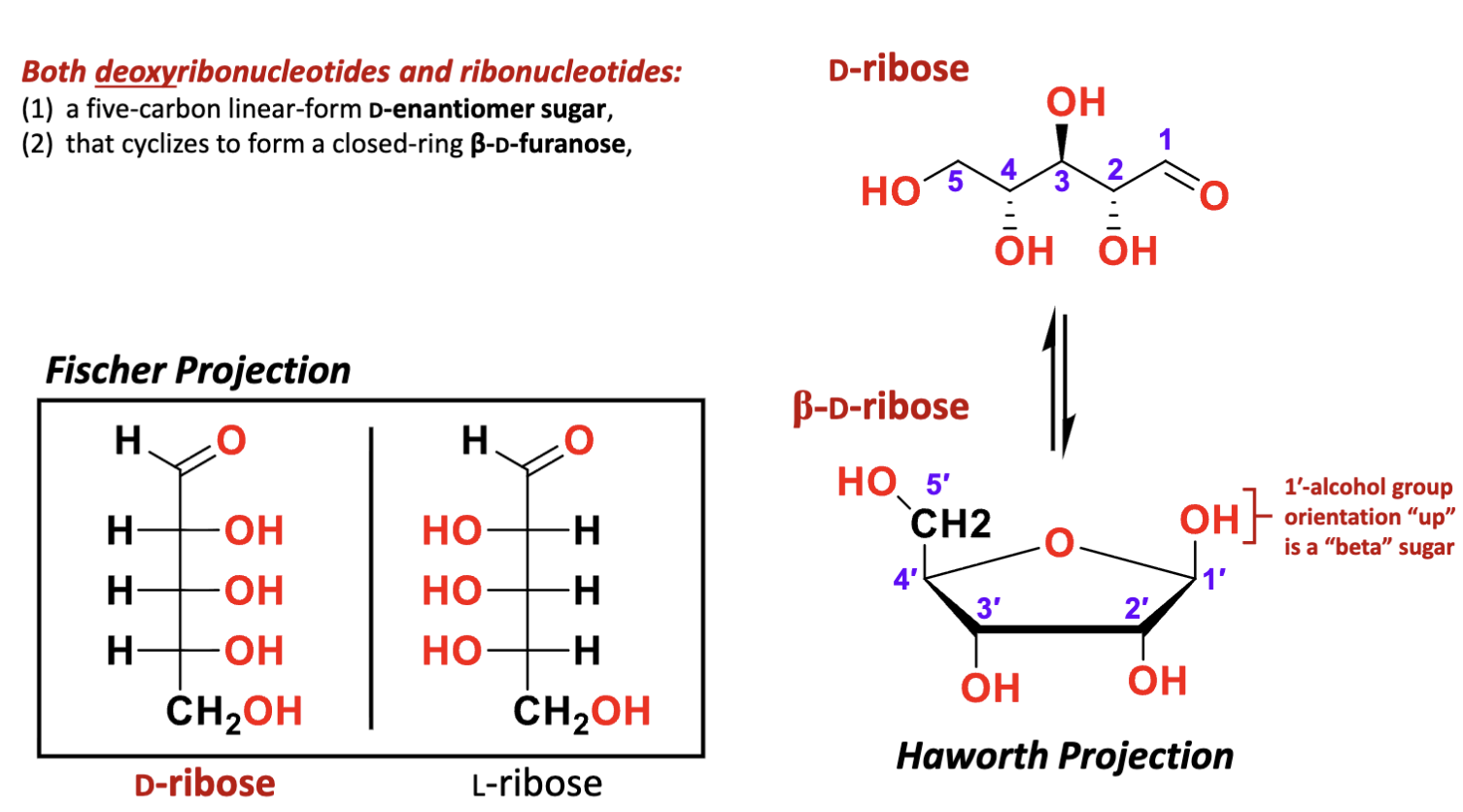
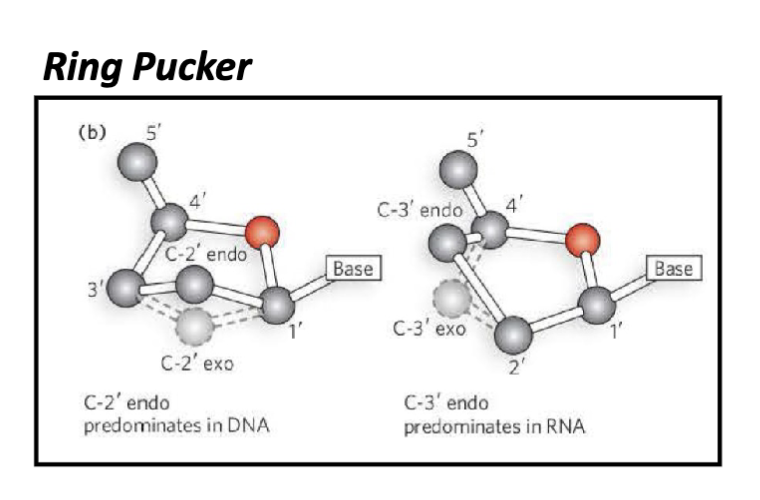
What are the features of the pucker in the pentose in RNA and DNA?
endo pucker:
the endo pucker has the 2’ out of the plane predominantly seen in DNA
the endo pucker has the 3’ out of the plane predominantly seen in RNA
What are the features of cyclization of (deoxy)ribose sugars by nucleophilic addition?
nucleophilic addition of the 4-hydroxyl to the 1 carbonyl
cyclization determines alpha or beta position of the sugar
because the attack came from underneath the OH group is displaced up
What is a nucleoside?
nucleosides sans phosphate
vs
nucleotides have the phosphate
What joins the nitrogenous base and the sugars (in RNA and DNA)?
glycosidic bonds
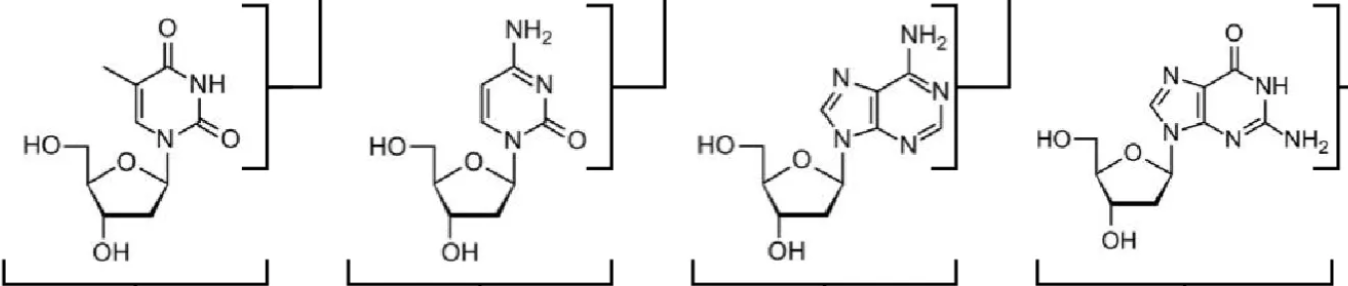
What are the names of these?
left to right: deoxythymidine (dT), deoxycytidine (dC), deoxyadenosine (dA), deoxyguanosine (dG)
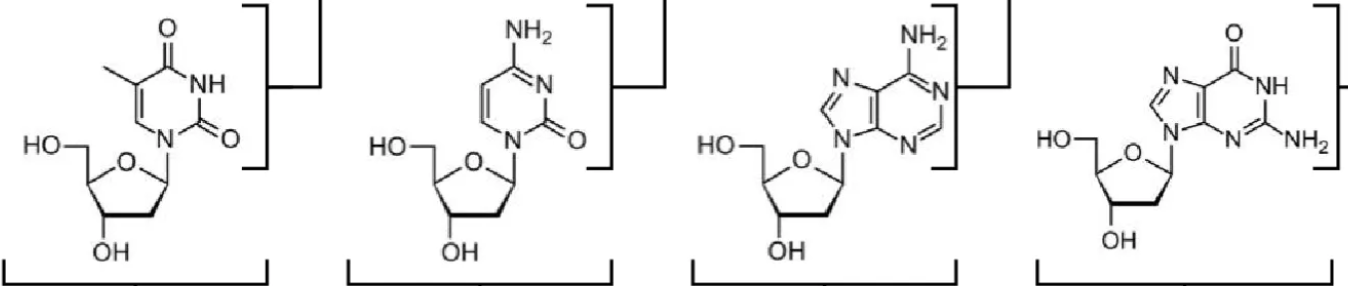
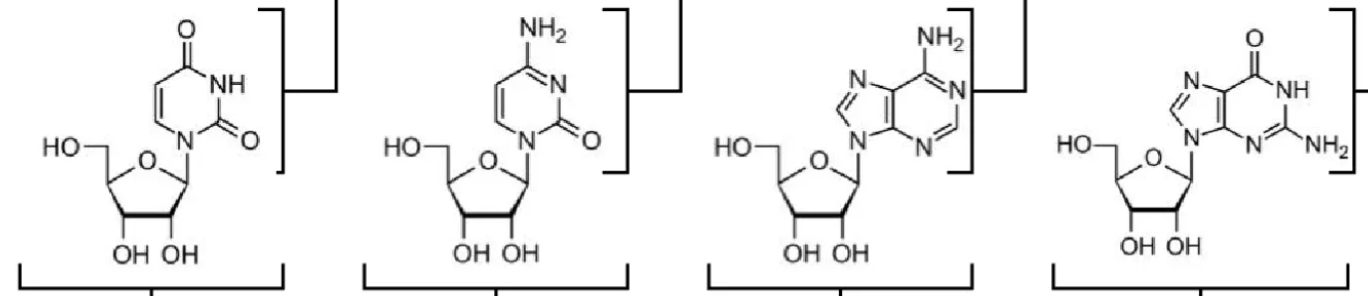
What are the names of these structures?
uridine (U), cytidine (C), adenosine (A), guanosine (G)
What are the features of the nitrogenous bases?
The monomers of DNA contain thymine, cytosine, adenine, and guanine nitrogenous bases.
The monomers of RNA contain uracil, cytosine, adenine, and guanine nitrogenous bases.
also energy storage molecules (due to the phosphate bonds)
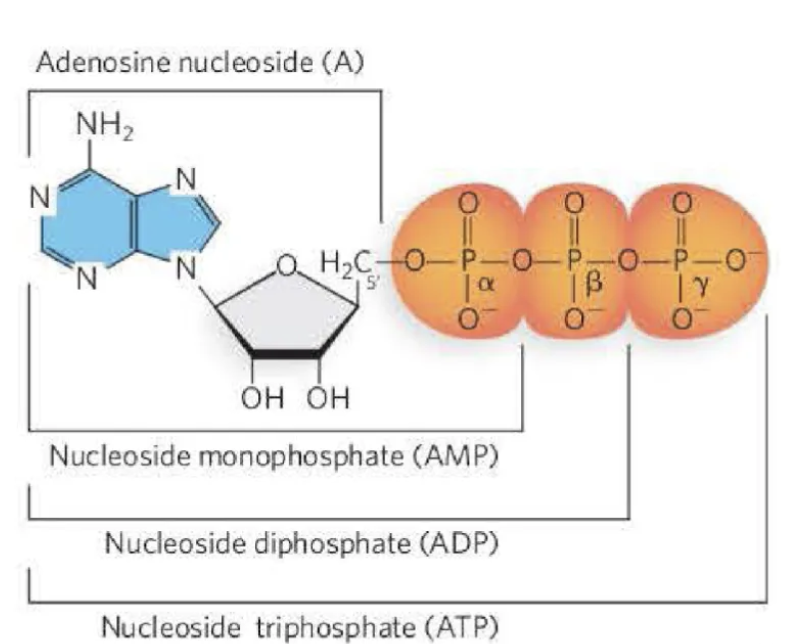
What are the features nucleosides?
The nucleosides that comprise the monomers of DNA
include the sugar β-D-deoxyribose covalently bonded
to the nitrogenous bases via a glycosidic bond.
The nucleosides that comprise the monomers of RNA
include the sugar β-D-ribose covalently bonded to the
nitrogenous bases via a glycosidic bond
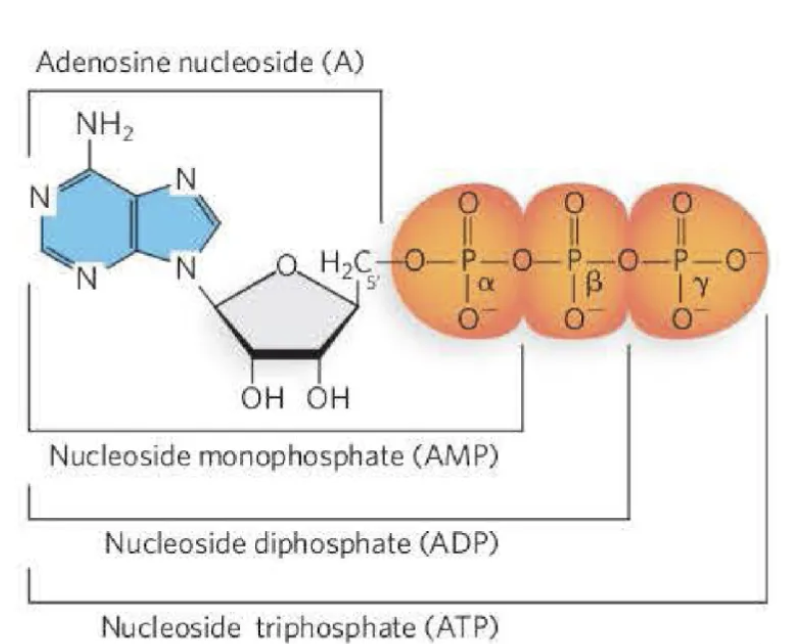
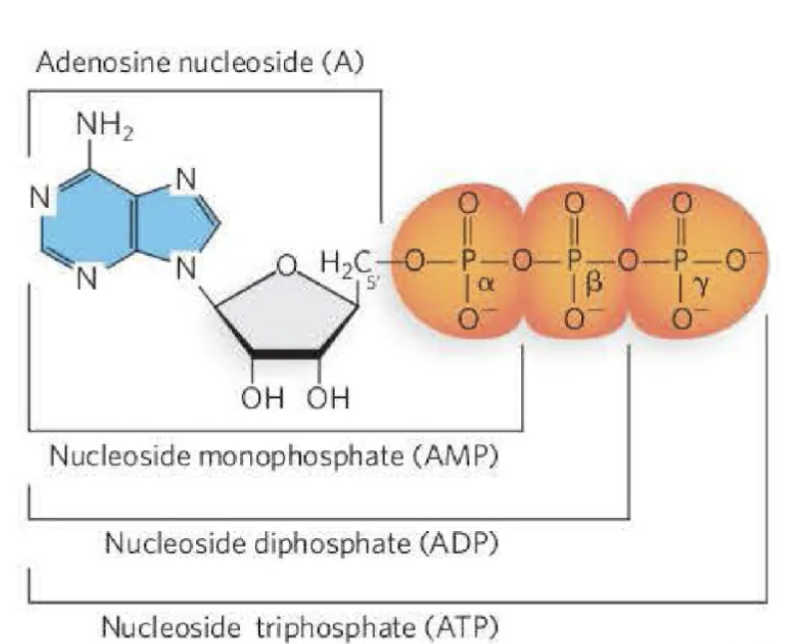
What are the features of nucleotides?
The nucleotides that comprise the monomers of DNA
include a nucleoside with a 5′ phosphate group.
The nucleosides that comprise the monomers of RNA
include a nucleoside with a 5′ phosphate group.
The nucleotide can have one- (mono), two- (di), or
three- (tri) phosphate groups. These are referred to as
the alpha (α), beta (β), and gamma (γ) phosphates.
Which position of the sugar is the phosphate attached?
nucleotides: 5’ spot of the sugar attached to the phosphate
How are nucleic acids formed?
formed by nucleotides joined by phosphodiester bonds
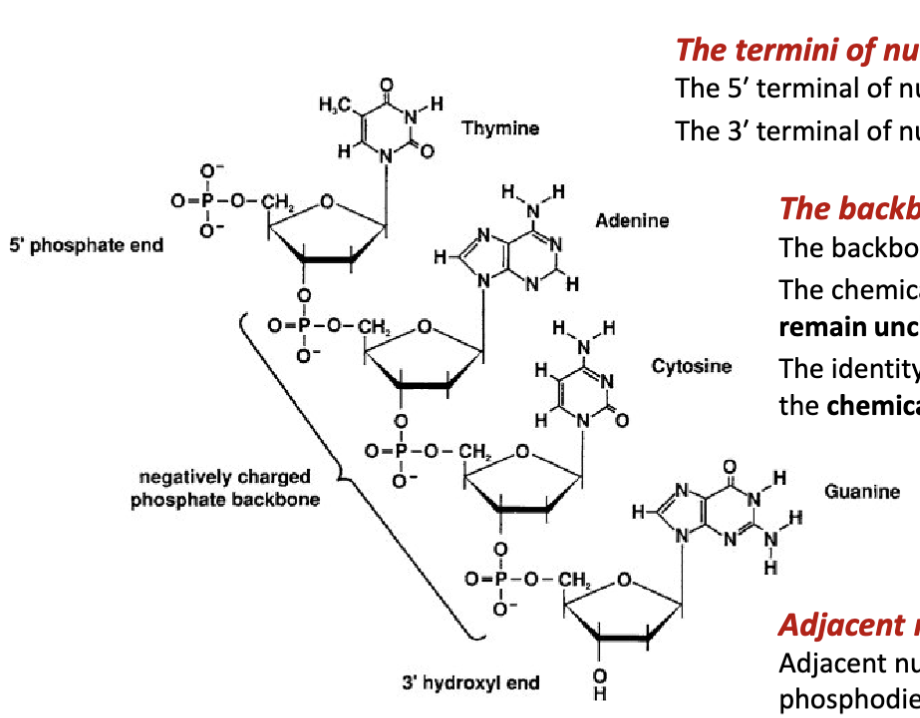
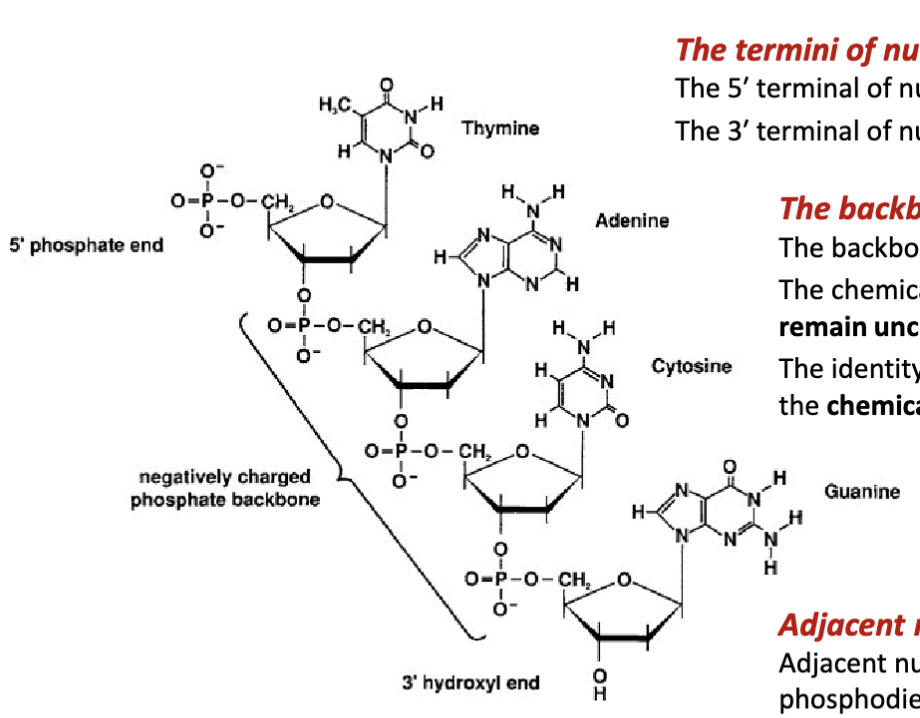
What are the features of the backbone of nucleic acids?
The backbone nucleic acids is negatively charged
The chemical structure of the phosphate group and sugar remain unchanged (fixed) for each nucleic acid residue.
The identity of the nucleic acid residue is determined by the chemical structure of its nitrogenous base.
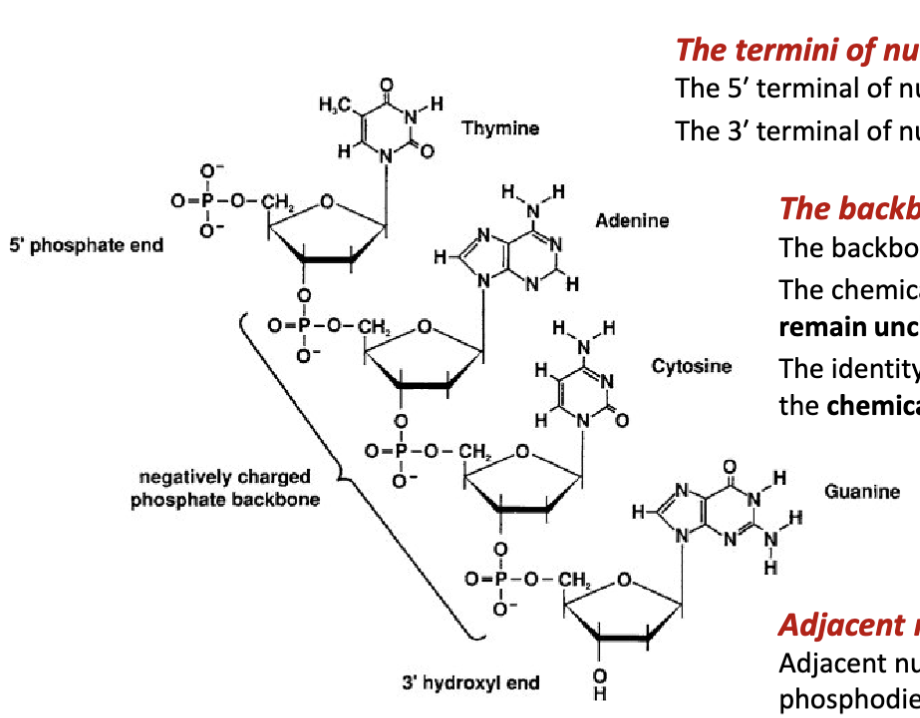
How are adjacent nucleotides joined together?
Adjacent nucleotides are covalently joined via a phosphodiester bond between the 3′ terminal alcohol (preceding nucleotide) and the 5′ terminal phosphate group (proceeding nucleotide.
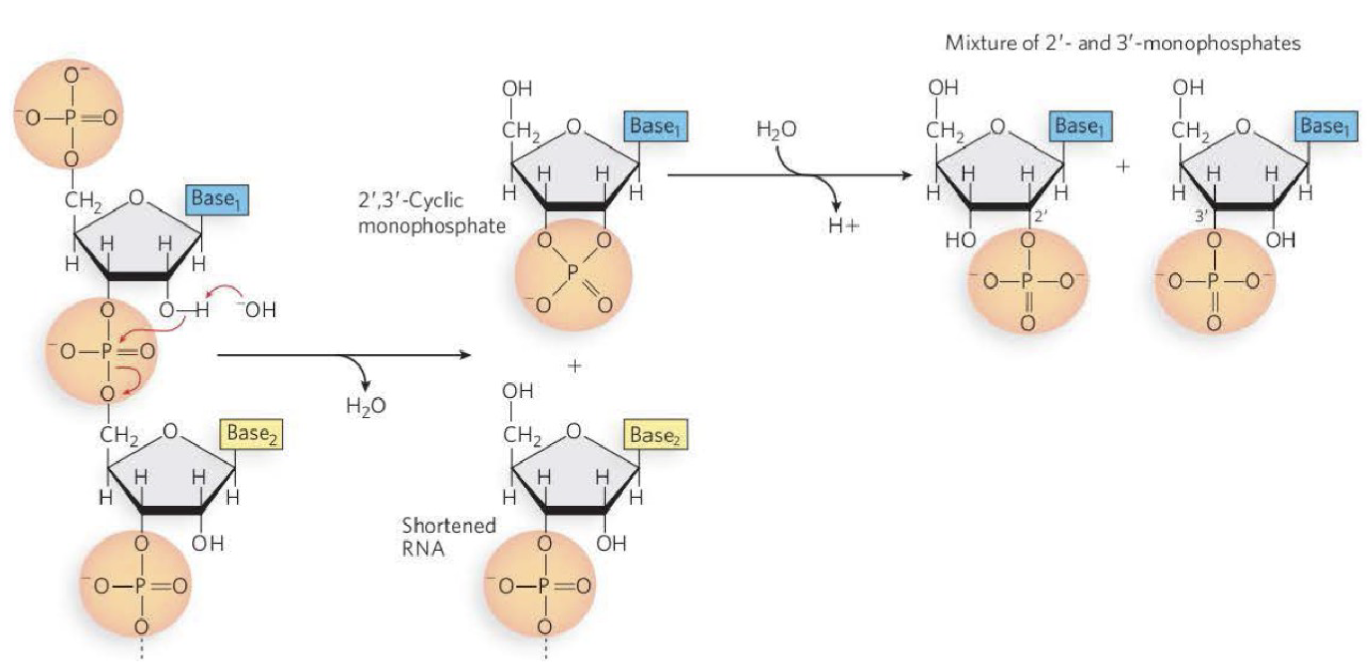
Is RNA or DNA more susceptible to hydrolysis? If so why?
The resonance structure of phosphate groups:
Phosphate groups are electrophiles and leaving groups.
This renders RNA susceptible to hydrolysis under basic conditions.
RNA is more susceptible to hydrolysis compared to DNA
2’ hydroxyl group makes the phosphodiester group susceptible to hydrolysis
-OH acts as the nucleophile will attack the oxygen which will attack the phosphate
the CH2 attached to a phosphate will end up with a OH attached instead of phosphate group after the hydrolysis process
the OH comes from water of the hydrolysis reaction (water used to break it apart)
What is name of the base and the nucleoside of A & dA?
A: adenine, adenosine
dA: adenine, deoxyadenosine
What is name of the base and the nucleoside of G & dG?
G:Guanine, guanosine
dG:Guanine, deoxyguanosine
What is name of the base and the nucleoside of C & dC?
C: Cytosine, cytidine,
dC: Cytosine, deoxycitidine
What is name of the base and the nucleoside of T & dT?
T: thymine, themidine
dT: thymine, deoxythymidine
What is name of the base and the nucleoside of U?
uracil, uridine
What are the features of nucleic acids?
The 5′ terminal of nucleic acids has a phosphate group. The 3′ terminal of nucleic acids does not have a phosphate group. Adjacent nucleotides are covalently joined via a phosphodiester bond
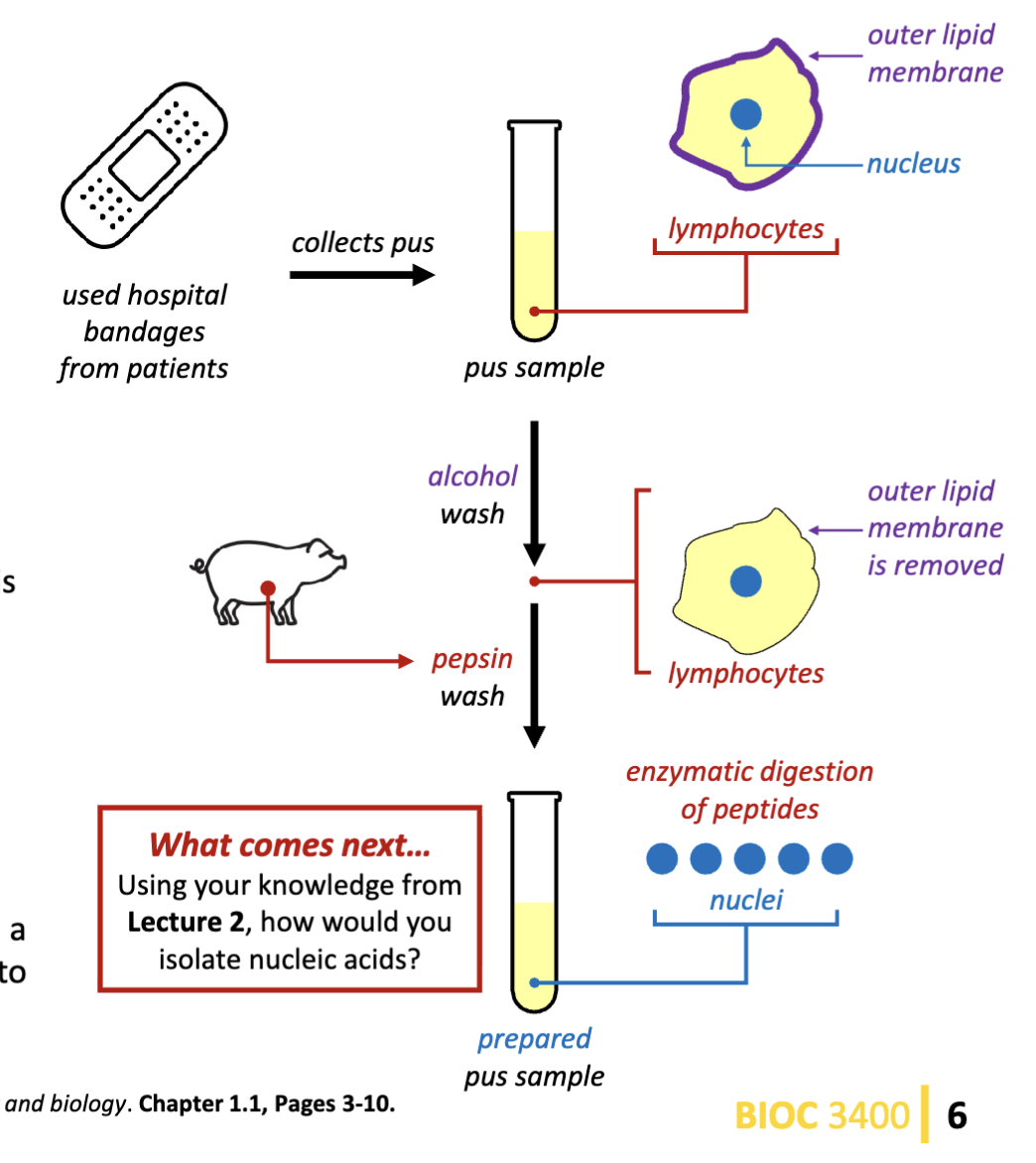
Who was the first scientist to isolate nuclein (nucleic acids)? How did they achieve this?
Friedrich Meisher
Uses an alcohol wash, then uses pepsin (protease that will degrade the peptides). This leaves the nucleic acids behind.
“nuclein” substance is present in yeast, kidneys, liver, testes, and nucleated RBCs.
base driven attack removing the phosphodiester bond
the ribose in RNA makes it more prone to hydrolysis (= most research done on RNA)
What is the evidence demonstrating that chromosomes contain nucleic acids?
Aniline dyes bind nucleic acids.
Chromosomes bind aniline dyes.
Therefore, chromosomes contain nucleic acids.
This work inspires: The hypothesis that DNA is involved in the chromosome theory of inheritance.
this work supports the notion that DNA is the molecular transporter of genetic information
Who discovered the evidence that chromosomes contain nucleic acid?
Altmann isolates the first protein-free nuclein materials and names them nucleic acids.
At this period in time: numerous aniline dyes had been developed. Their utility to stain cellular components inspired the systematic study of biological specimens and processes.
Flemming develops cell fixation techniques for cytology and applies aniline dyes to observe changes during mitosis in salamander embryos. (how we know the phases of mitosis)
Who identified the nitrogenous bases in nucleic acids?
Kossel adopts Altmann’s approach to nucleic acid isolation and
improves upon the process.
Kossel uses methods of hydrolysis and elemental analysis to
determine the chemical formula of the nitrogenous bases.
Awarded the 1910 Nobel Prize in Physiology or Medicine
“in recognition of cell chemistry made through his work on proteins,
including nucleic substance”
What are the features of early structural characterization of the nucleic acids?
Levene and Jacobs study and structurally characterize (1909) the
nucleosides produced by hydrolysis of isolated nucleic acids.
Levene adopts the use of enzymes to catalyze the hydrolysis of
nucleotides from DNA. This permits the first observation of
nucleosides containing the 2-deoxy-D-ribose sugar
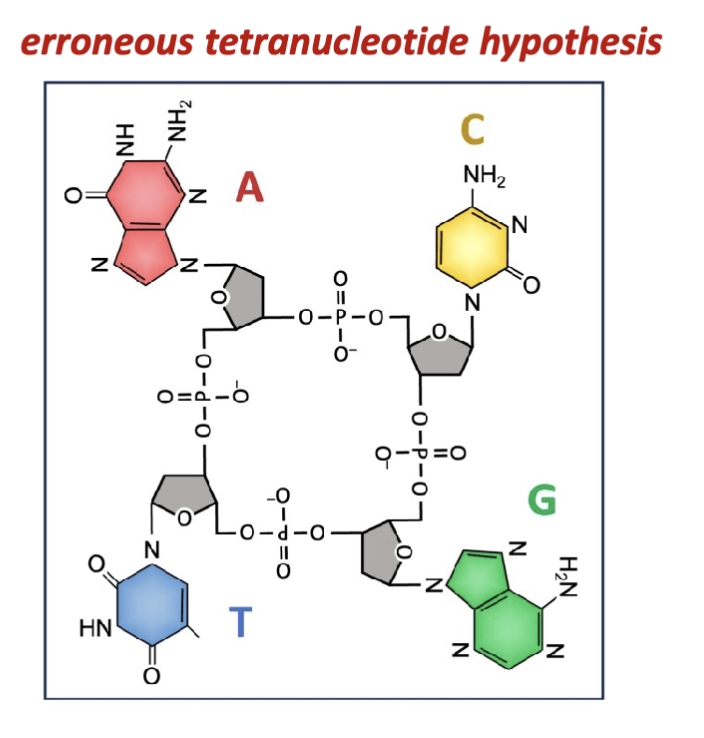
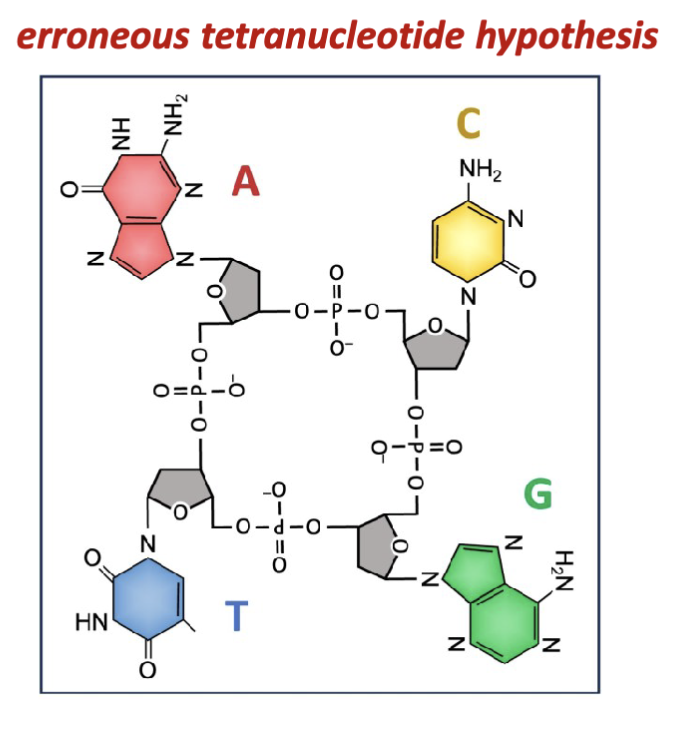
What is the error with Levene and Jacobs experimental results?
Isolates of nucleic acids consistently yielded all four nitrogenous
bases in equal relative proportions. This gave rise to the
erroneous “tetranucleotide hypothesis” which remained
accepted up until the late 1940s.
all 4 bases are not isolated in equal proportion (treats them like a 4 base cyclic structure)
the isolation procedures and hydrolysis and precipitation bias the equilibrium of all the nucleotides
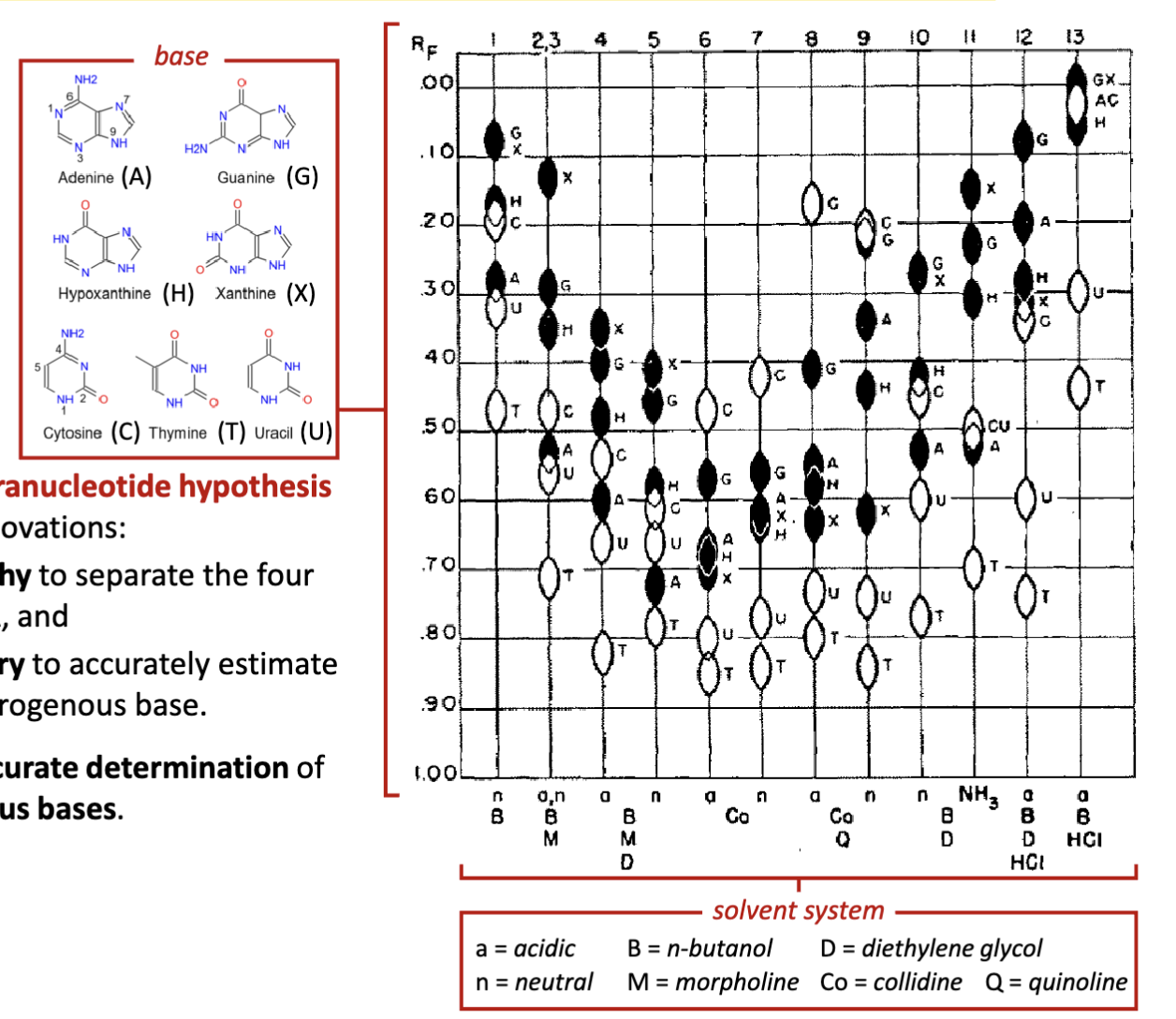
Who debunked the erroneous tetranucleotide hypothesis for nucleic acids? How?
Erwin Chargaff
Chargaff corrects the erroneous tetranucleotide hypothesis
by developing two experimental innovations:
1.developed paper chromatography to separate the four
nitrogenous bases found in DNA, and
2.employed UV spectrophotometry to accurately estimate
the relative amounts of each nitrogenous base. (aromatic: their aromatic orbitals can be excited)
Chargaff’s experiment allows for accurate determination of
the relative quantities of nitrogenous bases
What is Chargraff’s rule?
Chargraff’s rule: In any dsDNA molecule, the amount of adenine (A) is equal to the amount of thymine (T), and the amount of guanine (G) equals the amount of cytosine (C)
How was the structure of DNA determined?
Franklin developed a new X-ray chamber setup that could control humidity.
This innovation was necessary to produce better diffraction patterns
Franklin and Gosling’s data was critical to solving the structure of DNA and was published in Nature in the same issue with Watson and Crick’s structure
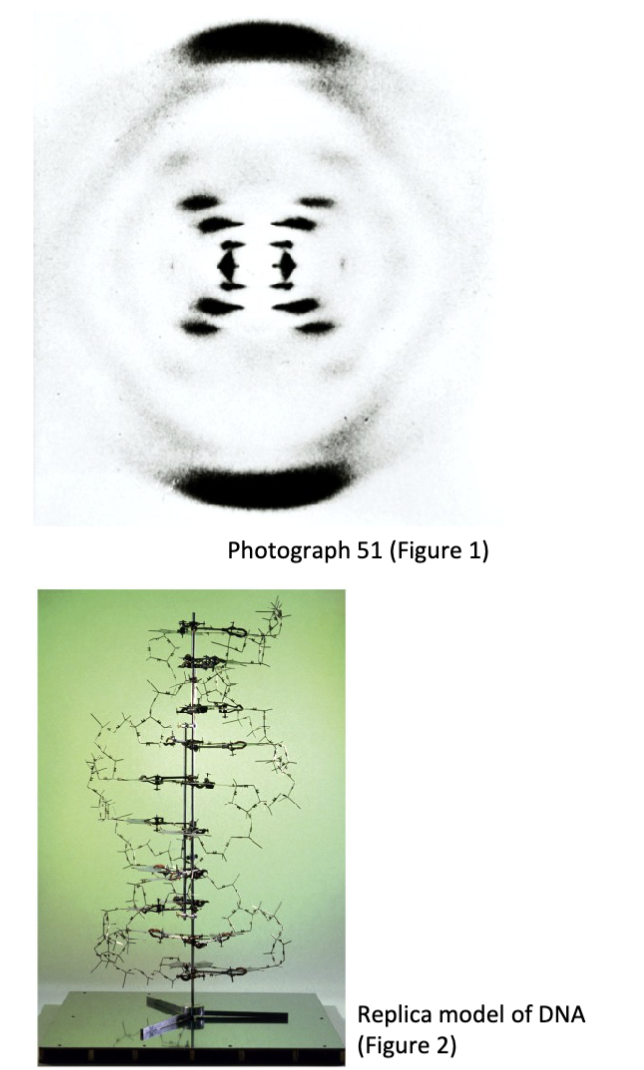
What are the structural rules and chemical rules of the DNA double helix?
The phosphate sugar backbone
Pairing of nitrogenous bases
Nitrogenous base molecular interactions
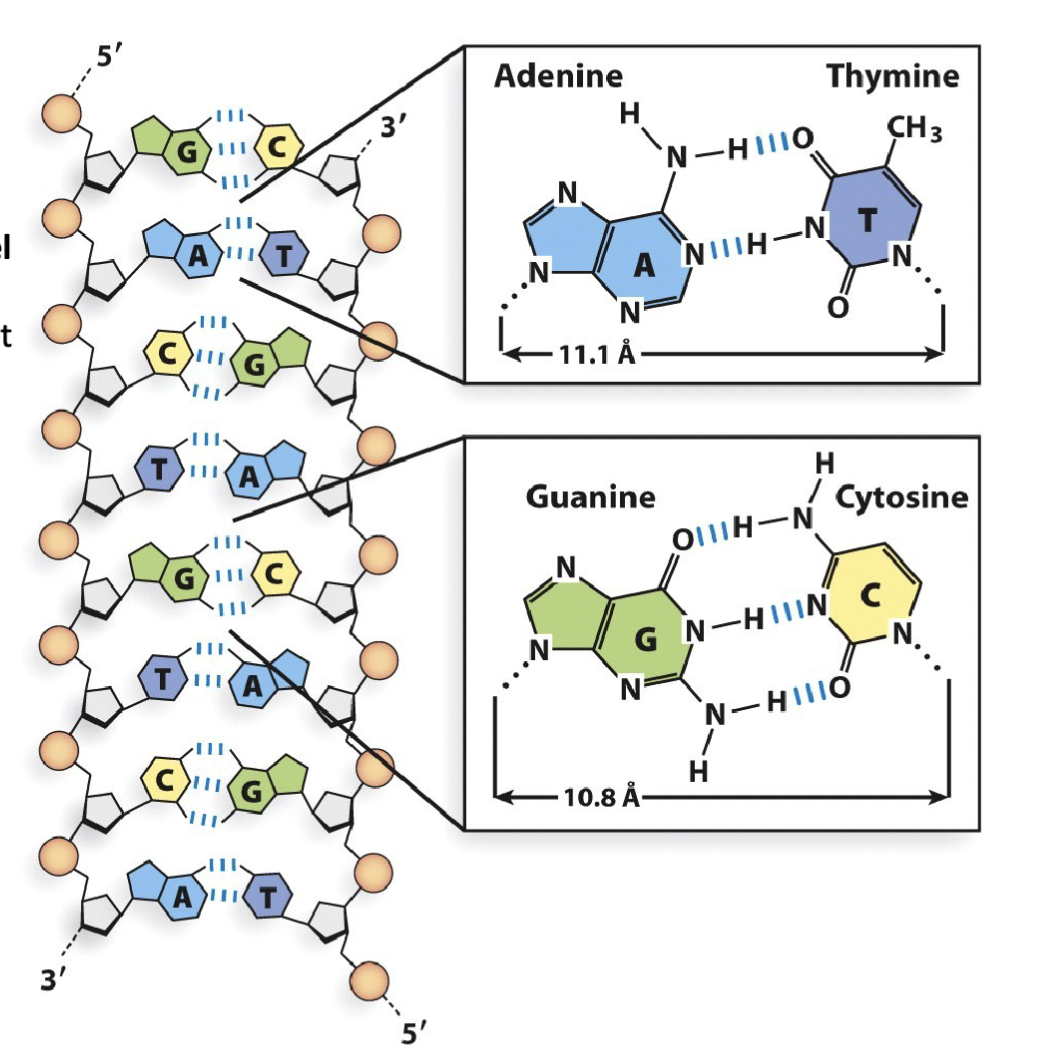
What are the features of the phosphate backbone in double helix DNA?
DNA is a double helix comprised of two single, unbranched nucleic acid polymers, referred to as “strands” of DNA.
These DNA strands are oriented in an anti-parallel arrangement, with the strand on the left running 5′-to-3′ top-to-bottom, and the strand on the right running 5′-to-3′ bottom-to-top.
Note: double-stranded RNA and DNA/RNA hybrids also have anti-parallel orientation
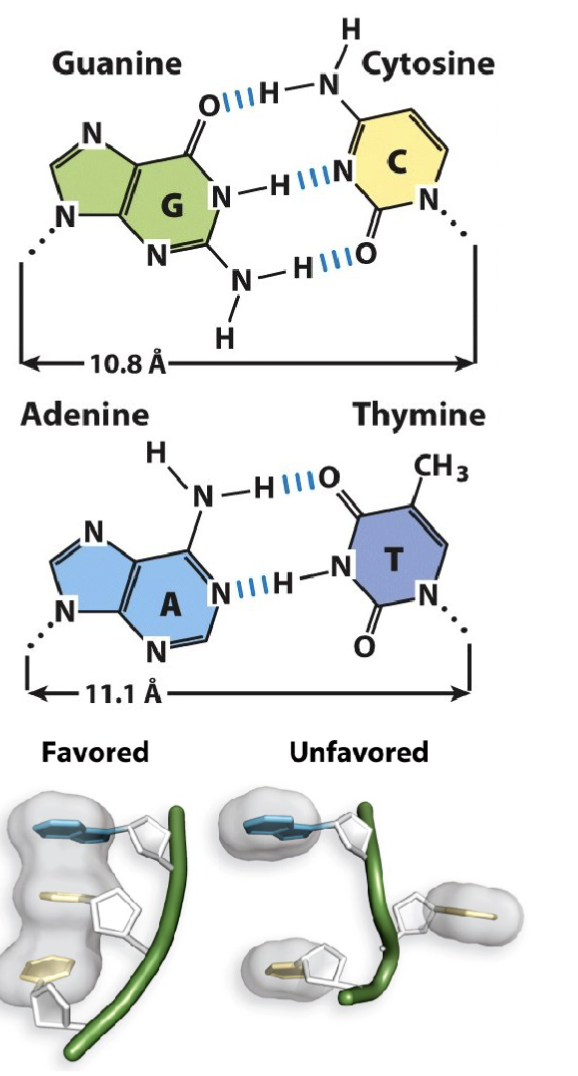
The DNA double helix has nearly-uniform width (10.8 – 11.1 Å) regardless of purine/pyrimidine pairing
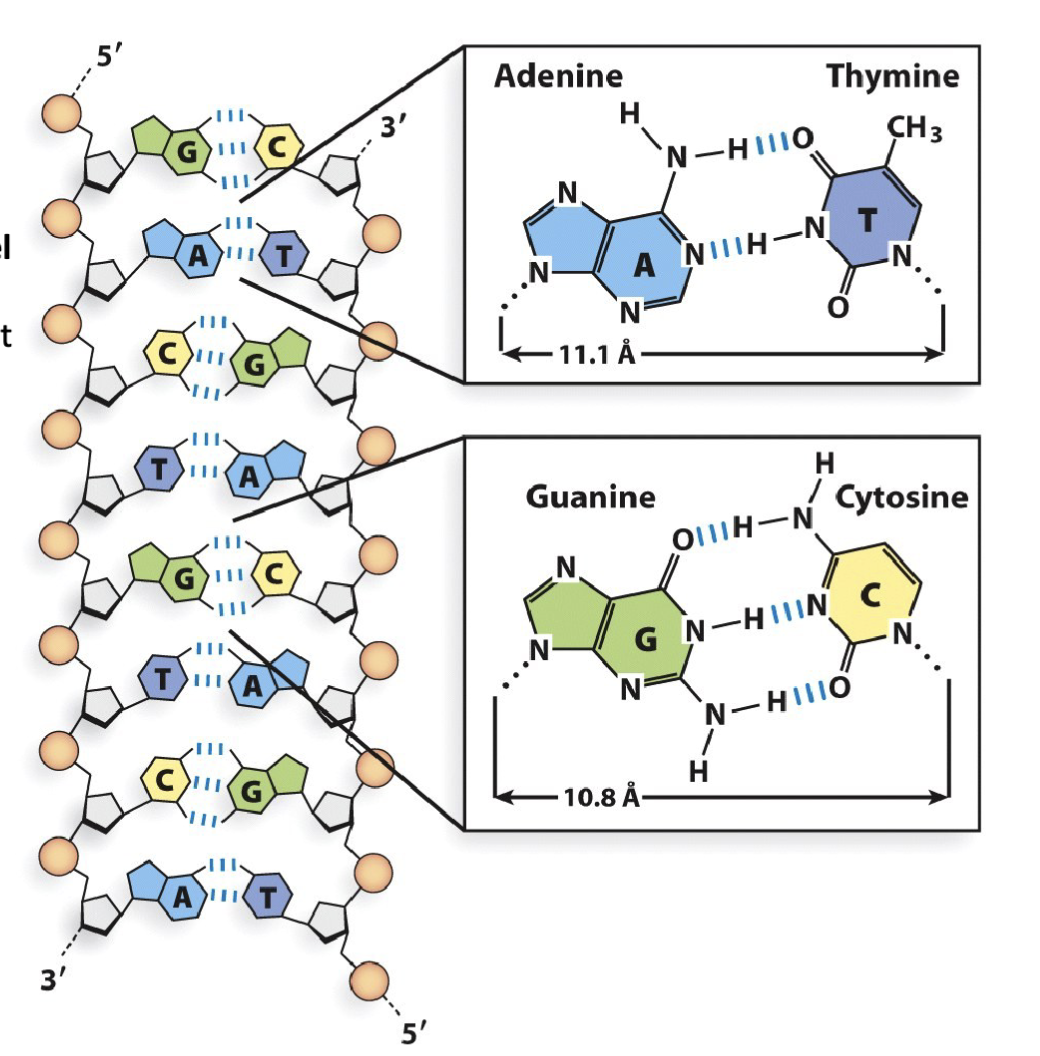
What are the features of nitrogenous base pairing?
Every instance of guanine (G) in a DNA strand is accompanied by a cytosine (C) in the anti-parallel counterpart DNA strand, and vice-versa
Every instance of adenine (A) in a DNA strand is accompanied by a thymine (T) in the anti-parallel counterpart DNA strand, and vice-versa
The structure adheres to Chargaff’s Rule
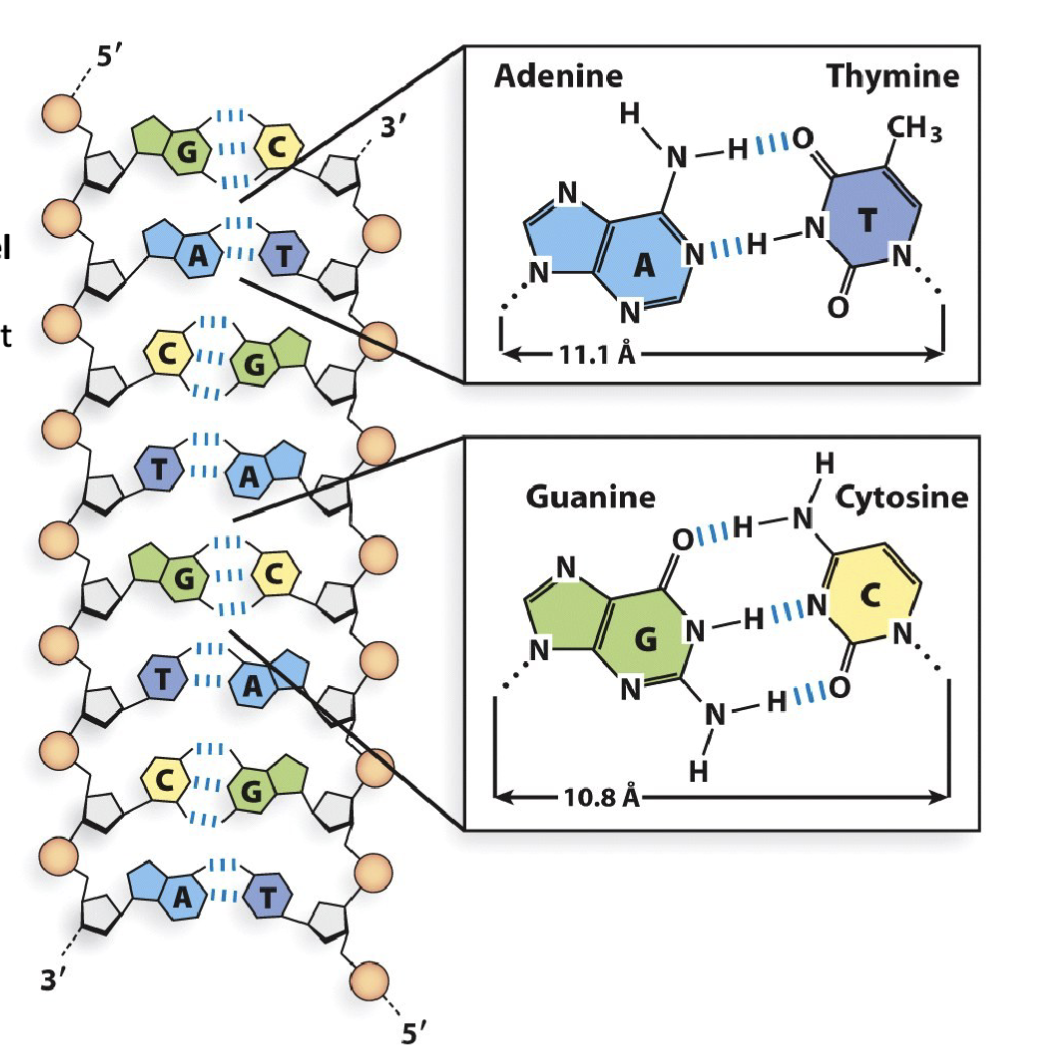
What are the features of Nitrogenous base molecular interactions in double helix DNA?
The nitrogenous base guanine (G) makes three hydrogen bond interactions with cytosine (C). The distance between their respective 1′-carbon is 10.8 Å
The nitrogenous base adenine (A) makes two hydrogen bond interactions with thymine (T). The distance between their respective 1′-carbon is 11.1 Å
Adjacent nucleotides in the same DNA strand share π-π stacking interactions (also referred to as base- stacking). These stacking interactions stabilize nitrogenous base orientation toward the inside of the double-stranded helix.
G-C have three bonds
A-T two bonds
one pi orbital above and below the sugar : creates electric field that allows for bases to stack upon each other
What are the three representations of double stranded DNA?
Ribbon representation depicts the phosphate-sugar backbone in blue and the nitrogenous bases as yellow bars
Stick representation illustrates the bonds between atoms, with carbons in grey, oxygen in red, nitrogen in blue, and phosphorous in yellow (or purple)
Space filling representation shows the proportional volume of atoms with the same corresponding colours in stick representation
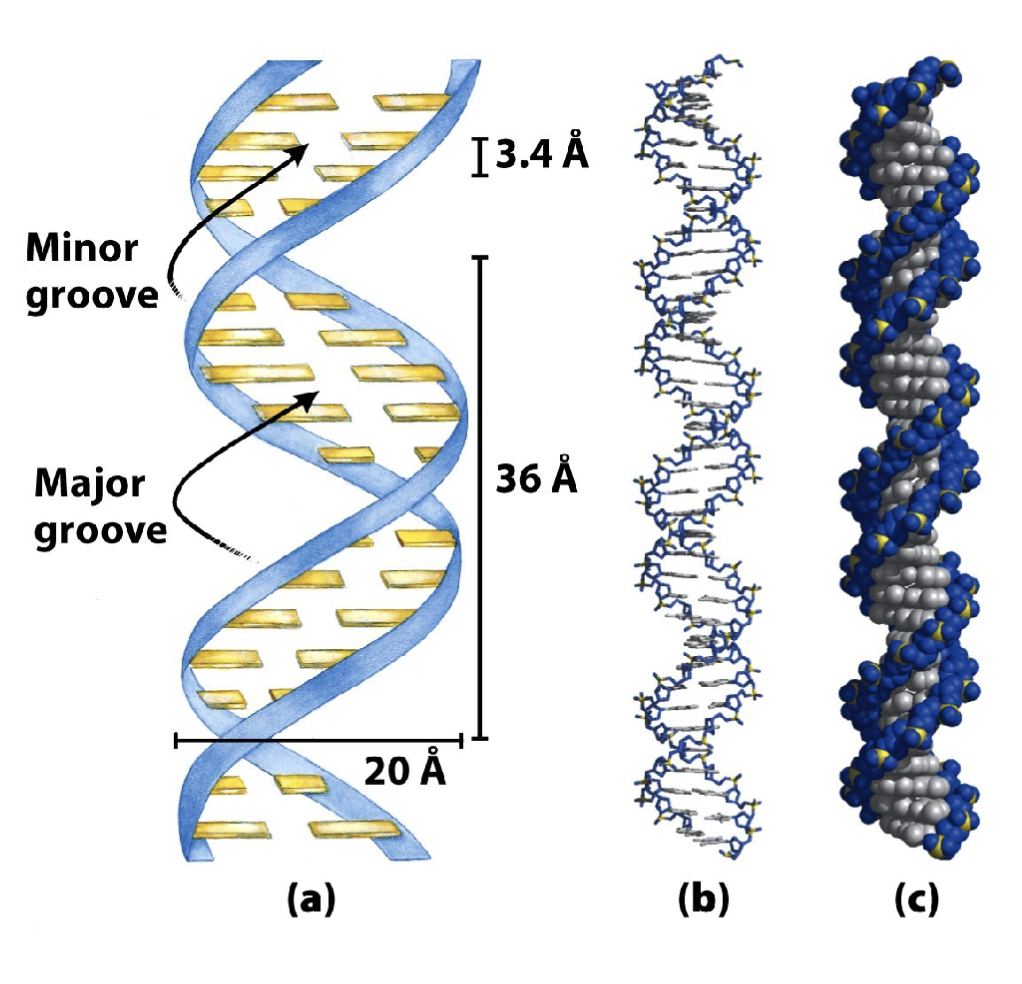
What are the features of the B-form of DNA?
The B-form of DNA (depicted here) has a right- handed helical turn following the direction (5′→3′) of each strand
Adjacent nitrogenous bases are spaced at intervals of 3.4 Å distance along the vertical axis (ideal for base pairing)
Each complete turn of the helix has 10.5 bases spanning 36 Å distance along the vertical axis
There are two grooves (major and minor) in each helical turn
The most common form of DNA under normal conditions (salt and
humidity)
Has a right-handed helical turn
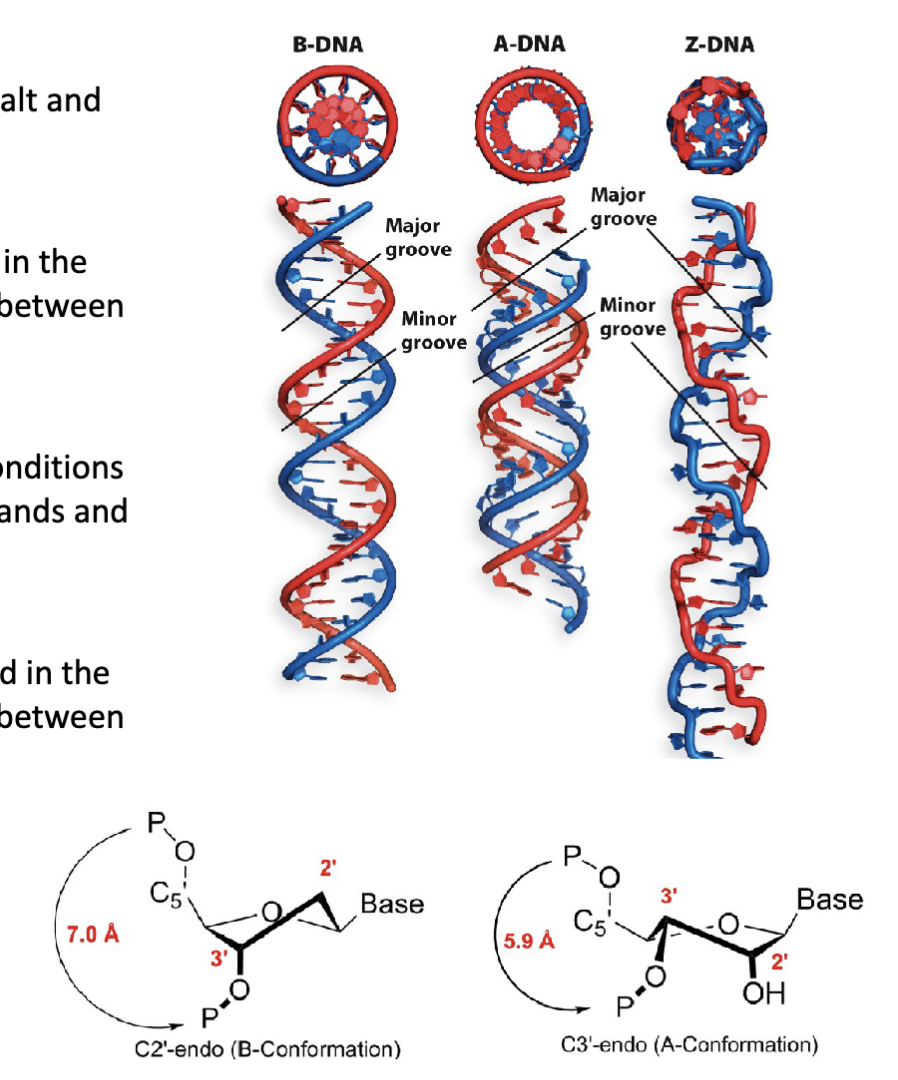
What are the three structural conformations (forms) of DNA?
B-form DNA conformation (B-DNA)
A-form DNA conformation (A-DNA)
Z-form DNA conformation (Z-DNA)
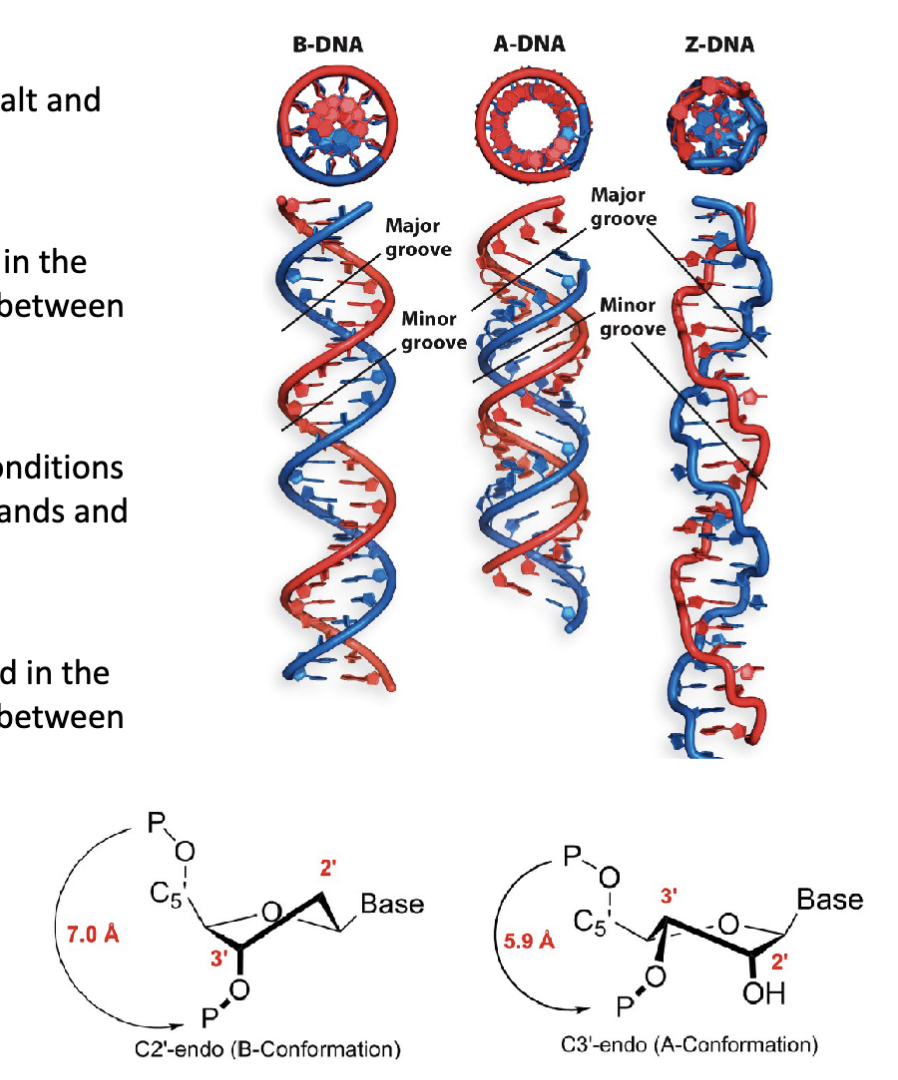
How does the endo pucker affect the B-form of DNA?
The pucker of the backbone β-d-deoxyribose sugar is found in the
C2′-endo conformation. This affects the backbone distance between
adjacent nucleotides.
What are the features of the A-form of DNA?
The “length-compacted” conformation under alternative conditions
(high salt and low humidity). Present in DNA/RNA hybrid strands and
the active site of DNA polymerase.
Has a right-handed helical turn
The pucker of the backbone β-d-(deoxy)ribose sugar is found in the
C3′-endo conformation. This affects the backbone distance between
adjacent nucleotides
A squishes the distance between the phosphates
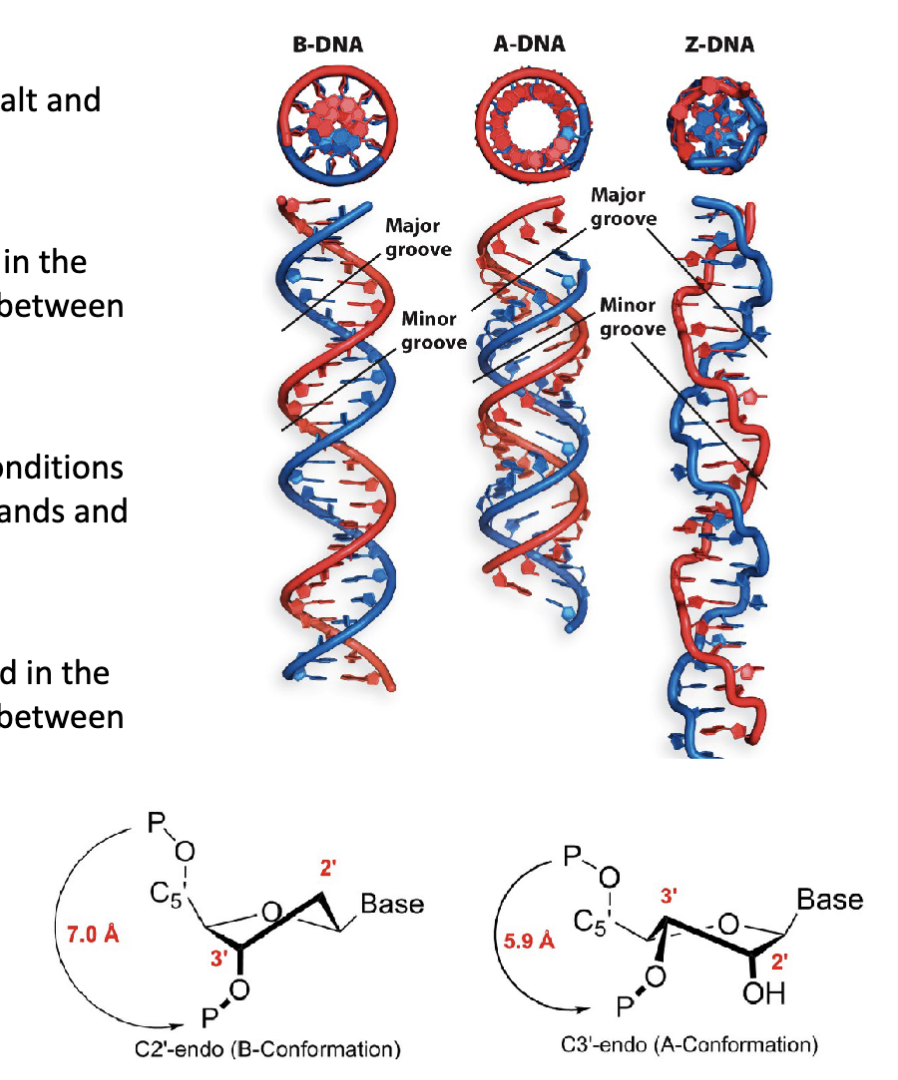
What are the features of the z form of DNA?
Formed from DNA sequences with alternating purines and pyrimidines (e.g., 5′−GCGCGCGCGC−3′)
Has a left-handed helical turn
What is the molecular structure of DNA and how does this explain its function?
DNA is double stranded
Crick recognizes that the structure of DNA confers the ability for DNA to serve as its own template
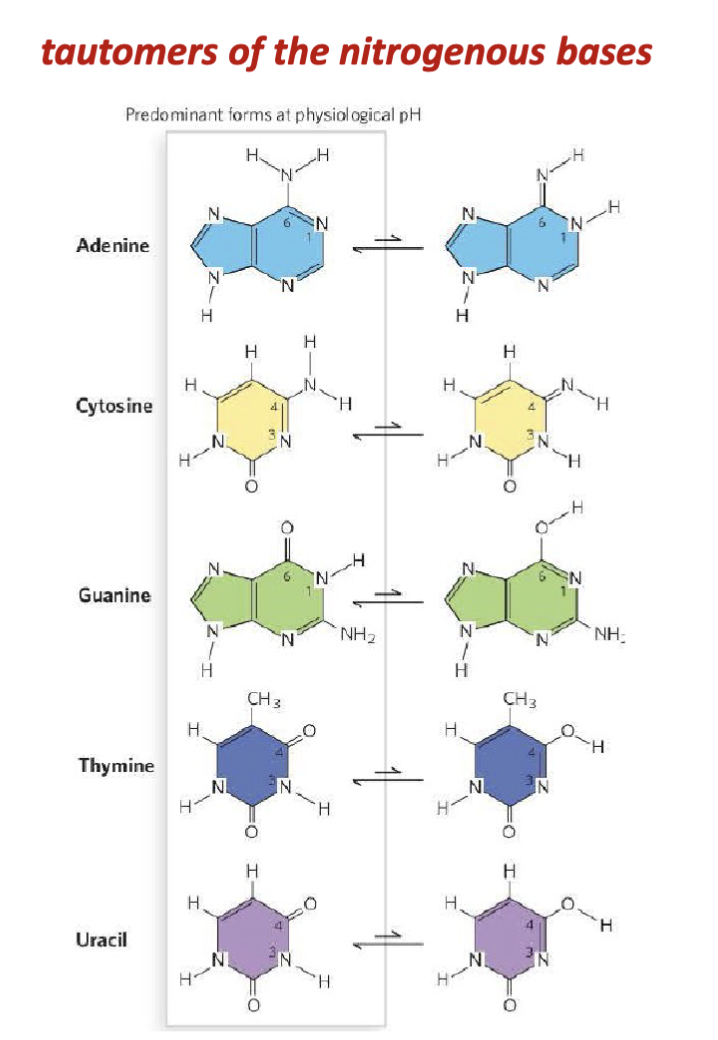
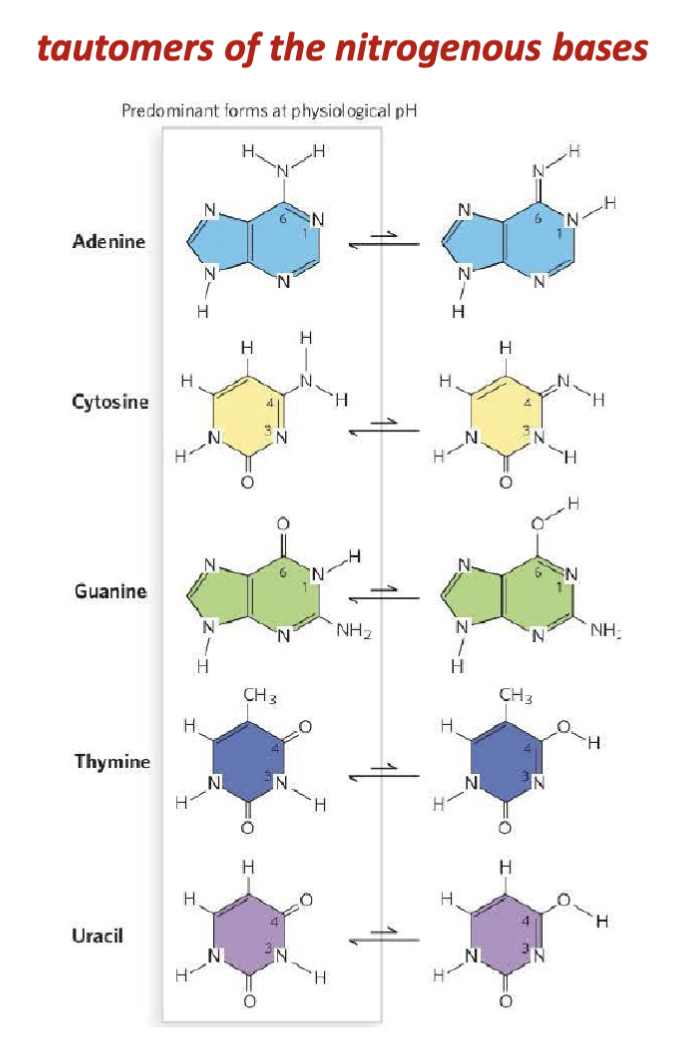
What are the features of the tautomers of nucleic acids?
When they were characterizing the nucleic acid bases (Kossel, Levene, Jacobs), they would be extracting the tautomers and not the common form in the body.
tautomers: lone pairs can abstract one of the protons on the adjacent amino group
shifting of a hydrogen atom and a double bond within the base’s ring system. This is a type of proton transfer + resonance rearrangement.
What are the features of Watson and Crick’s proposed model of DNA?
Their model is consistent with multiple experimental observations:
1: the nucleic acid strands are linear polymers of nucleotides joined by phosphodiester bonds
2: the nitrogenous bases have molecular interactions (purine and pyrimidine H-bonded pairs) that are consistent with Chargaff’s Rule
(equal proportions of A:T and G:C)
3:the double helix fits x-ray diffraction data
4: complimentary DNA strands can serve as template
x-ray data: two repeating patterns, repeating helical turns, diffraction data could not make a physical structure
How does the model structure for DNA predict is replication process?
Semi-conservative one strand copies of the other to ensure a complete replication is achieved of both halves of the helix.
If the structural model developed by Watson and Crick is accurate, it is expected that DNA replication should be semiconservative
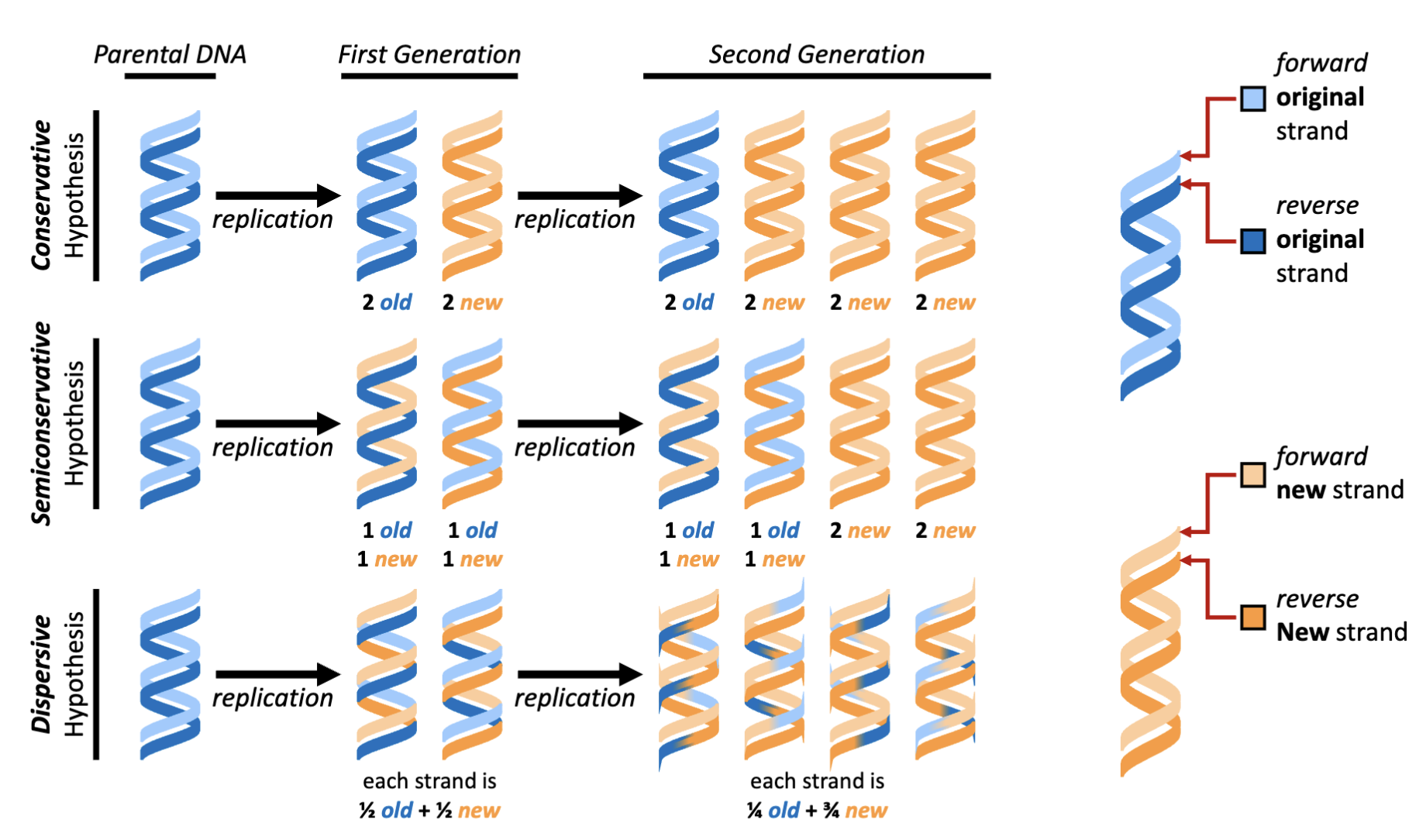
What are the hypotheses of DNA replication models?
Conservative, semi-conservative (works with the Watson and Crick model of DNA), dispersive (new DNA is incorporated piece wise)
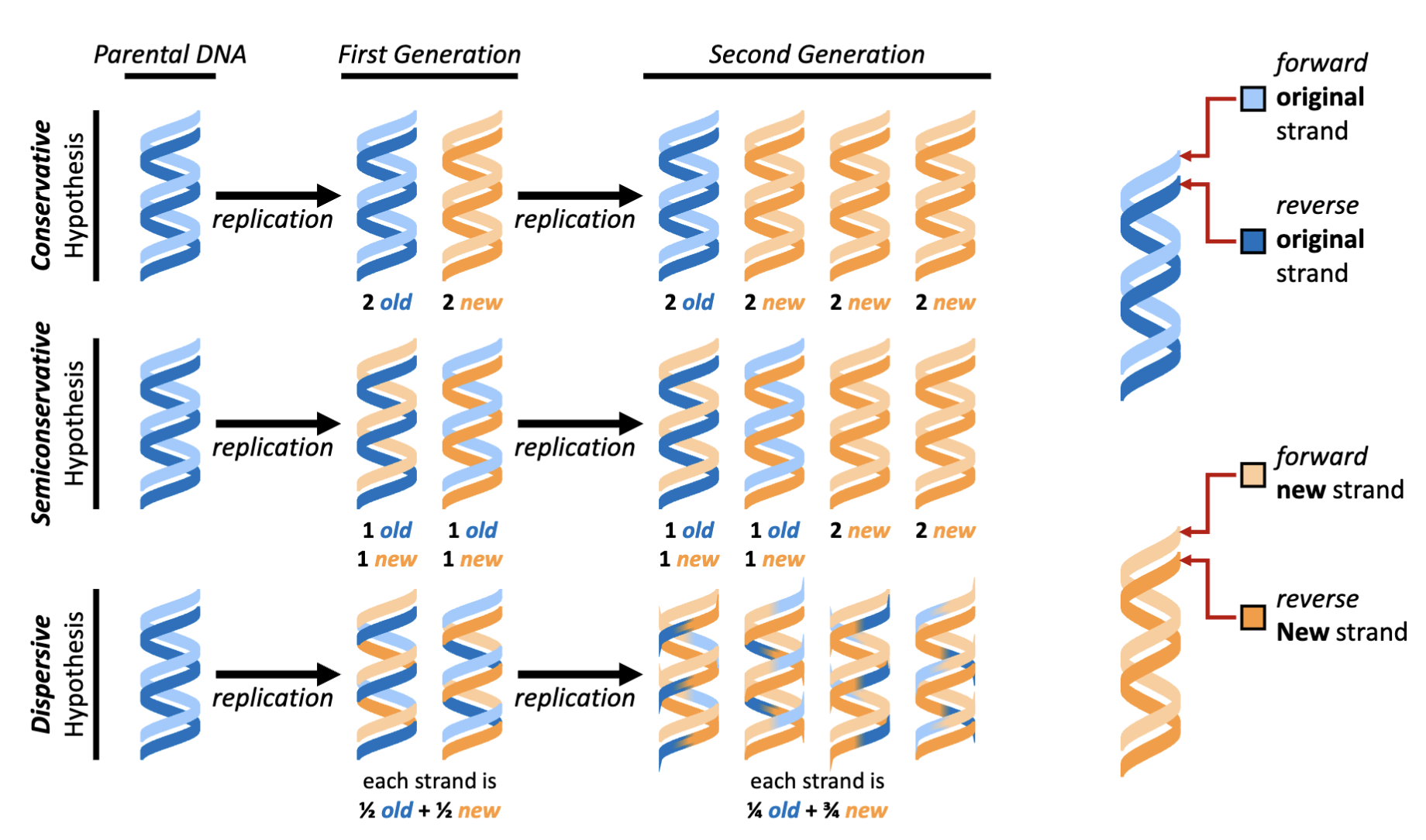
How was the hypothesis of DNA replication tested?
Meselson and Stahl design a simple test of this hypothesis, called “the beautiful experiment”
Meselson and Stahl grow Escherichia coli in a medium containing 15NH4Cl as the sole nitrogen source.
14N – the abundant isotope typically in DNA (light)
15N – the isotope for labelling DNA experiment (heavy)
DNA is 17% nitrogen resulting in 15N labelled DNA having a 1.2% increase in mass relative to 14N labelled DNA.
This mass difference can be separated by centrifugation using a cesium chloride (CsCl) density gradient
take parental bacteria allow it to divide with N14 source will result in the production of new DNA of components that are lighter
perform acid isolation of nucleic acids (last class)
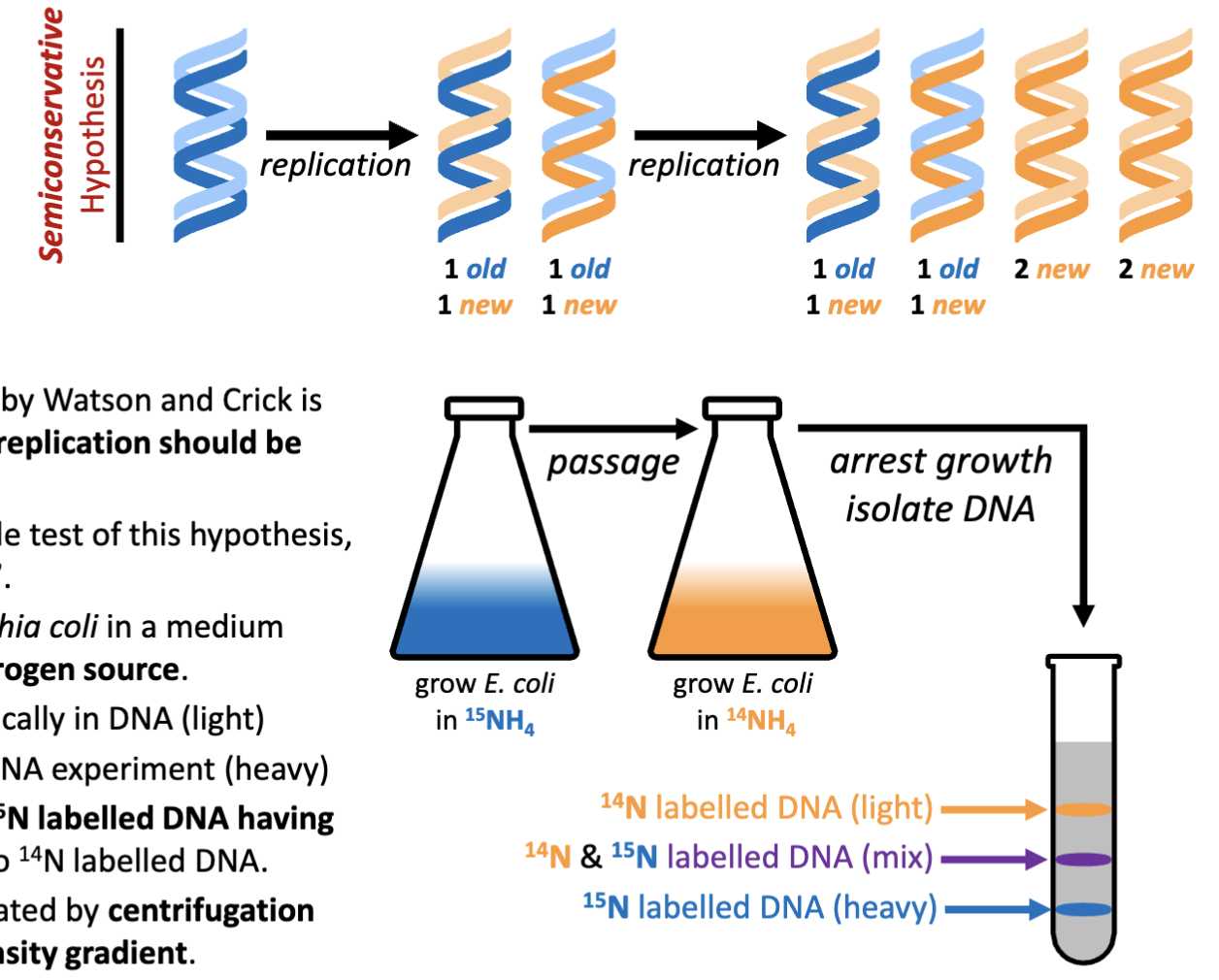
What would be the results of the “beautiful experiment” if DNA was replicated by all three hypothesized methods?
Semi-conservative was the outcome observed
native gradient: DNA is allowed to form double helix
alkaline: separate the two chains
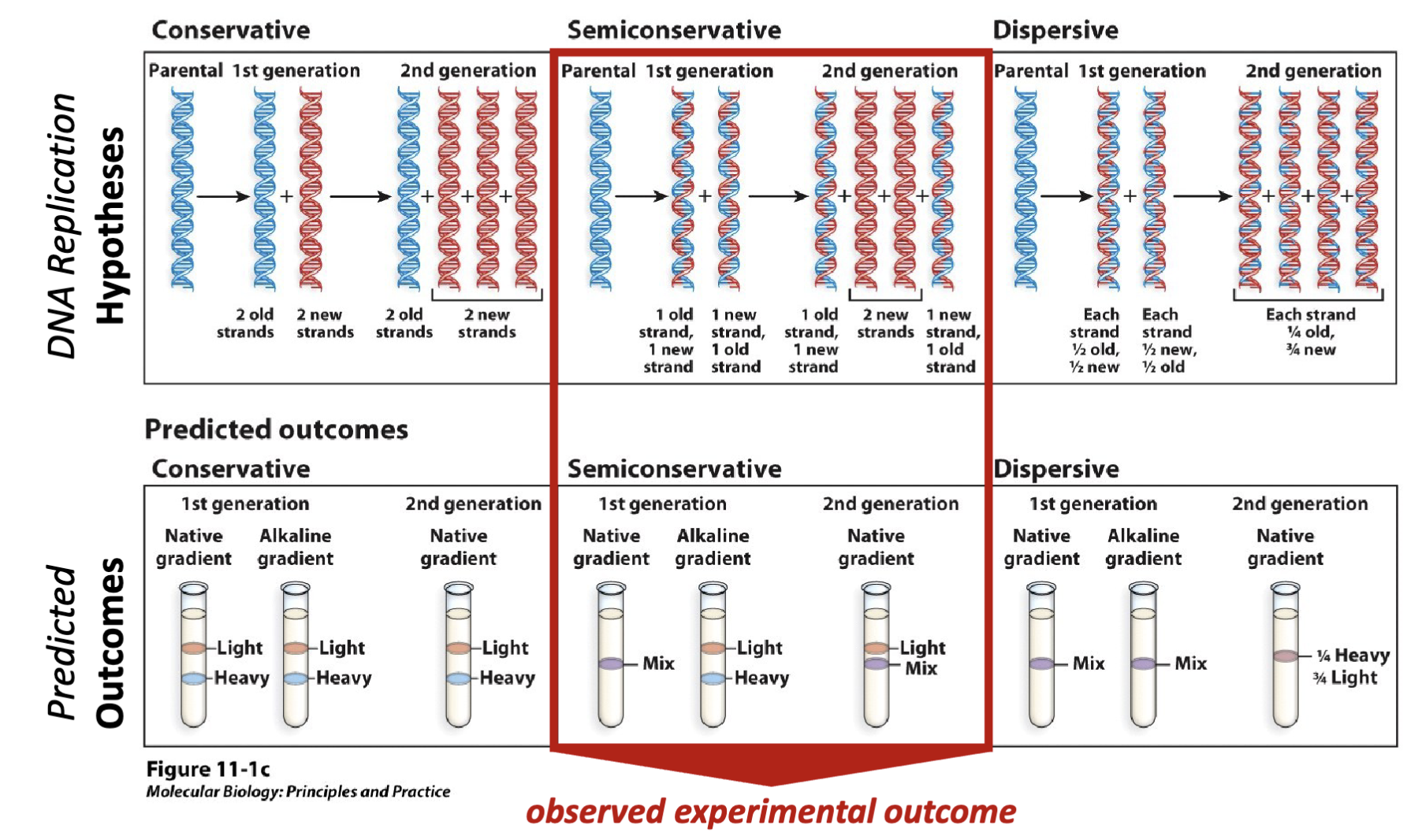
What were the results of the beautiful experiment?
First generation experimental results- Centrifugation with an alkaline (i.e., single-stranded DNA) CsCl density gradient yields a 50:50 mixture of light and heavy DNA.
Second generation experimental results- Centrifugation with a native (i.e., double stranded DNA) CsCl density gradient yields a 50:50 mixture of light and mixed (light + heavy) DNA
Experimental conclusion- Meselson and Stahl demonstrate that the structural model developed by Watson and Crick is consistent with the
observation that DNA replication is semiconservative
alkaline/ denaturing conditions: equal portions of heavy and light in first generation
natured condition: duplex condition- two parts of old and new and two parts of new DNA
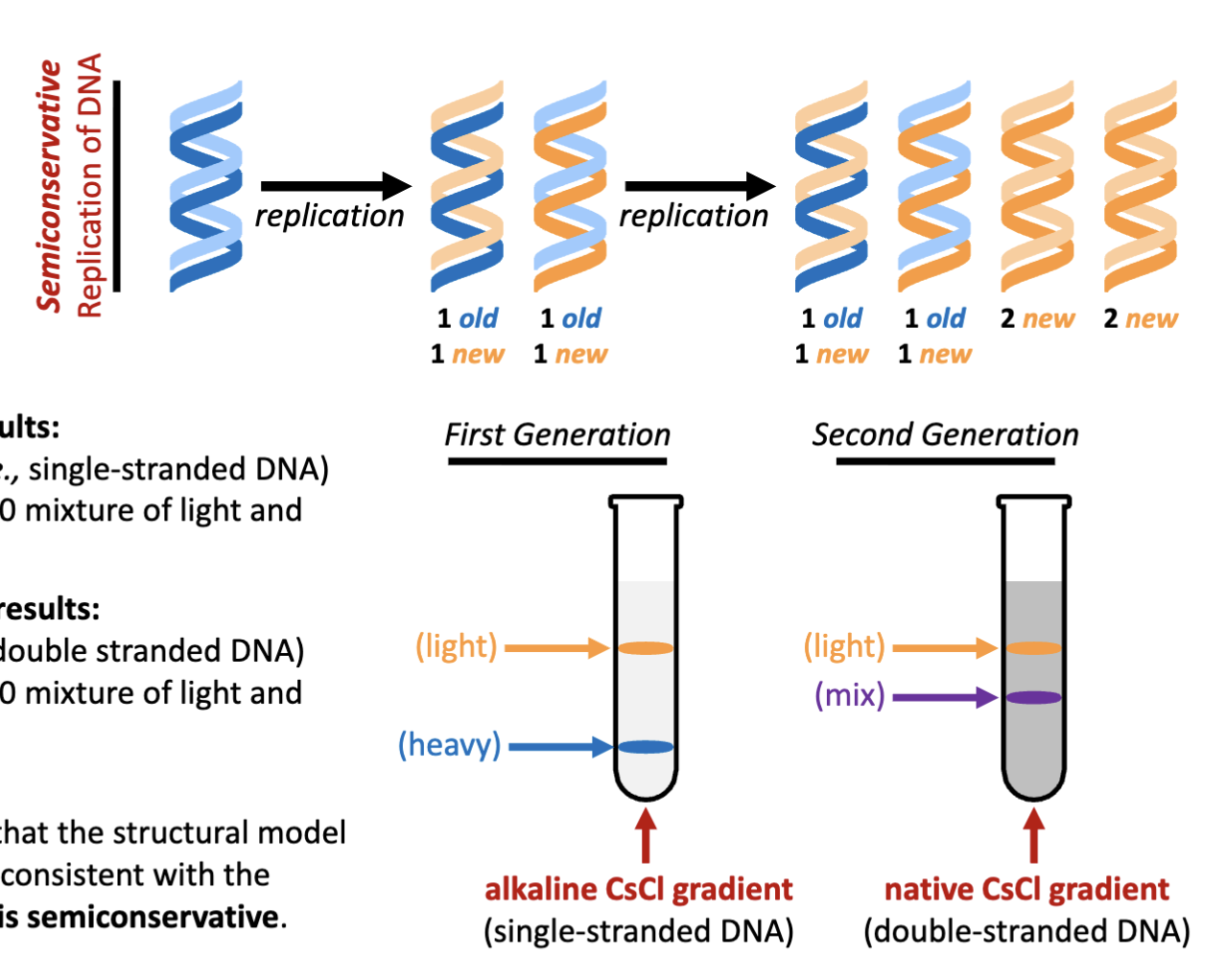
How was the replication of DNA first viewed? How was this done?
autoradiography and electron microscopy
rectangles are radiolabelled,
wash it with emulsion of silver halide (left to sit on the sample)
the radiation (phosphorus) from the sample interacts with silver releasing Ag+ cations
only silver metal will condense from the emulsion,
wash away excess and only left with biological sample with silver metal on it
electron microscope: visualize silver metal
follows the semi conservative hypothesis
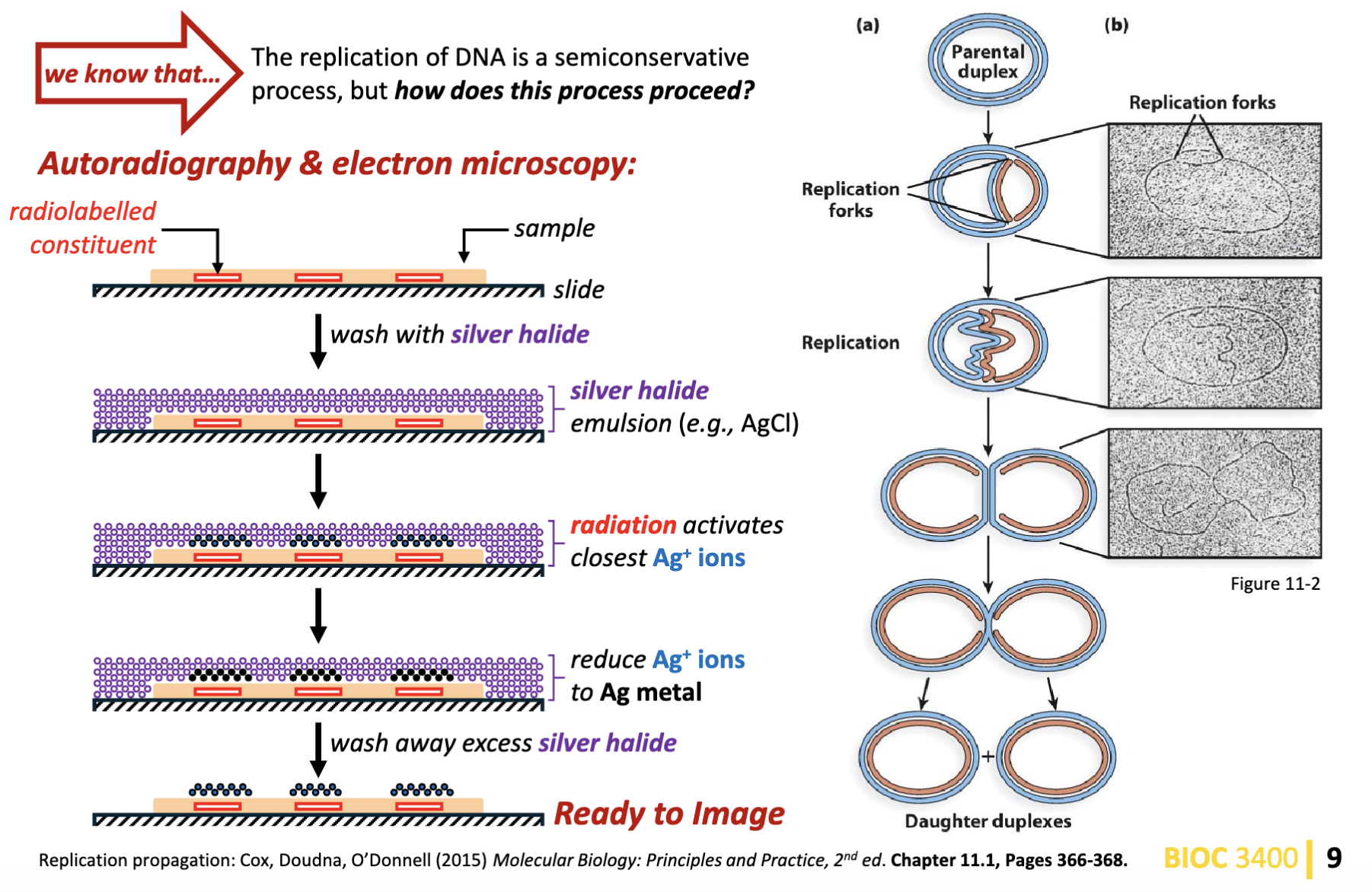
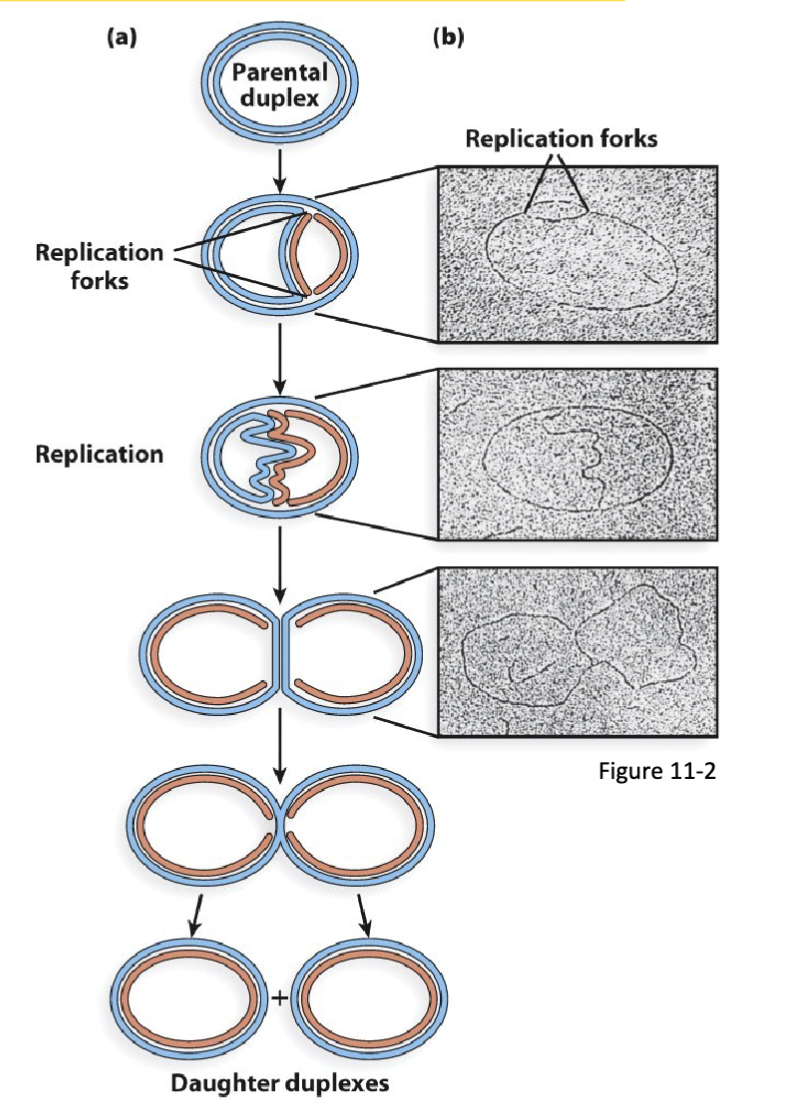
What is the process of circular DNA replication? How is the process of semiconservative DNA replication propagated?
Autoradiography & electron microscopy of circular DNA with one replication origin:
circular DNA (e.g., bacterial genomes, bacterial plasmids, viral DNA, mitochondria and chloroplast DNA) has one origin of replication from which replication is initiated
bidirectional replication of the parental duplex DNA proceeds from the origin of replication
this results in the formation of two replication folks moving in opposite directions (one clockwise, the other counterclockwise)
DNA replication terminates when the two replication folks meet on the plasmid location opposite to the origin of replication
replication of circular plasmid DNA is continuous
in rare cases, DNA replication is unidirectional
can have circular bundles of DNA in human body
in the replication fork goes up one side and down in one other
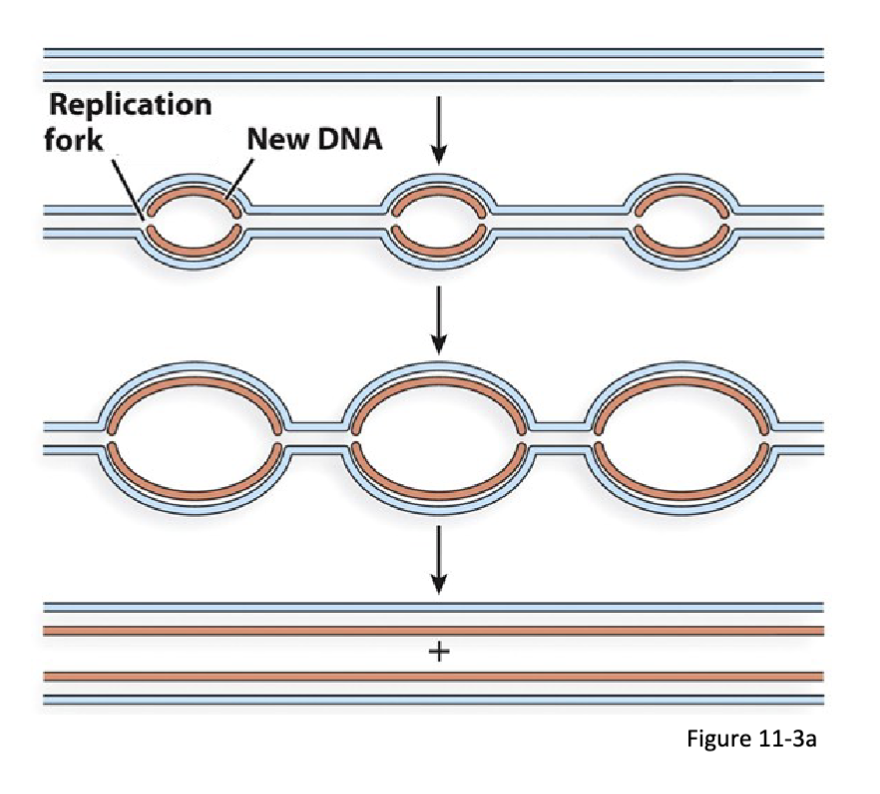
What is the process of the replication of linear DNA?
Multiple replication origins:
typical of eukaryotic nuclear chromosomes
bidirectional replication at multiple sites starting from each origin
this results in the formation of two replication folks moving in opposite directions
pairs of neighbouring replication folks move toward each other and terminate once they meet
replication of plasmid DNA is semidiscontinuous
Eurkaryotic replications folks are considerably
slower than those of bacteria (hence the evolution
of multiple origins of replication)
starts in multiple sites and origins= formation of two forks heading in opposite directions (will not meet up again)
semidiscontinuous: DNA only goes in one direction, one of the strands needs to go backwards (section by section)
Is DNA unwound before replicated in either circular or linear DNA?
in both circular and linear DNA replication processes, the DNA is not
completely unwound before it is replicated
In what directions is DNA replicated in?
5’ to 3’ end
What happens when unwinding DNA?
It causes torsional strain
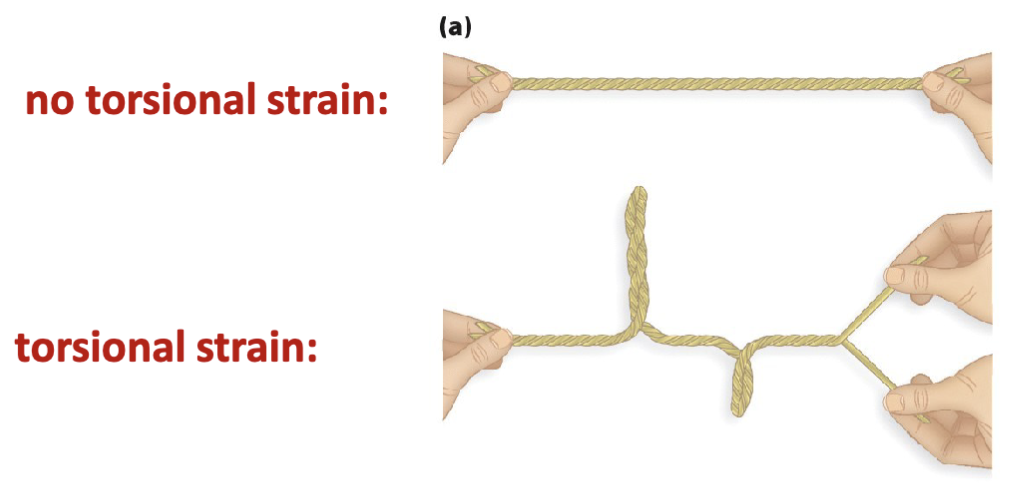
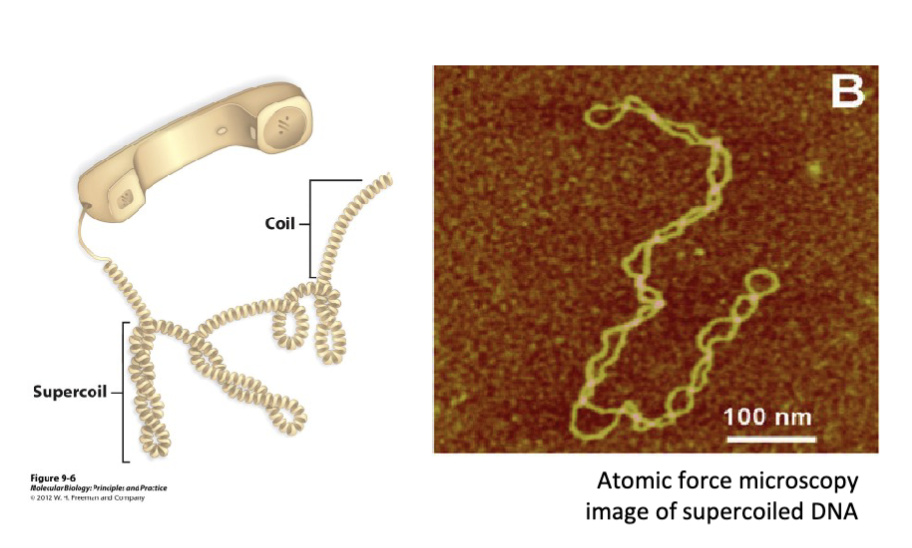
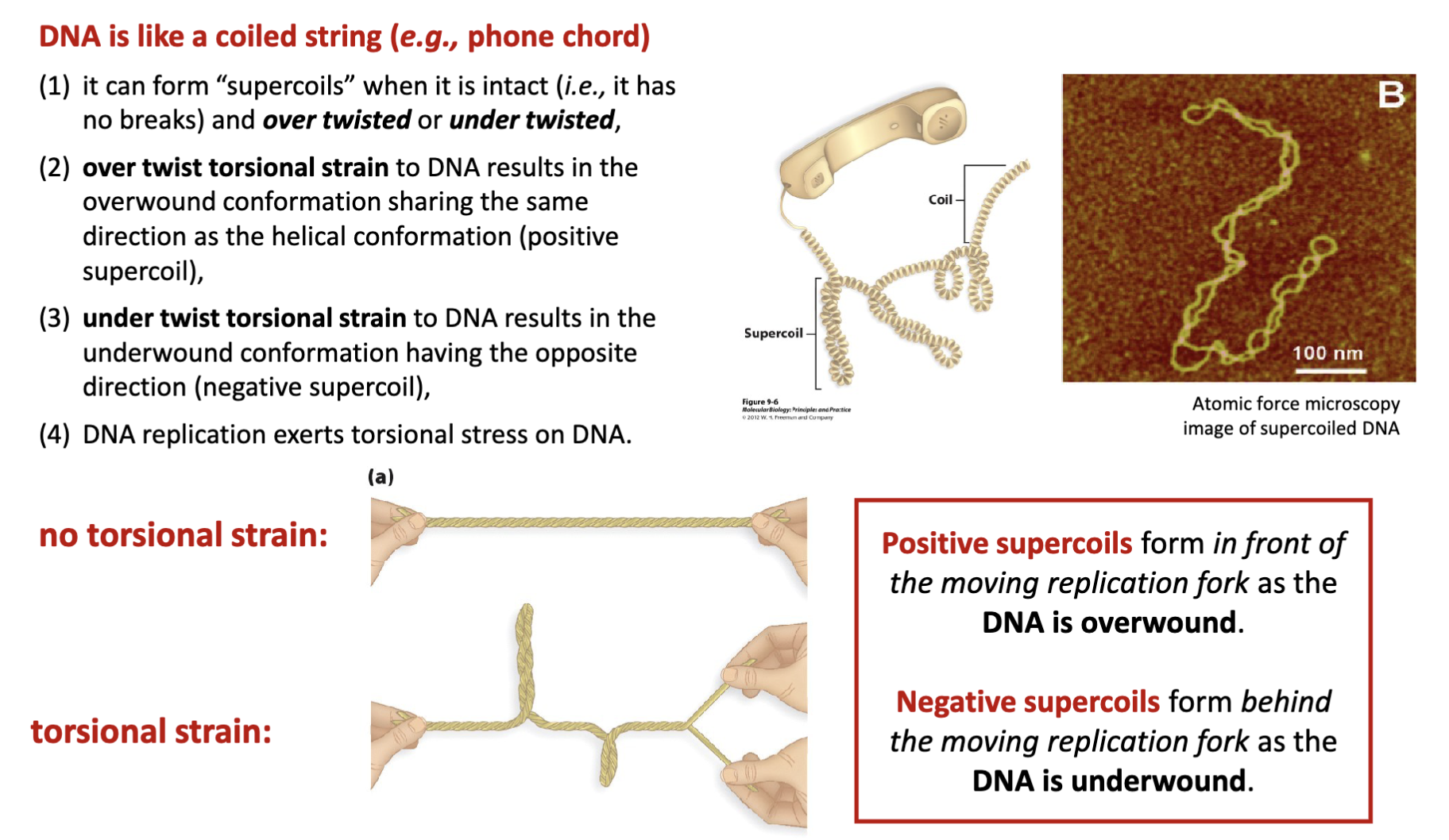
How is DNA like a coiled string (e.g., phone chord)?
it can form “supercoils” when it is intact (i.e., it has no breaks) and over twisted or under twisted
over twist torsional strain to DNA results in the overwound conformation sharing the same direction as the helical conformation (positive supercoil)
under twist torsional strain to DNA results in the underwound conformation having the opposite direction (negative supercoil)
DNA replication exerts torsional stress on DNA
What are negative and positive supercoils?
Positive supercoils form in front of the moving replication fork as the
DNA is overwound
Negative supercoils form behind the moving replication fork as the
DNA is underwound
over twisted (positive strain), under twist (negative strain)
over twist B DNA form supercoil loops, if under twist try to unravel that would be
B form DNA right hand helix, DNA infront of replication is positively strained
DNA being under strained is after the replication (or inside the replication bubble) creates left handed supercoils
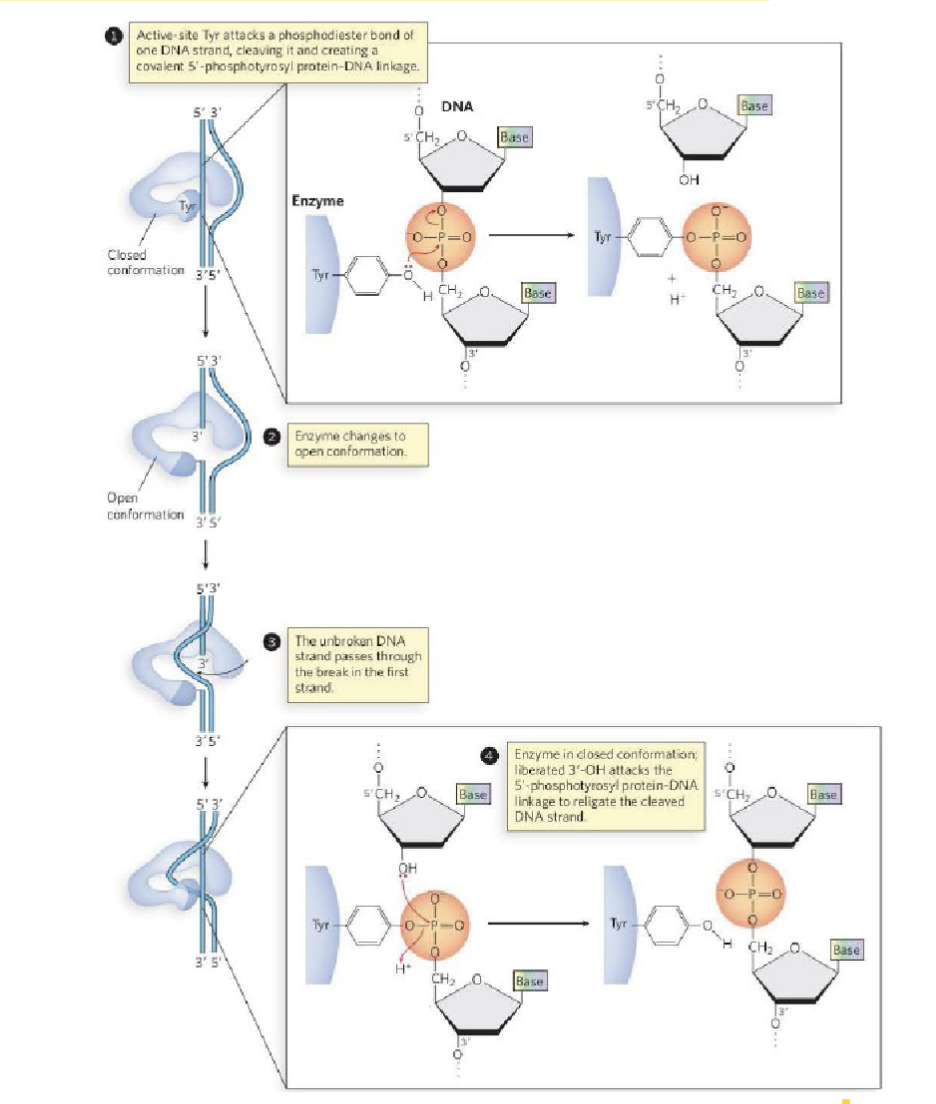
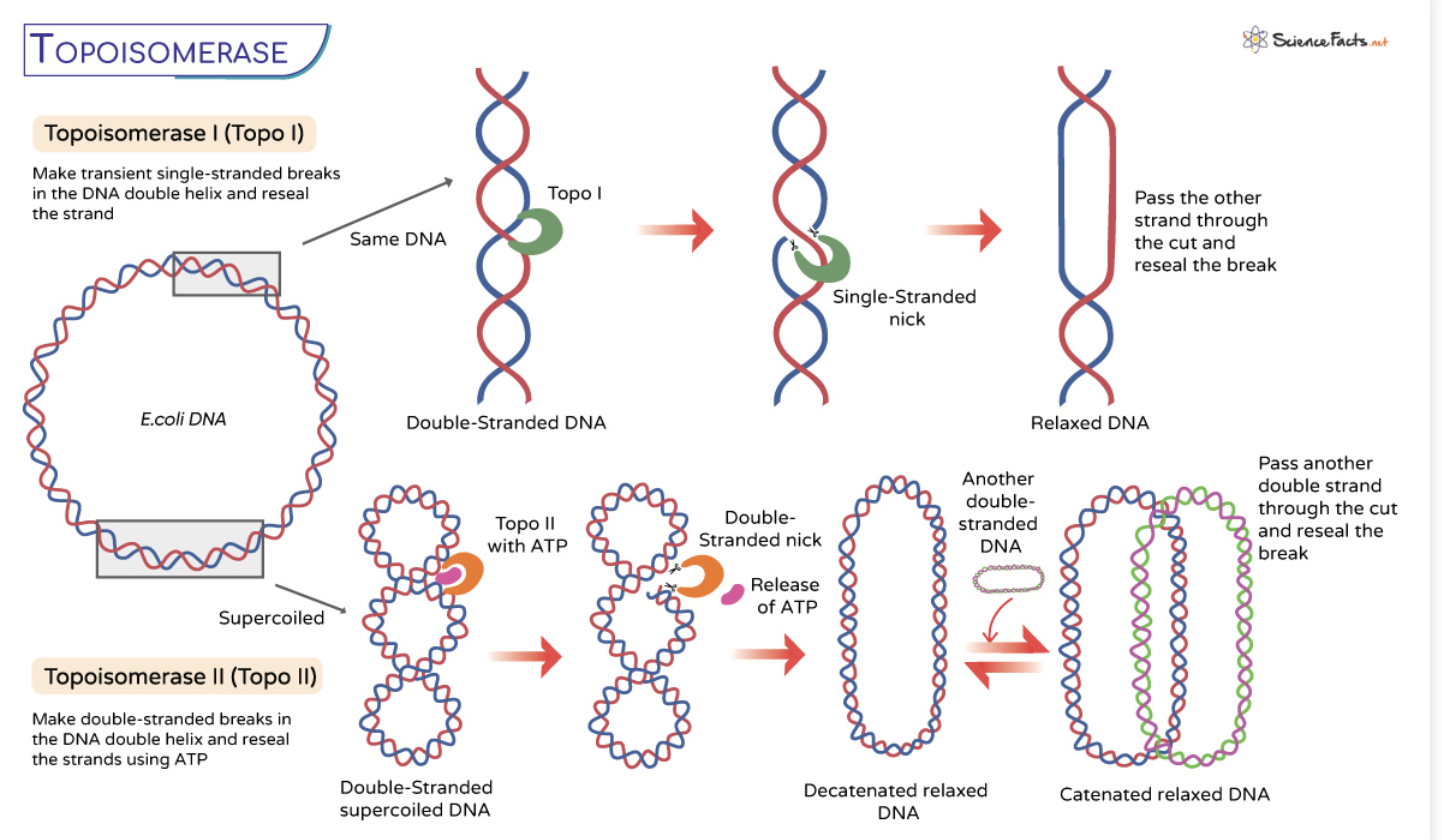
What is the element that alleviates torsional strain?
Topoisomerase I
topoisomerase is recruited to sites of DNA strain
the enzyme adopts a closed conformation where it then catalyzes the cleavage of one DNA strand
this forms a protein-DNA covalent bond
the enzyme then adopts an open conformation
this permits the unbroken, second DNA strand to pass through the break of the first, broken DNA strand (covalently bonded to topoisomerase)
topoisomerase then readopts its closed conformation to rejoin the broken, first DNA strand
thereby alleviating the torsional strain of the replication bubble
topoisomerase goes from closed to open configuration that then allows the enzyme to thread the torsional strain it cuts it and undoes the loop and the brings the DNA back together (reverse) forms the phosphodiester bond but without the torsional strain the replication bubble previously had
this happens multiple times (when nucleotides are being made 5’ to 3’)
What does topoisomerase II do?
cut both DNA strands
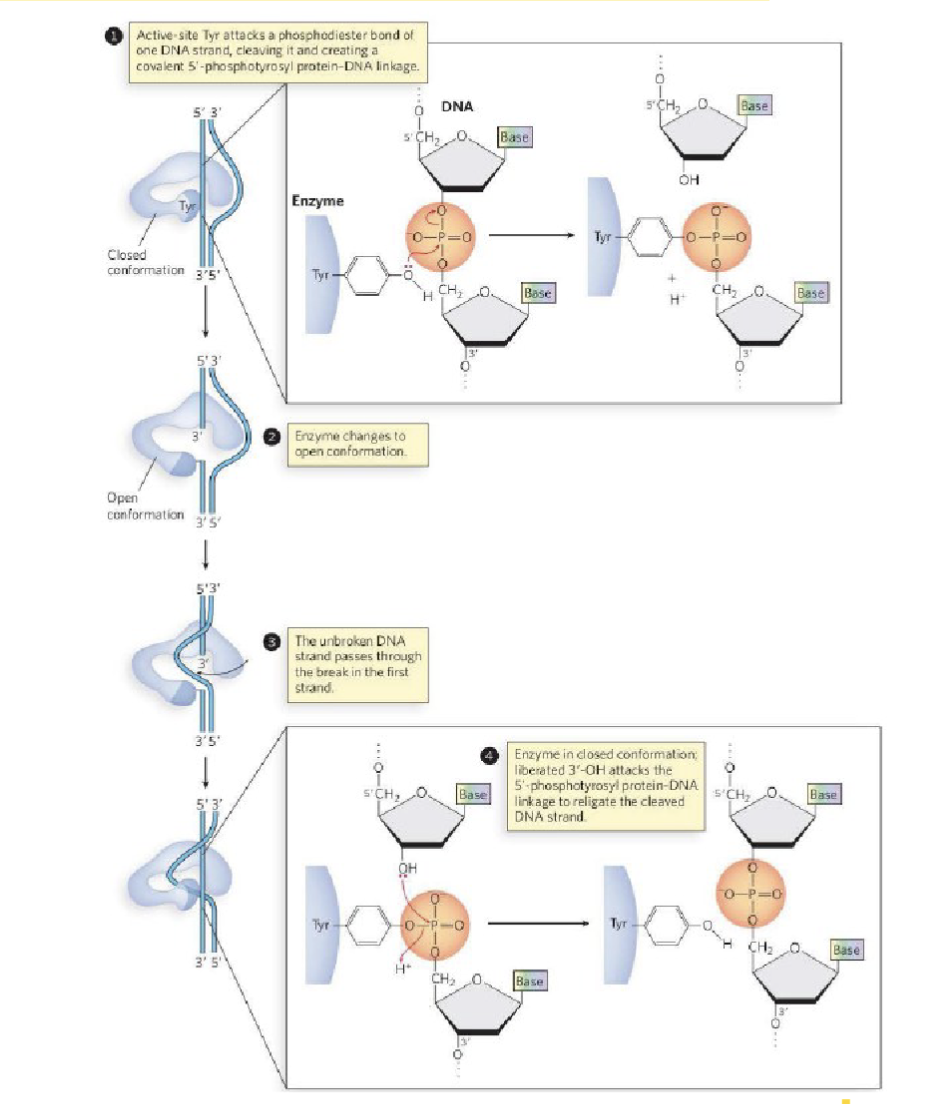
What are the details of the bond that topoisomerase breaks?
involves the hydrolysis of the nucleic acid backbone
D form sugar (alcohol pointing down)
phosphodiester bond is at the 5’ carbon (upstream)
phosphate is an electrophile
tyrosine-OH (base would come along and give an H ) → tyrosine-O- the charge will cleave the phosphodiester bond and cleave it on the 3’ alcohol : makes protein covalent linkage where topoisomerase is bonded to the 3’ end of the nucleic acid
broken strand left open, deoxyribose sugar( 3’ O-, picks up the proton from base)
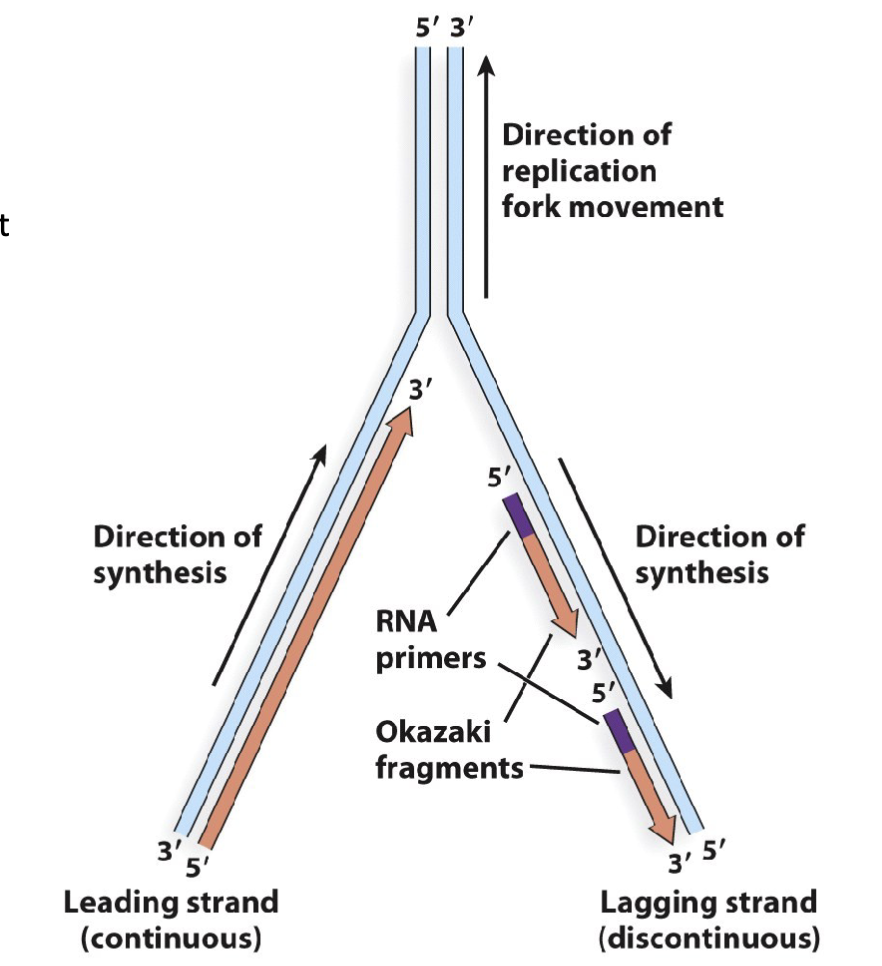
What are the features of Semidiscontinuous replication: DNA is synthesized in a 5′ to 3′ direction?
Linear DNA replication is semidiscontinous:
replication of linear DNA involves the formation of a replication bubble with two replication forks
the parent DNA strand is separated from its compliment
stranddaughter DNA strands are synthesized by DNA polymerase in a 5′ to 3′ direction
this produces a leading strand consisting of continuous
DNA daughter strand and a lagging strand consisting of discontinuous segments of daughter DNA (referred to as Okazaki fragments)this replication behaviour is distinguished from circular DNA replication in that it is semidiscontinous
linear DNA where both strands are copied at the same time has one continuous and multiple discontinuous
daughter strand (newly made)