week 7: pedigrees and complicating factors
1/43
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
44 Terms
how are autosomal dominant disorders recognized in pedigrees?
what is achondroplasia?
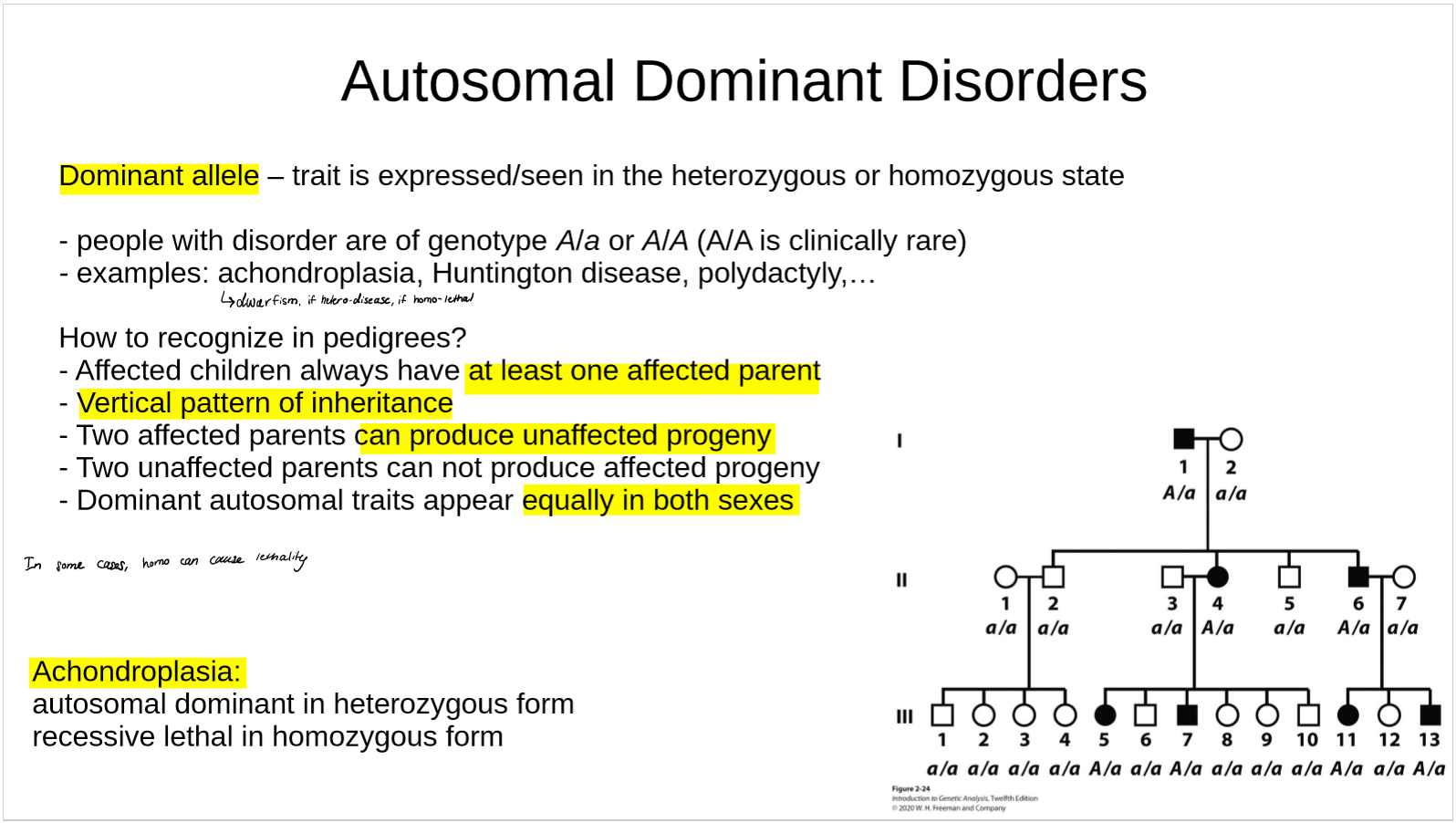
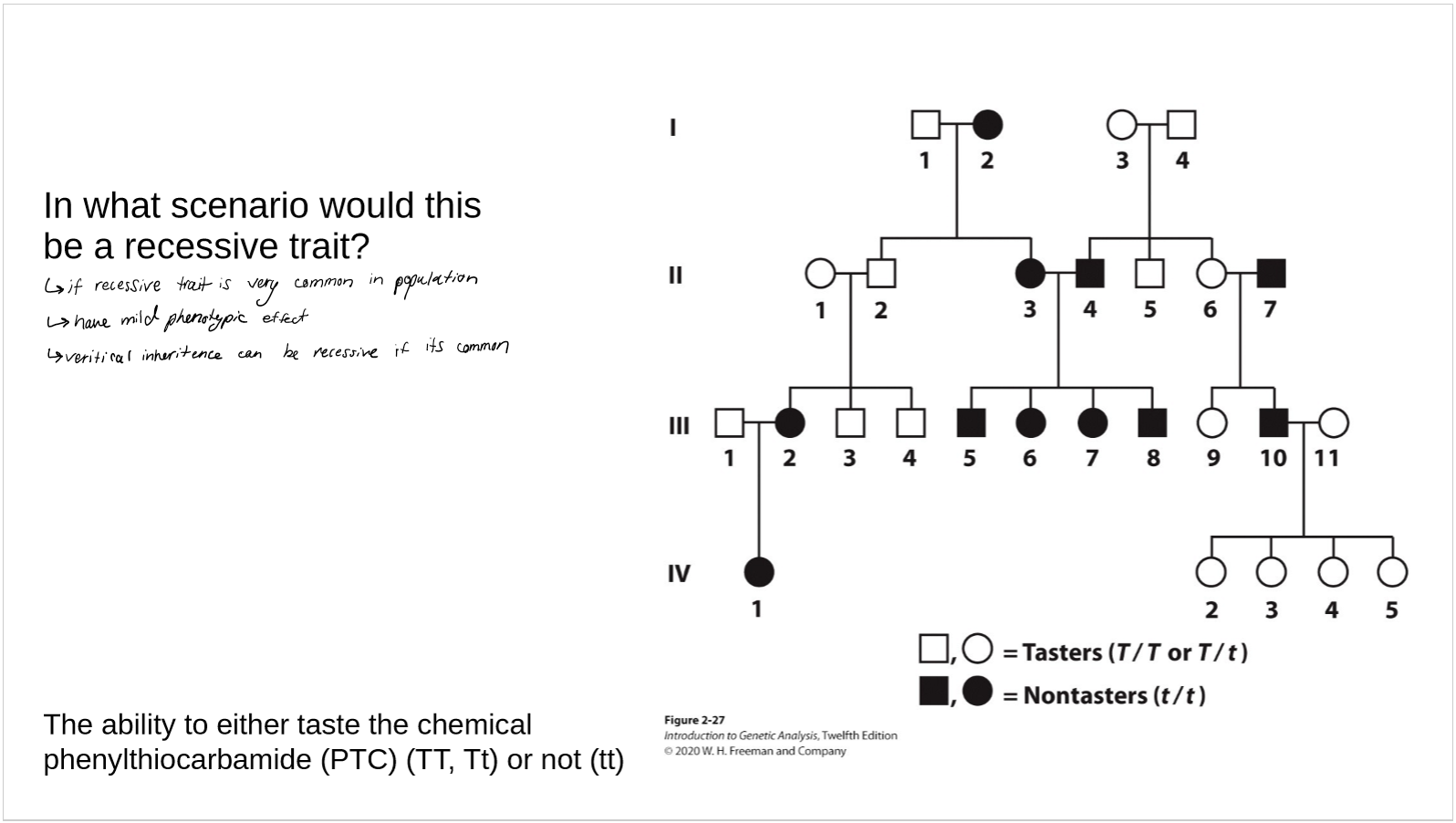
when can vertical inheritance be recessive (rather than the usual case of dominance)?
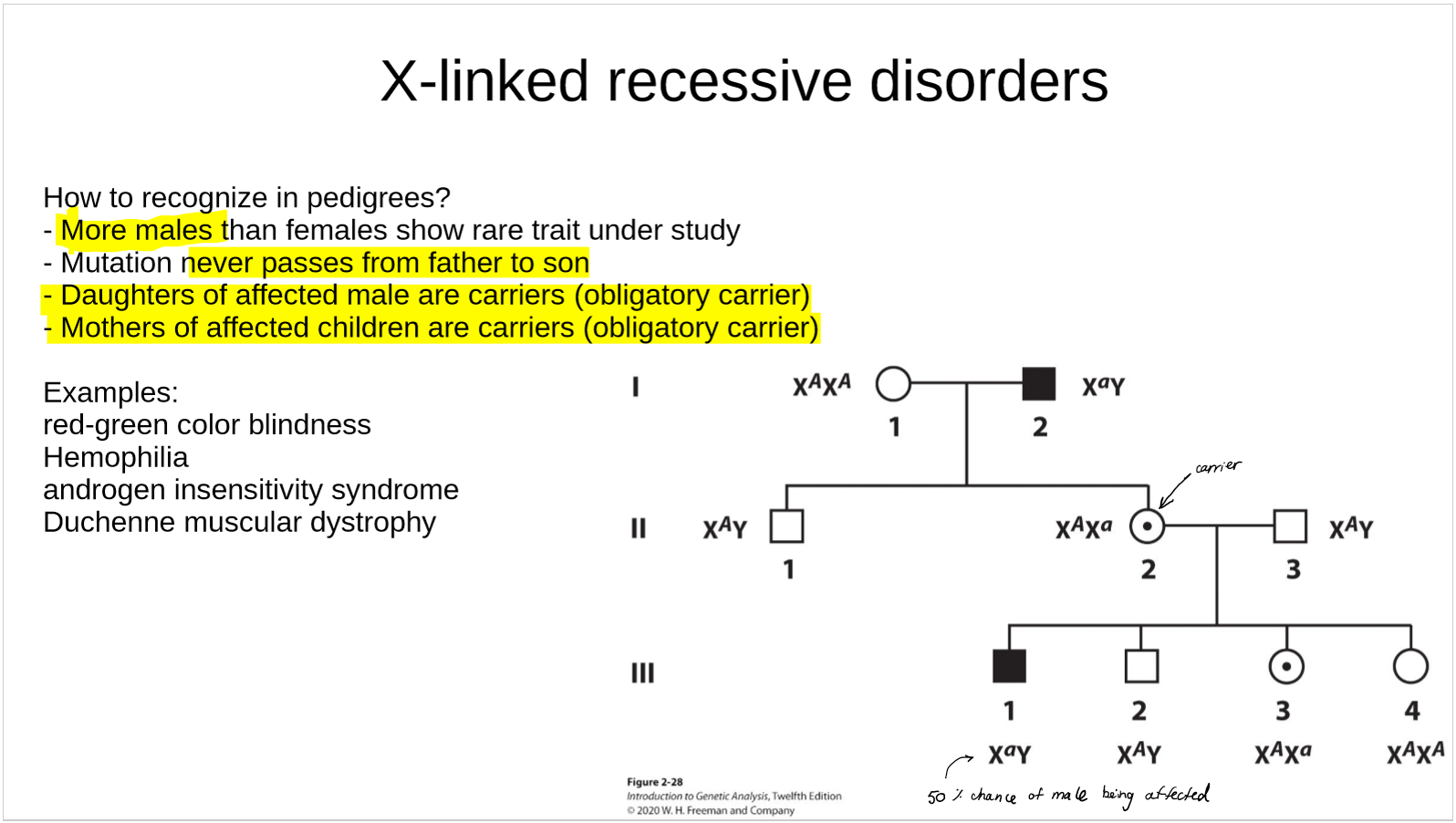
how are X-linked recessive disorders recognized in pedigrees?
what are some examples of X-linked recessive disorders?
how are X-linked dominant disorders recognized in pedigrees?

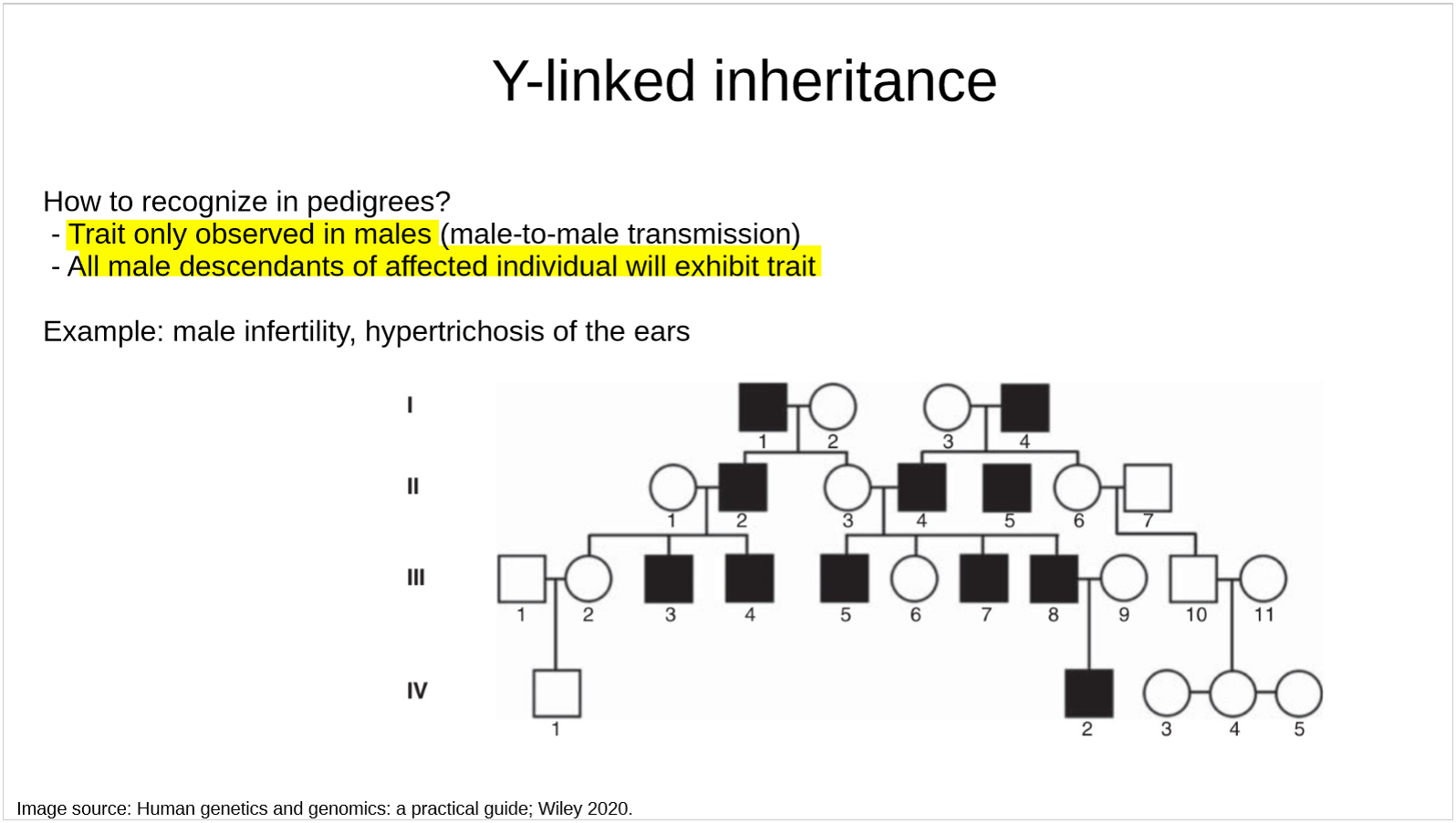
how are Y-linked disorders recognized in pedigrees?
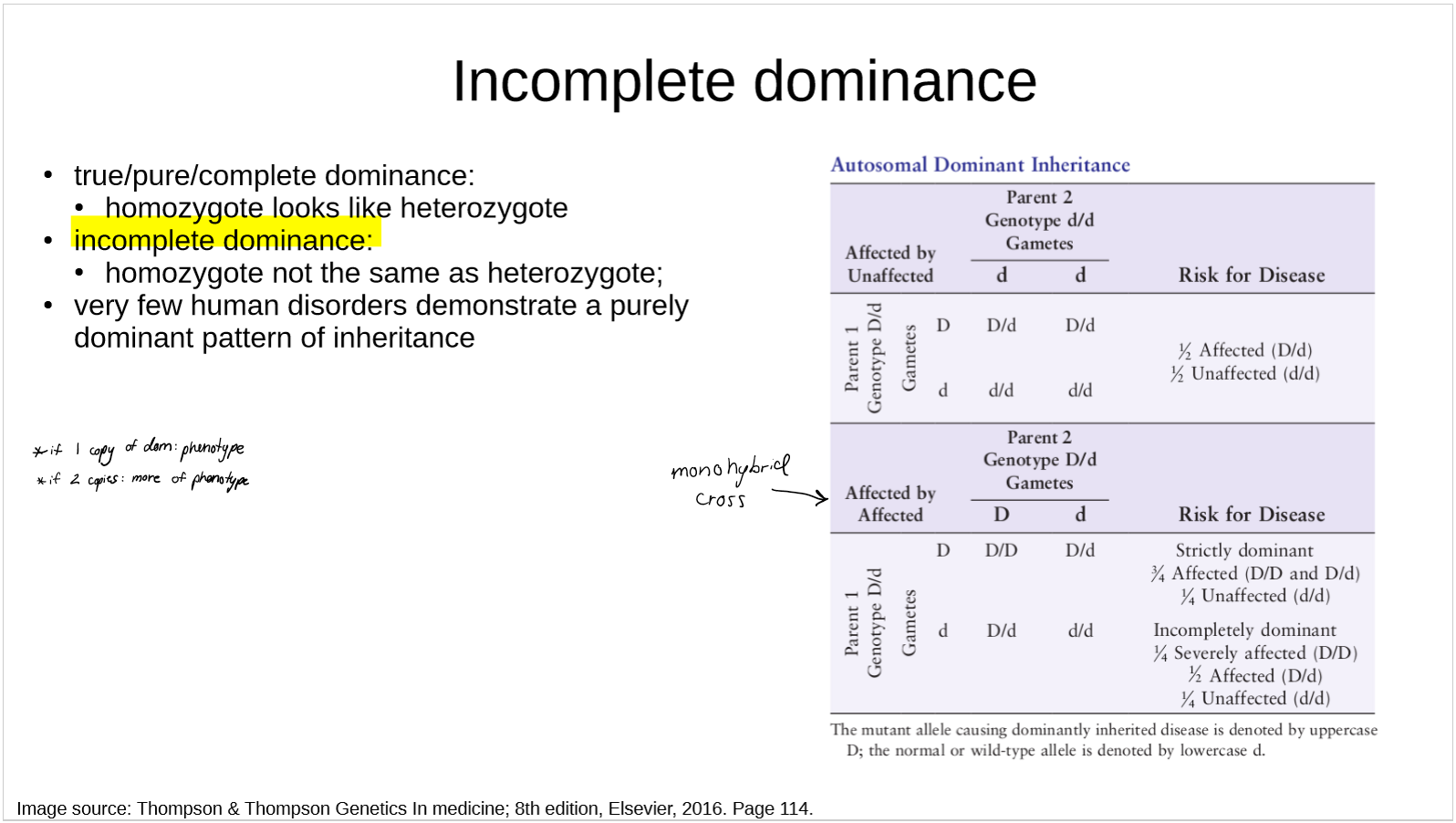
compare complete and incomplete dominance
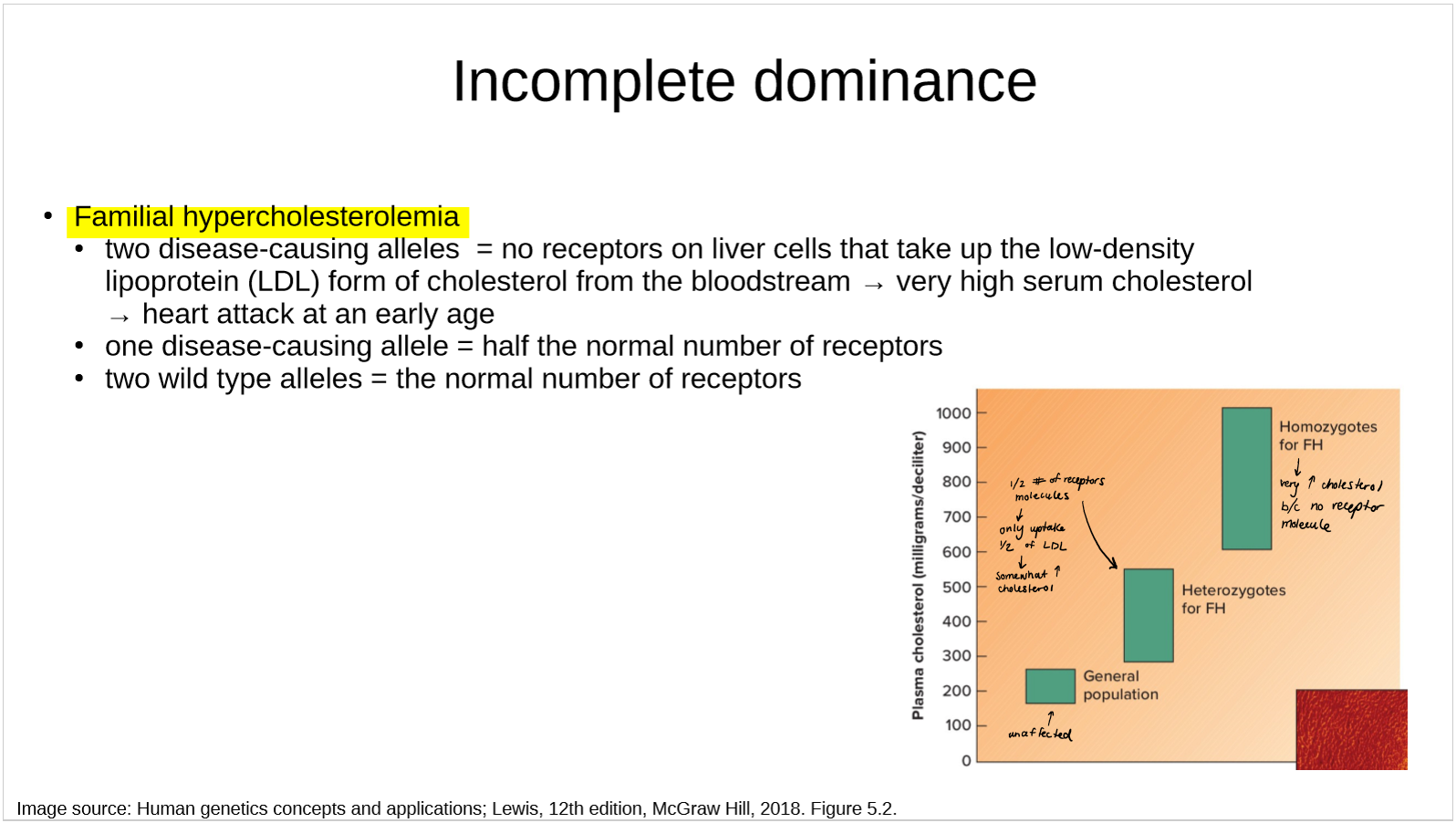
describe the genetics of familial hypercholesterolemia
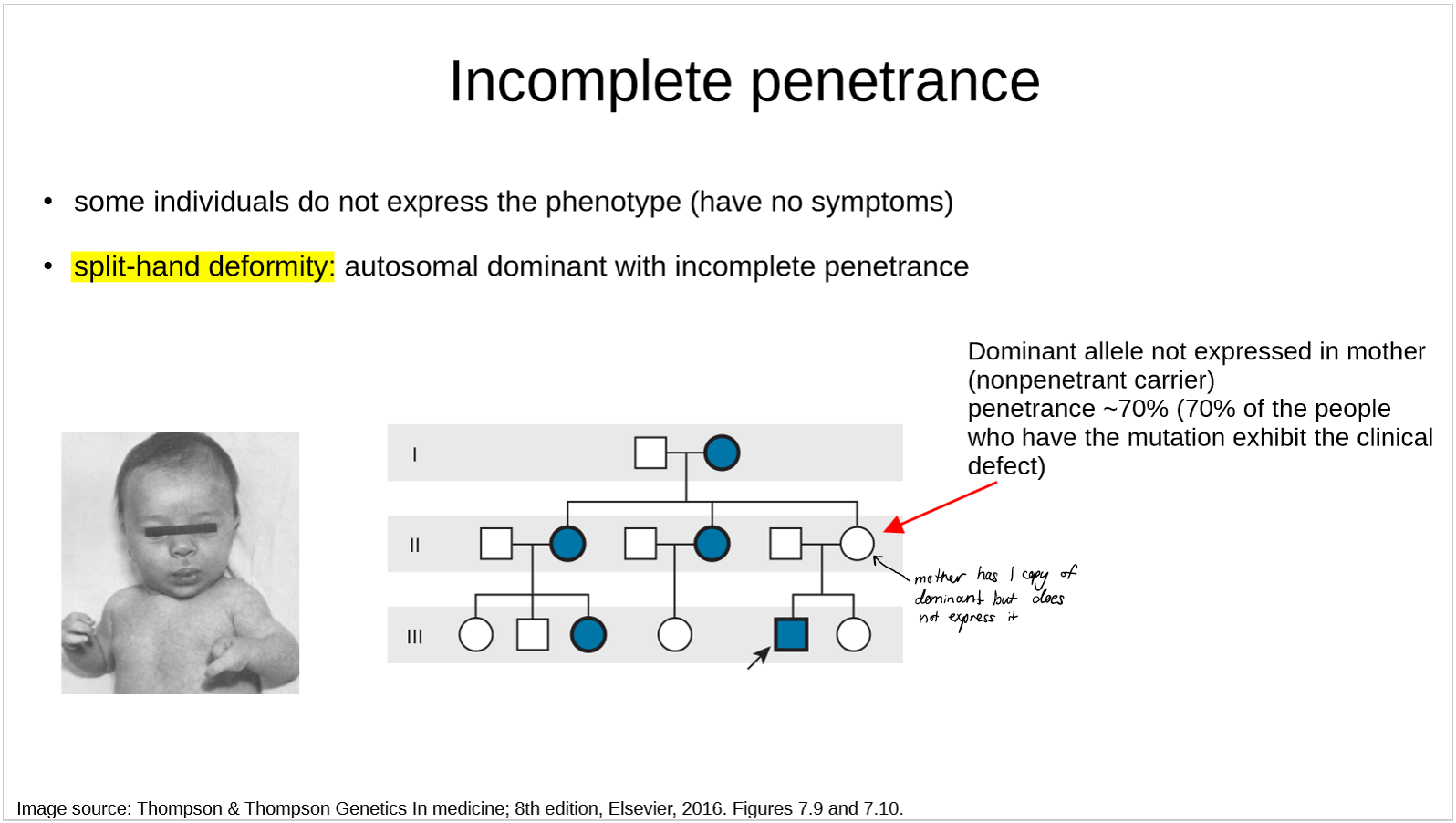
describe the genetics of split-hand deformity
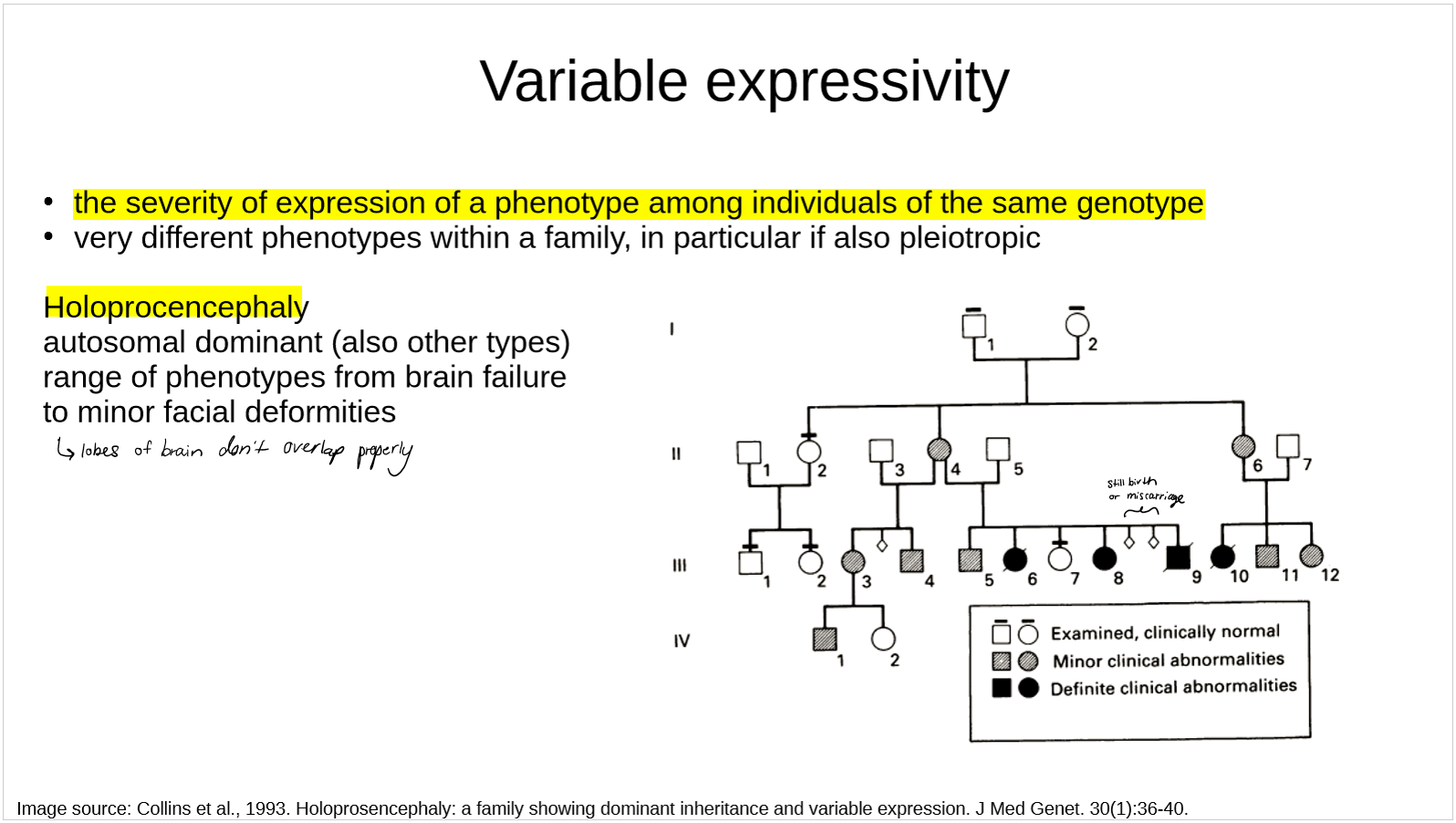
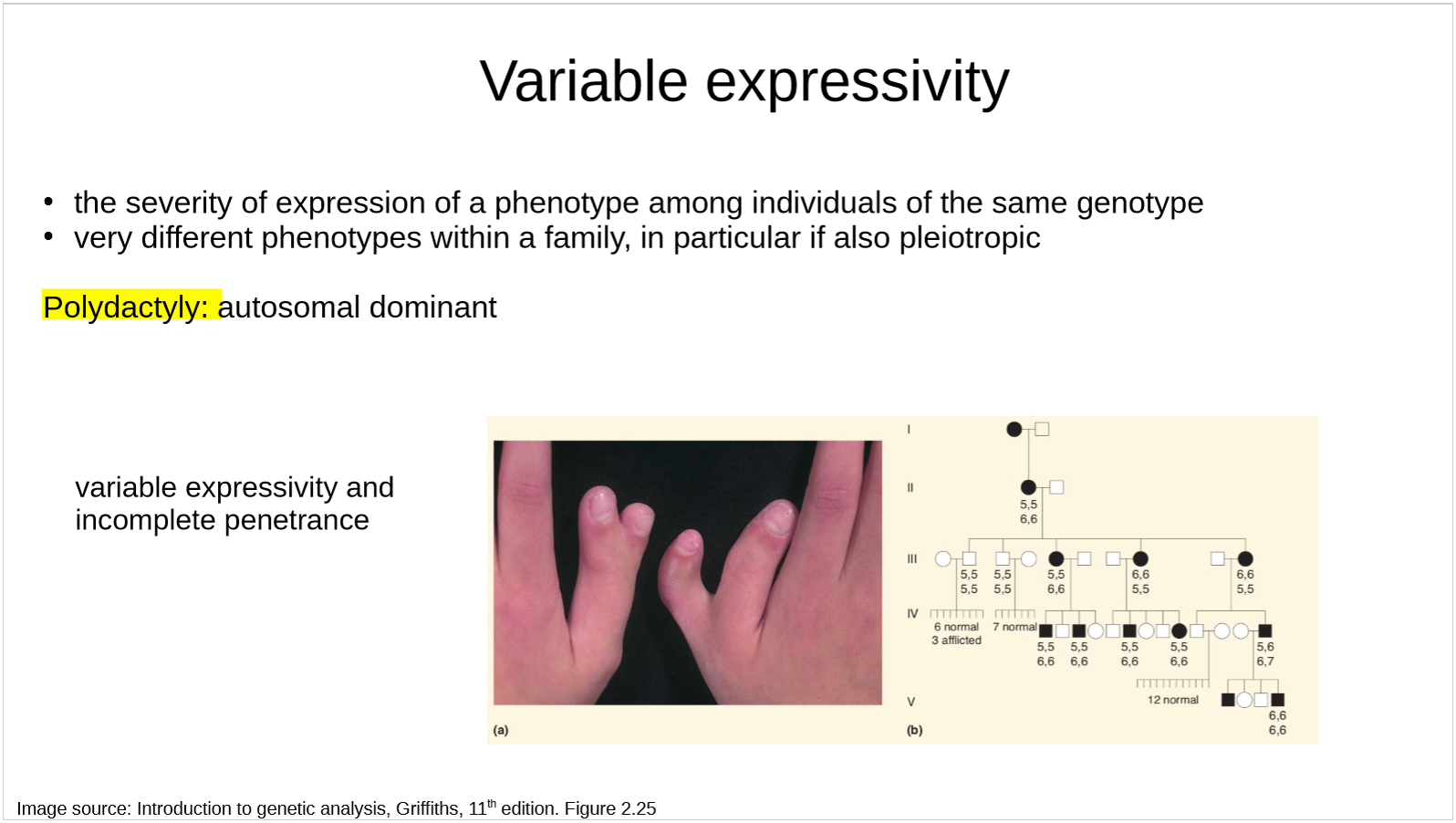
what is variable expressivity?
describe the example of holoprocencephaly
is polydactyly an example of variable expressivity or incomplete penetrance or both?
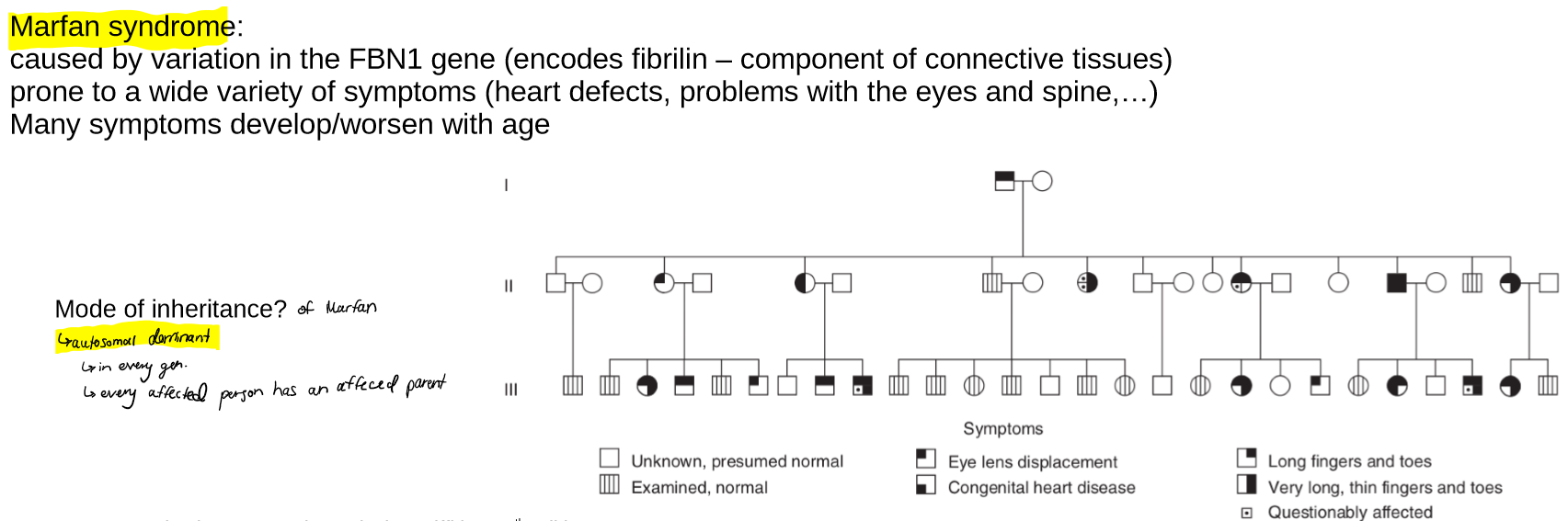
describe pleiotropy

describe Marfan syndrome in the context of pleiotropy
what is its mode of inheritance
epistasis
one gene masks or otherwise affects the phenotype of another (interaction between different genes, not alleles of same gene)
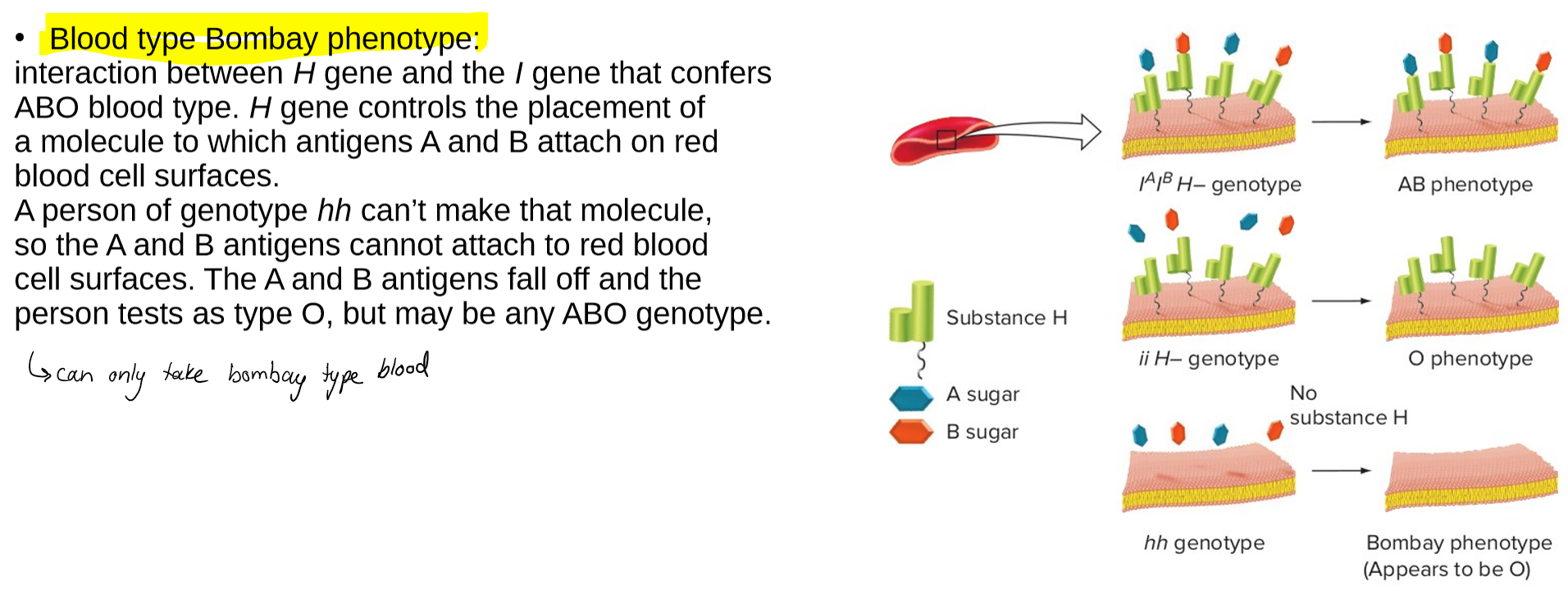
describe the epistasis of the blood type Bombay phenotype
what are modifier genes?

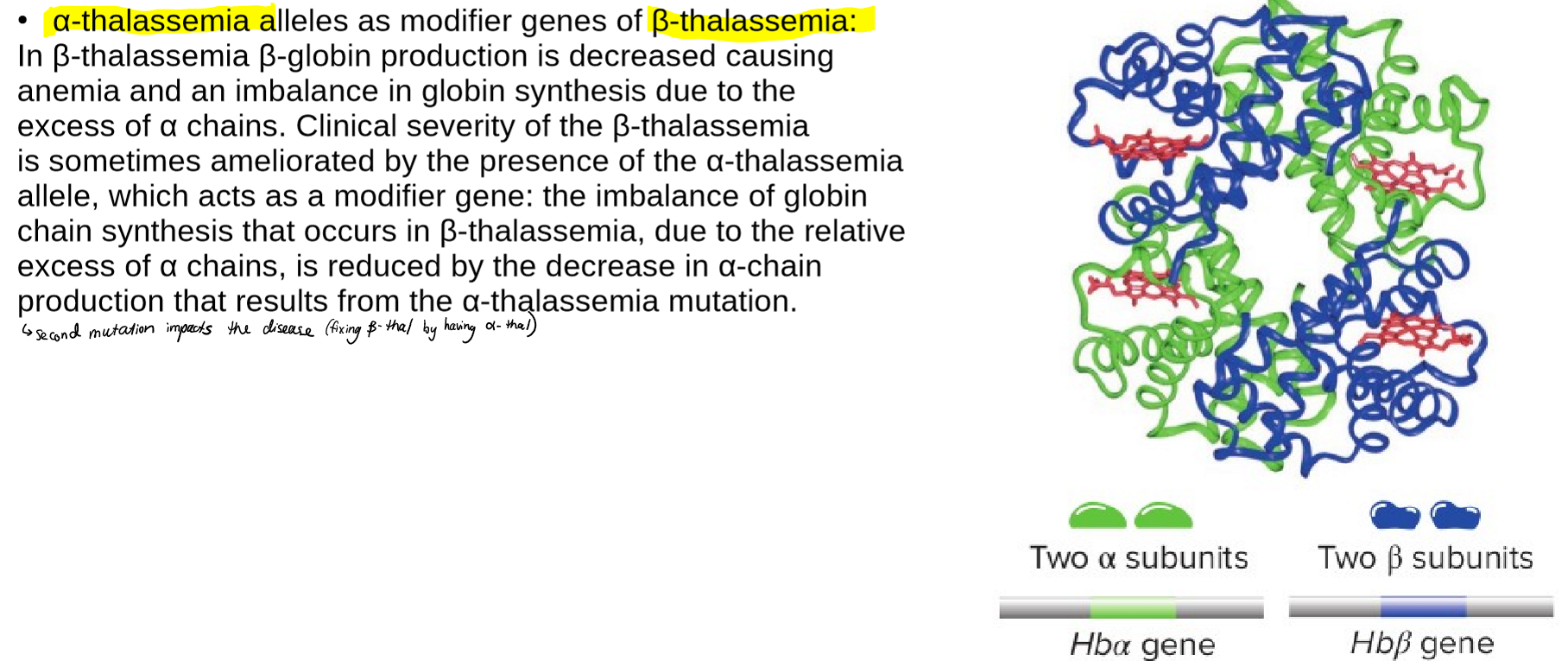
describe the how modifier genes are related to beta-thalassemia
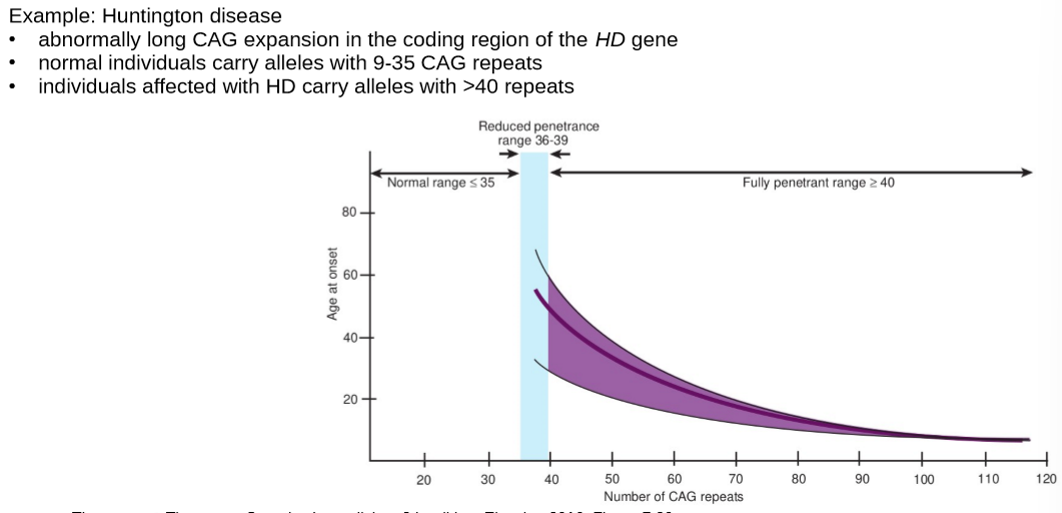
describe dynamic mutations

describe Huntington disease dynamic mutations
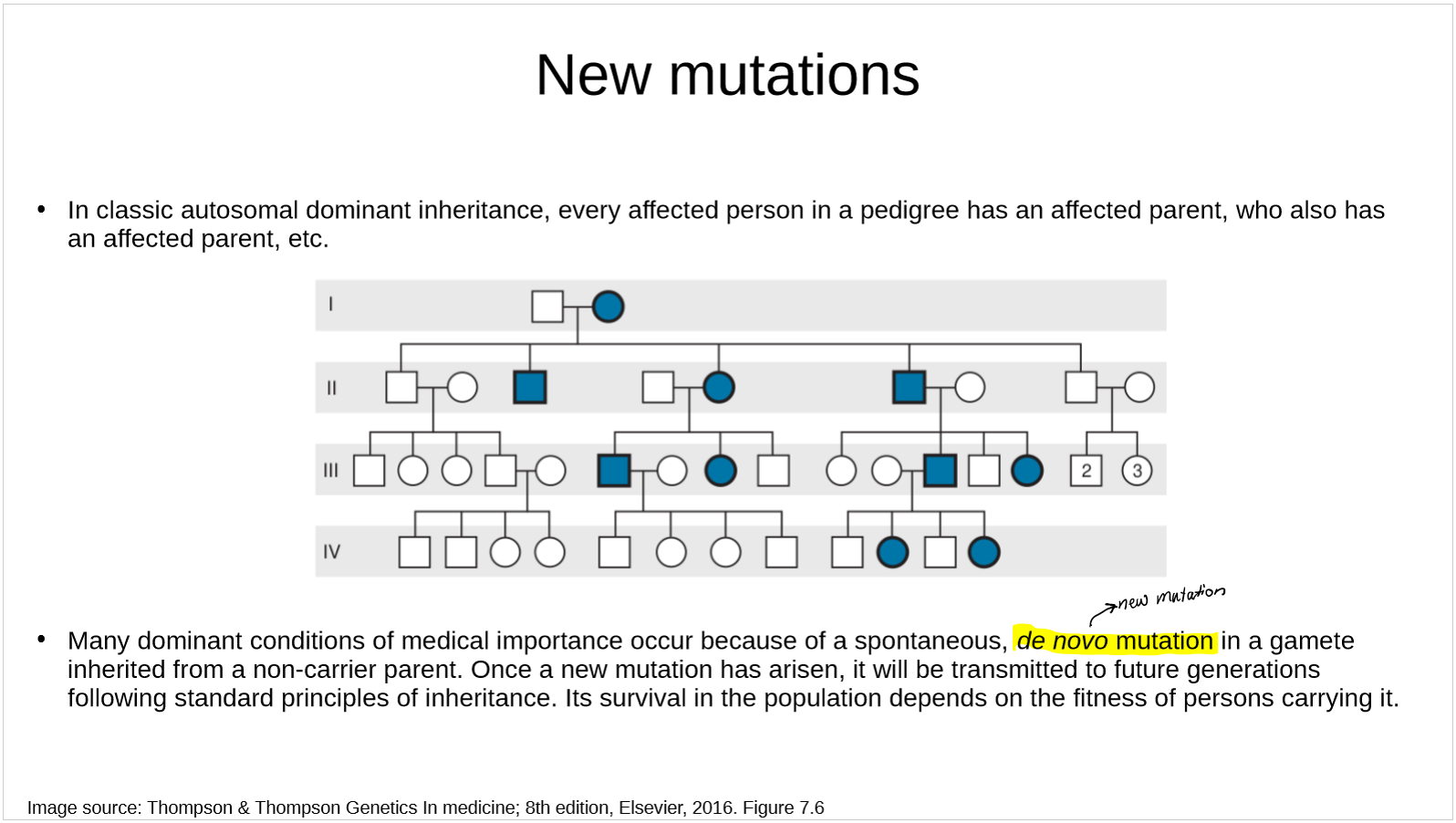
describe new mutations
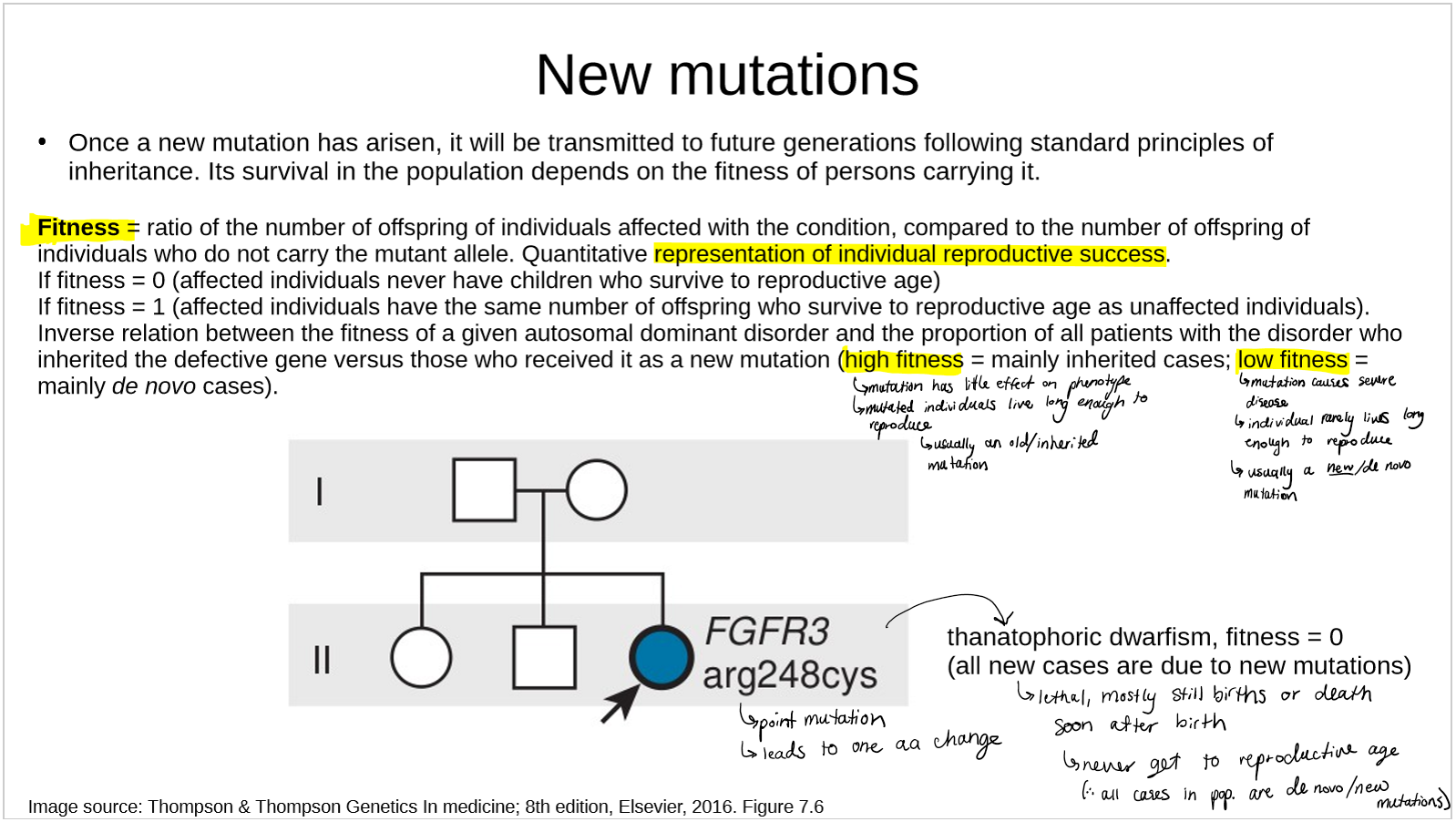
describe how new mutations relate to fitness
describe sex-influenced phenotypes

describe sex-limited phenotypes
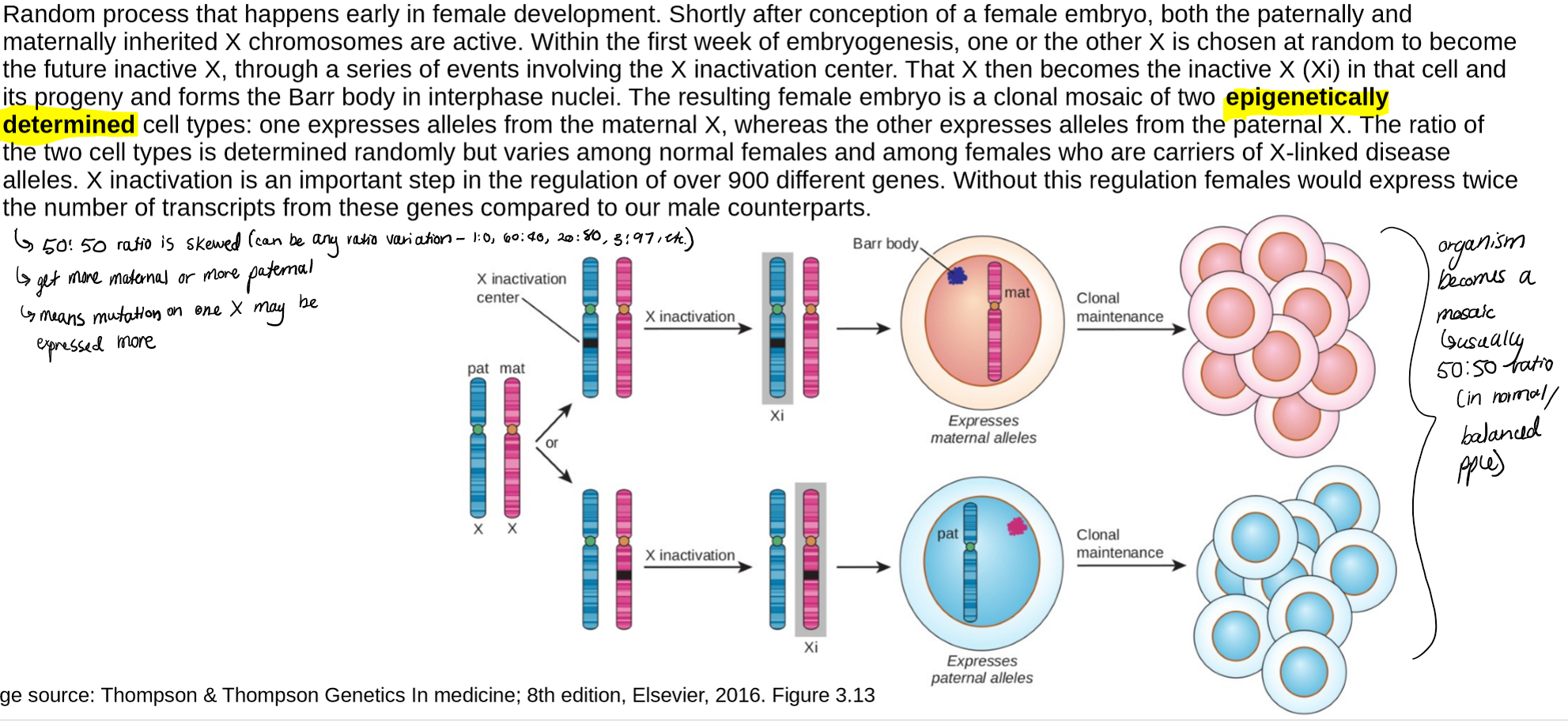
describe normal X-inactivation
normal female cells (46, XX) undergo random X inactivation, resulting in a mosaic of 2 cell populations in which either the maternal or paternal X is the inactive X (Xi)
in phenotypically normal females, the ratio of the 2 cell populations has a 50:50 mode, but with variation observed in the population, some with an excess of cells expressing alleles from the parental X and some with an excess of cells expressing alleles from the maternal X
how does X-inactivation relate to clinical presentations/phenotypes?
depending on the pattern of random X inactivation of the 2 X chromosomes, 2 female heterozygotes for an X-linked disease may have very different clinical presentations because they differ in the proportion of cells that have the mutant allele on the active X in relevant tissue
describe X-linked disorder penetrance in female heterozygotes
incomplete penetrance varies as a function of X inactivation patterns
many X-lined disorders are incompletely penetrant in female heterozygotes
ALL DEPENDS on X inactivation patterns
when are X-linked recessive disorders observed in females?

how does X-inactivation relate to X-linked single gene disorders?

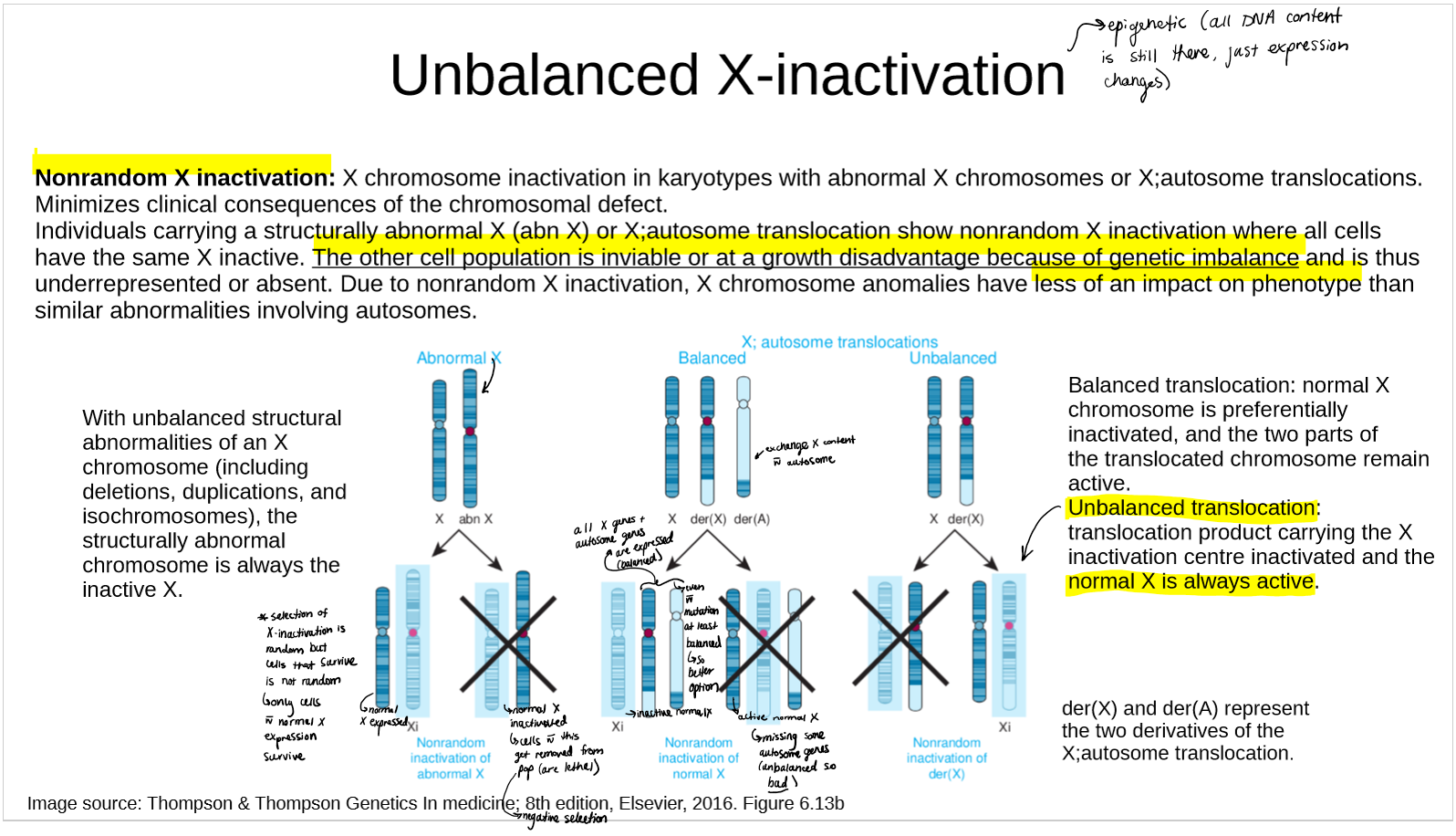
describe nonrandom X inactivation
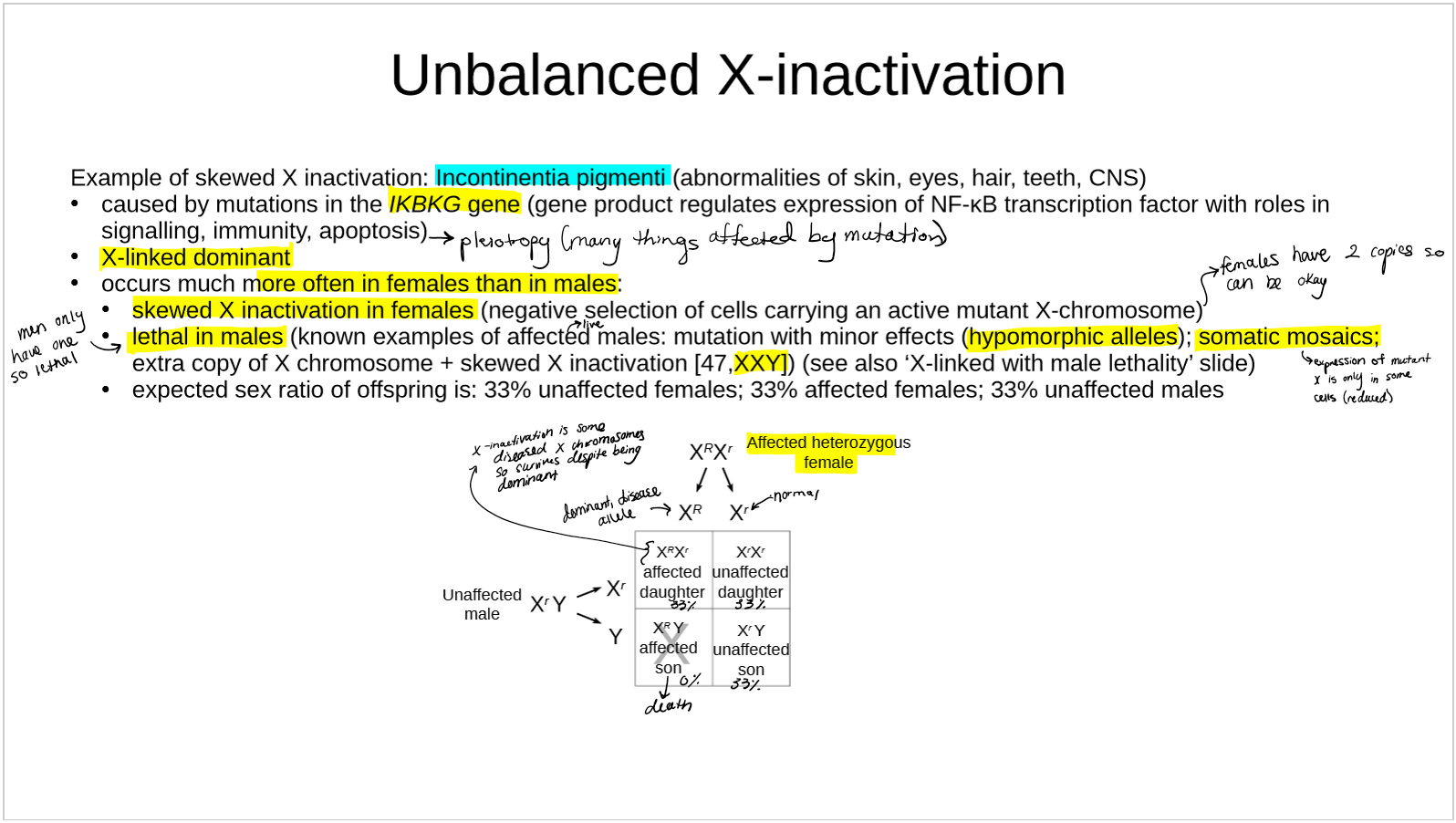
describe Incontinentia pigmenti as an example of unbalanced X-inactivation
what is X-inactivation escape?
Biallelic instead of monoallelic expression of some X-linked genes.
Partial expression of genes from the inactive X chromosome.
Escape genes: ~ 15% of genes on the X chromosome are expressed not only from the active X, but also to some extent from the silent X. Escape gene expressed at 10 - 30% of the level of transcripts made by the homologous allele on the active X (= increased dosage of escape genes)
describe how X-inactivation escape plays a role in autoimmune disorders
e.g. lupus, rheumatoid arthritis, multiple sclerosis more common in females (70-80% of cases are female)
● many genes on the X chromosome are involved in immune responses
● expression of certain genes on the inactive X is partly responsible for the sex differences in some cases of
autoimmune diseases
● the extra activity may provide females with faster immune responses with higher amplitude, better protection
against infectious agents and faster clearance of infection
● the extra activity may make females more susceptible to autoimmune disorders
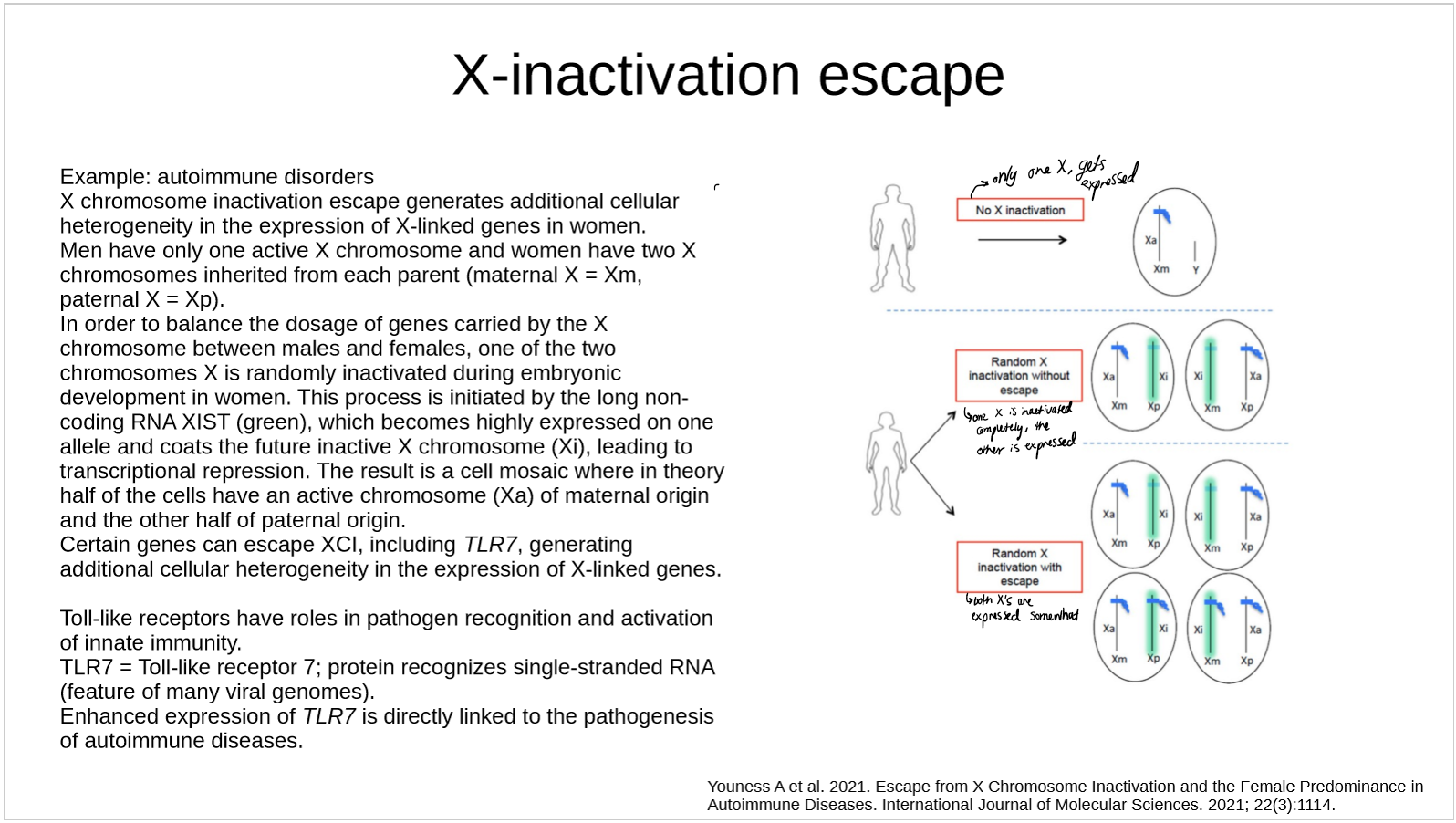
describe X-linked male lethality
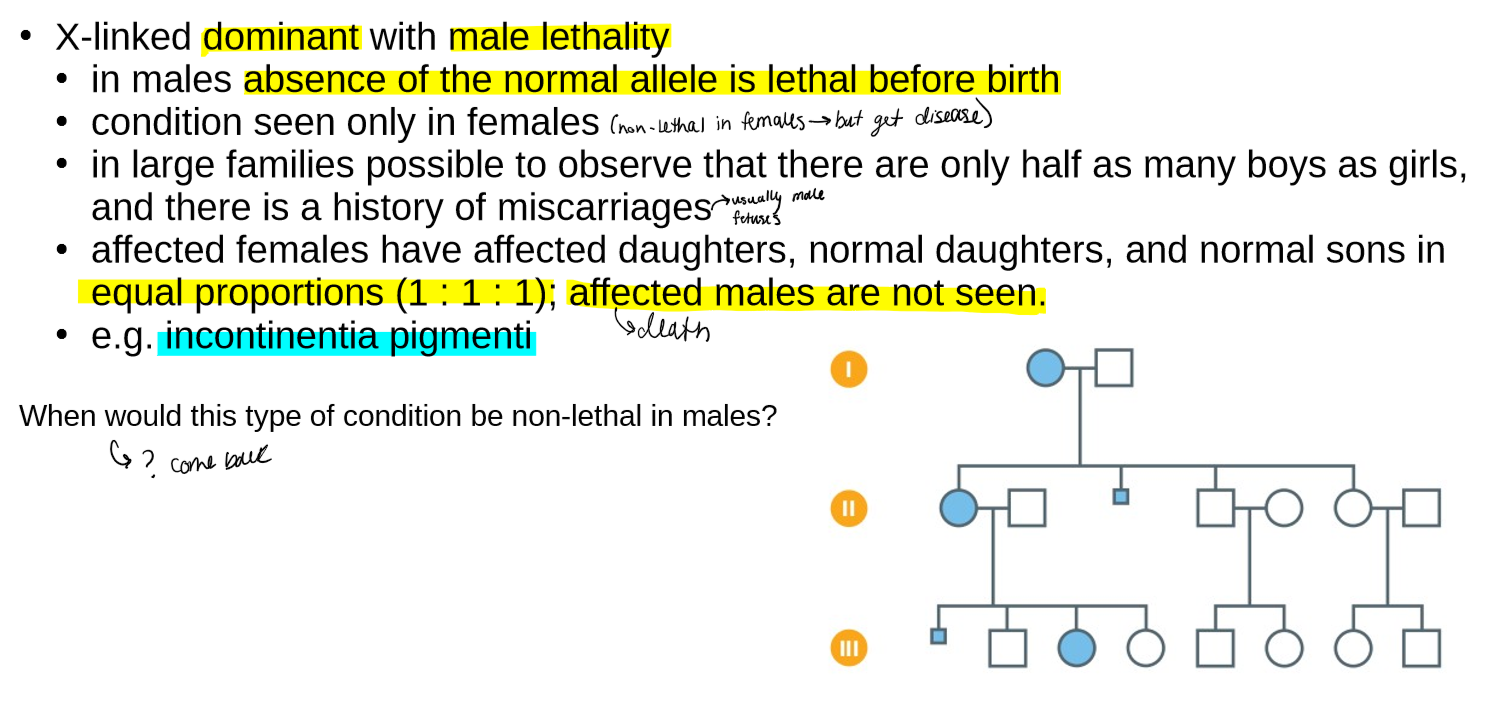
describe X-linked male sparing
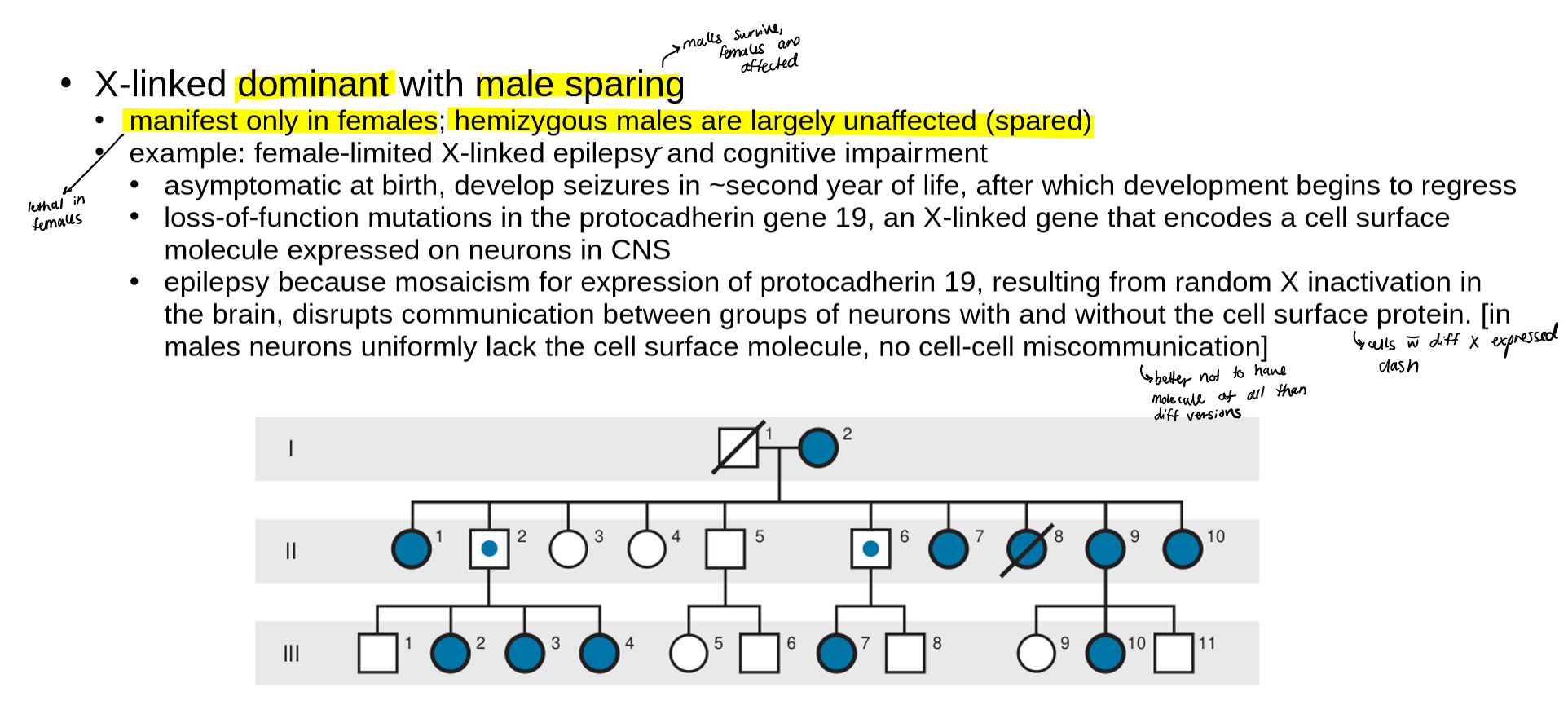
what is pseudo autosomal inheritance?
describe an example
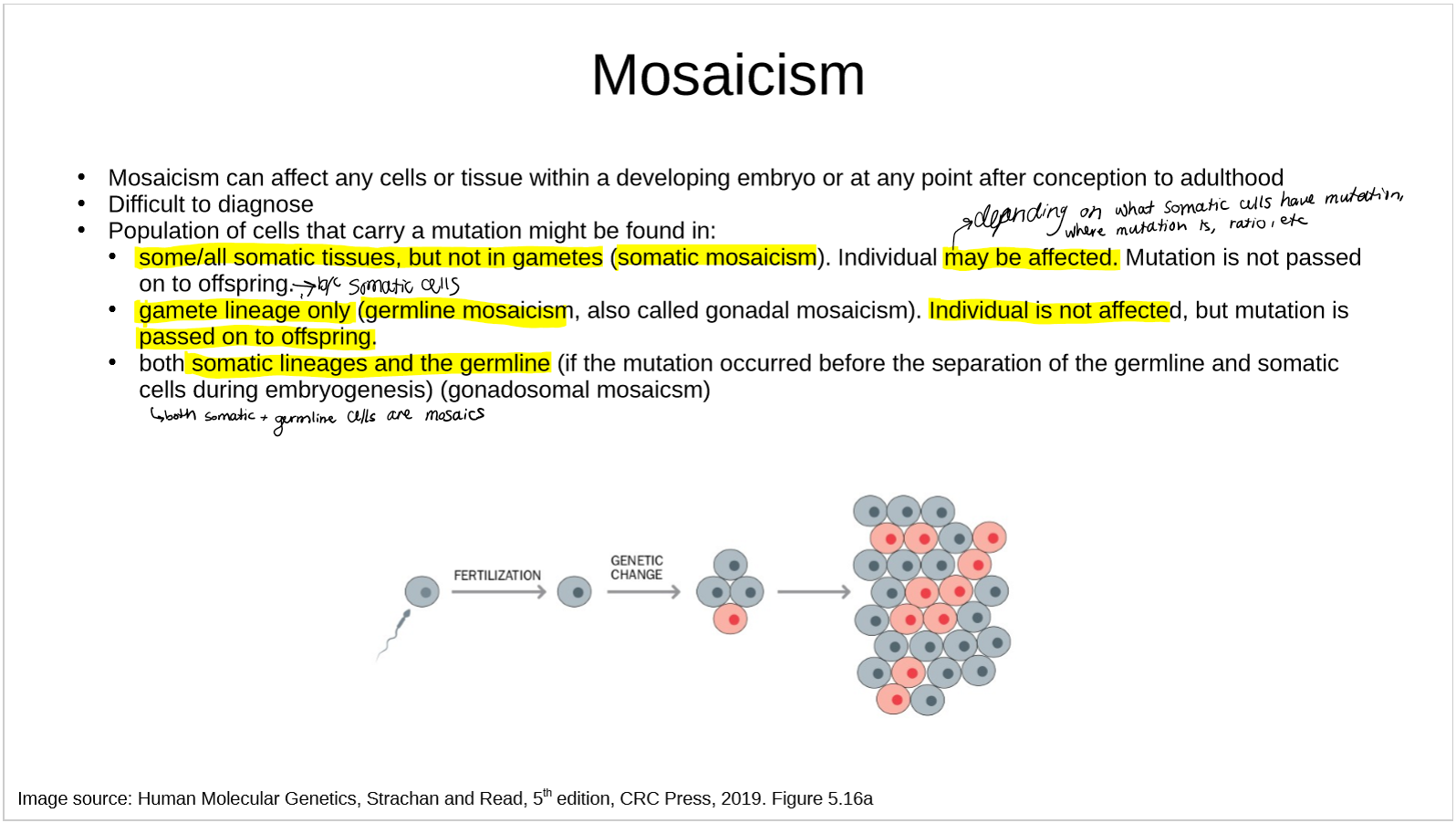
what is mosaicism?
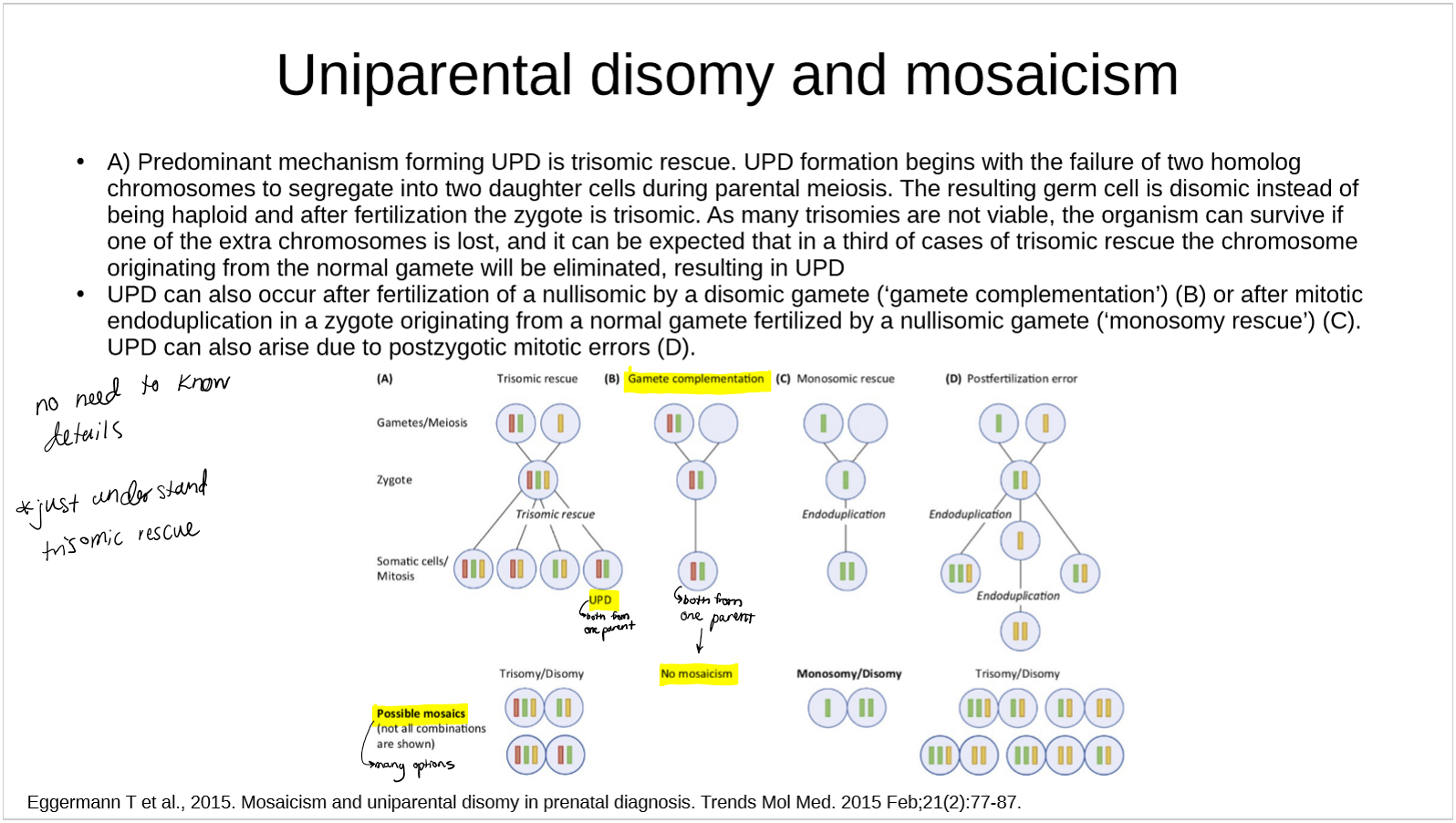
how is Uniparental disomy related to mosaicism
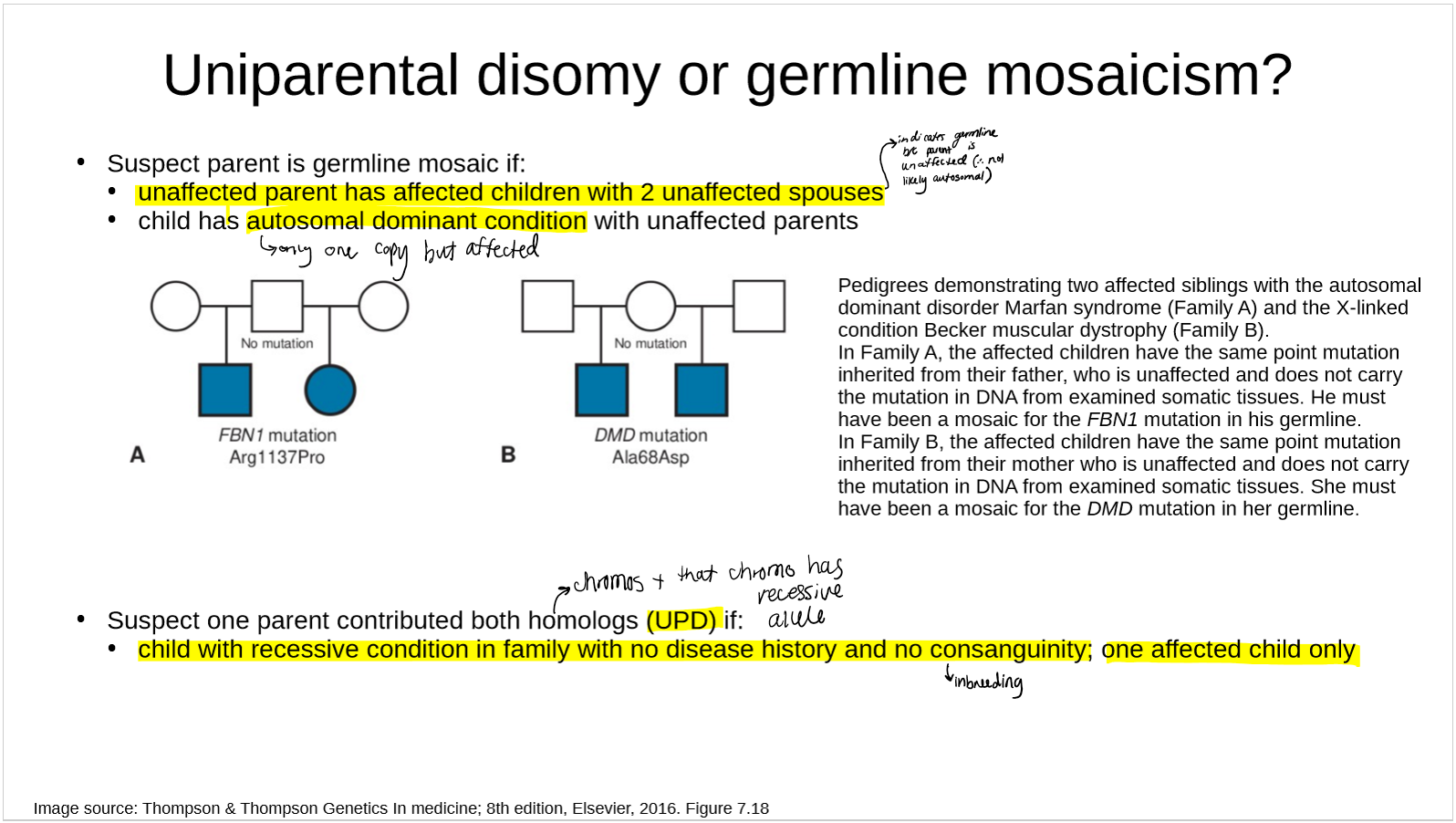
how can you tell if the individual is a consequence of UPD or germline mosaicism?
what is genomic imprinting?
Methyl (CH3) groups cover a gene or several linked genes and prevent
them from being expressed.epigenetics
Chromatin is modified in maternal/paternal DNA and passed down to
offspring in a sex-dependent pattern.different genes get imprinted in females and males
>150 imprinted genes in the human genome
how is it passed down? what happens in mitosis?
imprinting pattern is passed from cell to cell in mitosis, but not
from individual to individual through meiosis.When silenced DNA is replicated during mitosis, the pattern of
blocked genes is exactly placed, or imprinted, on the new DNA, covering the same genes as in the parental DNA.In the figure, maternally methylated genes are shown in red and paternally methylated genes in black.
In germ-line cells during meiosis, somatic cell methylation marks are erased, and new sex- specific methylation marks are established (these are specific to the organism’s own sex and will be transmitted to the next generation
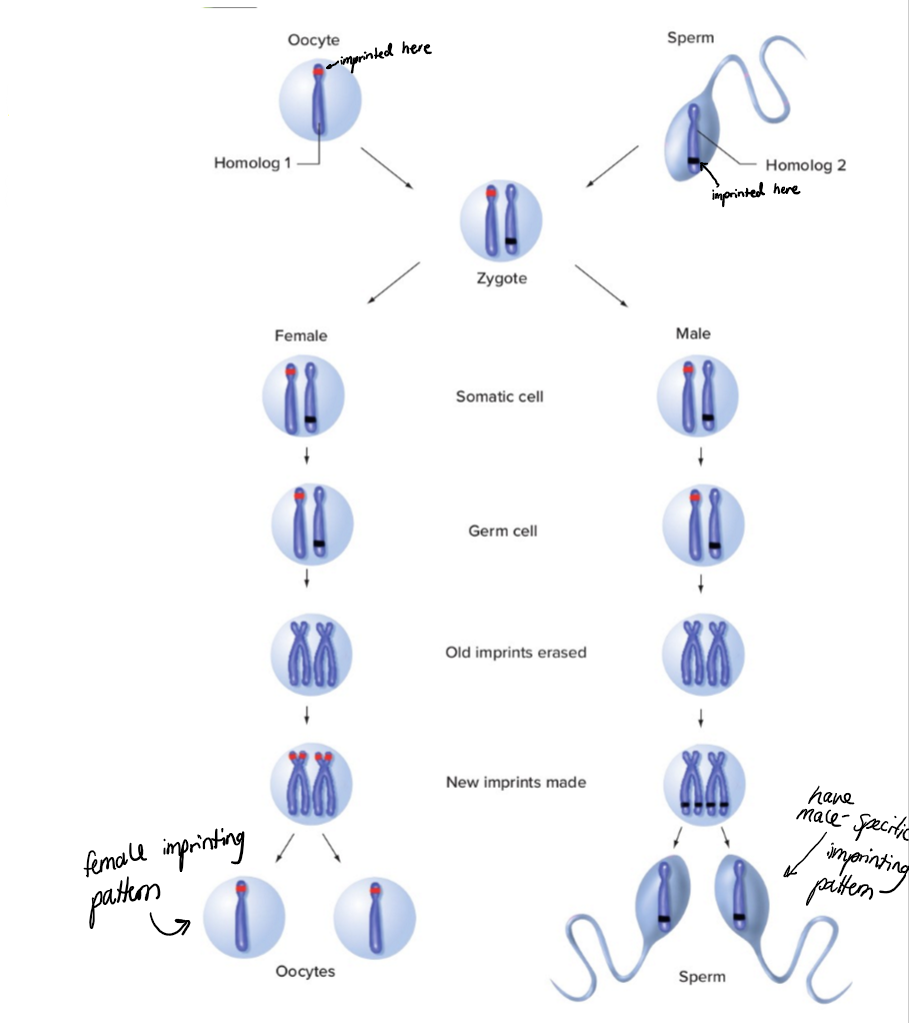
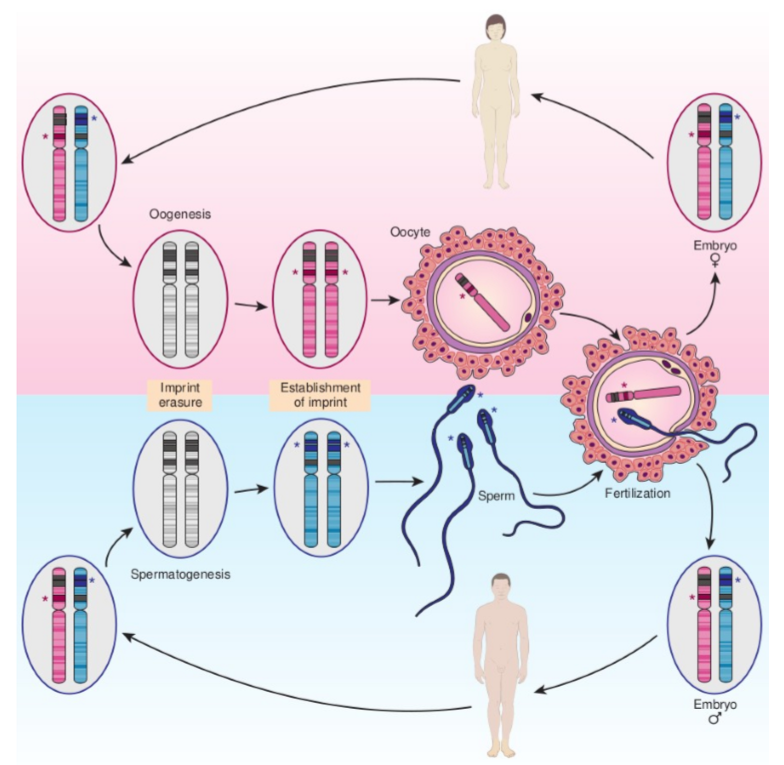
describe Genomic imprinting and conversion of maternal and paternal imprints during passage through male or female gametogenesis
Within a hypothetical imprinted region on an pair of homologous autosomes, paternally imprinted genes are indicated in blue, whereas a maternally imprinted gene is indicated in red. After fertilization, both male and female
embryos have one copy of the chromosome carrying a paternal imprint and one copy carrying a maternal imprint.During oogenesis (top) and spermatogenesis (bottom), the imprints are erased by removal of epigenetic marks, and new imprints determined by the sex of the individual are established within the imprinted region.
Gametes thus carry a monoallelic imprint appropriate to the parent of origin, whereas somatic cells in both sexes carry one chromosome of each imprinted type
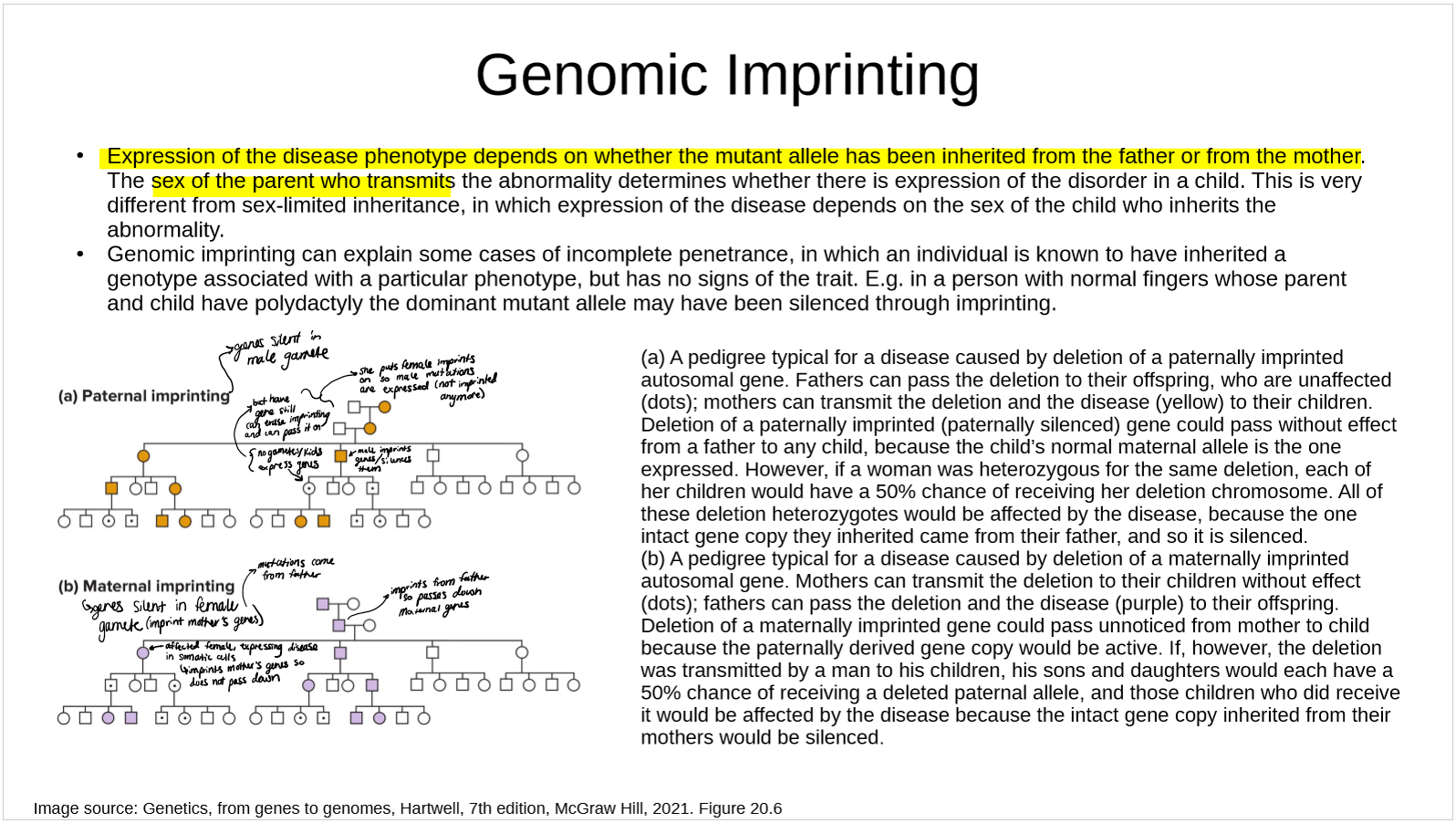
how does genomic imprinting affect disease phenotype
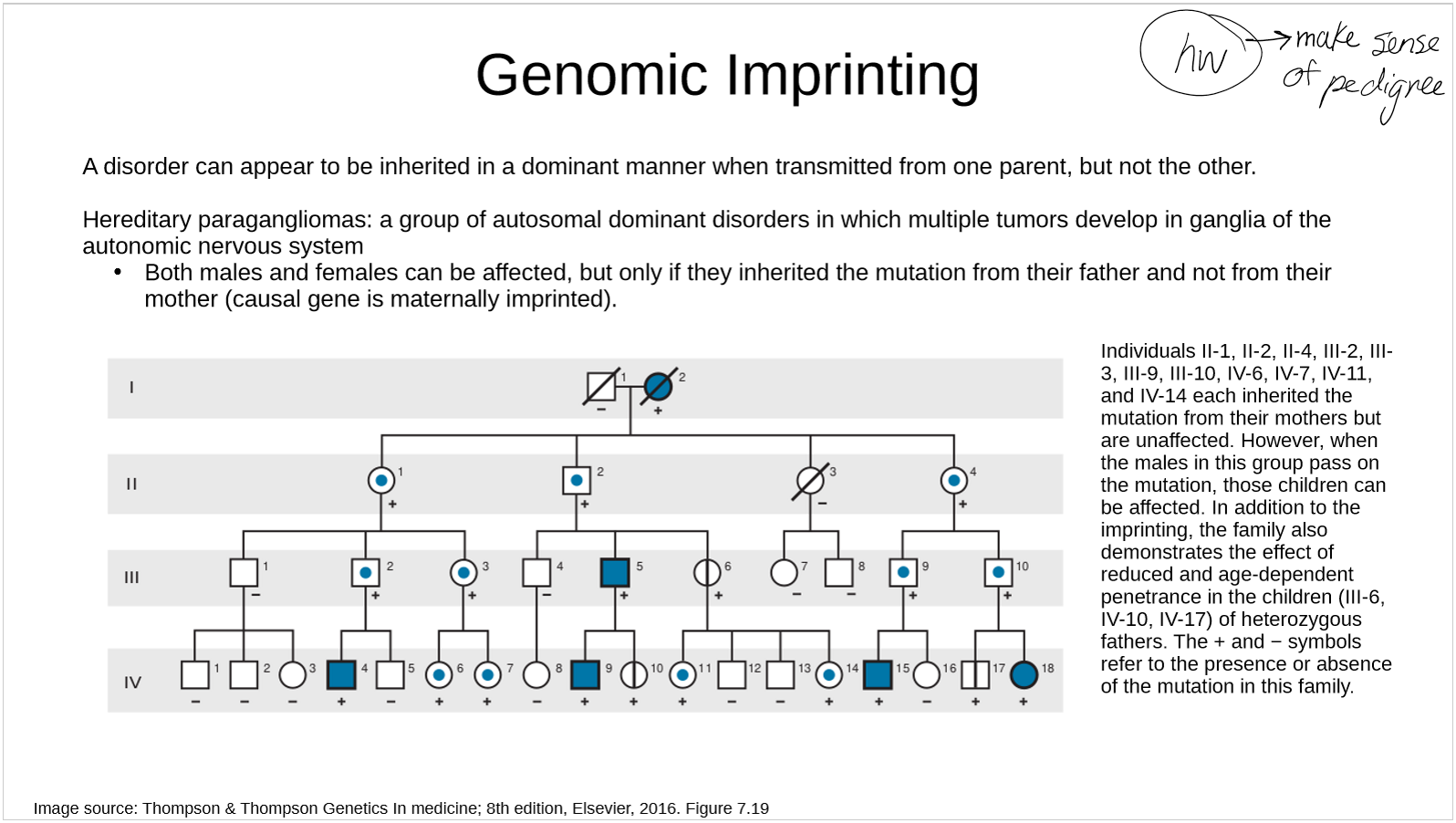
describe herediatary paragangliomas in terms of genomic imprinting
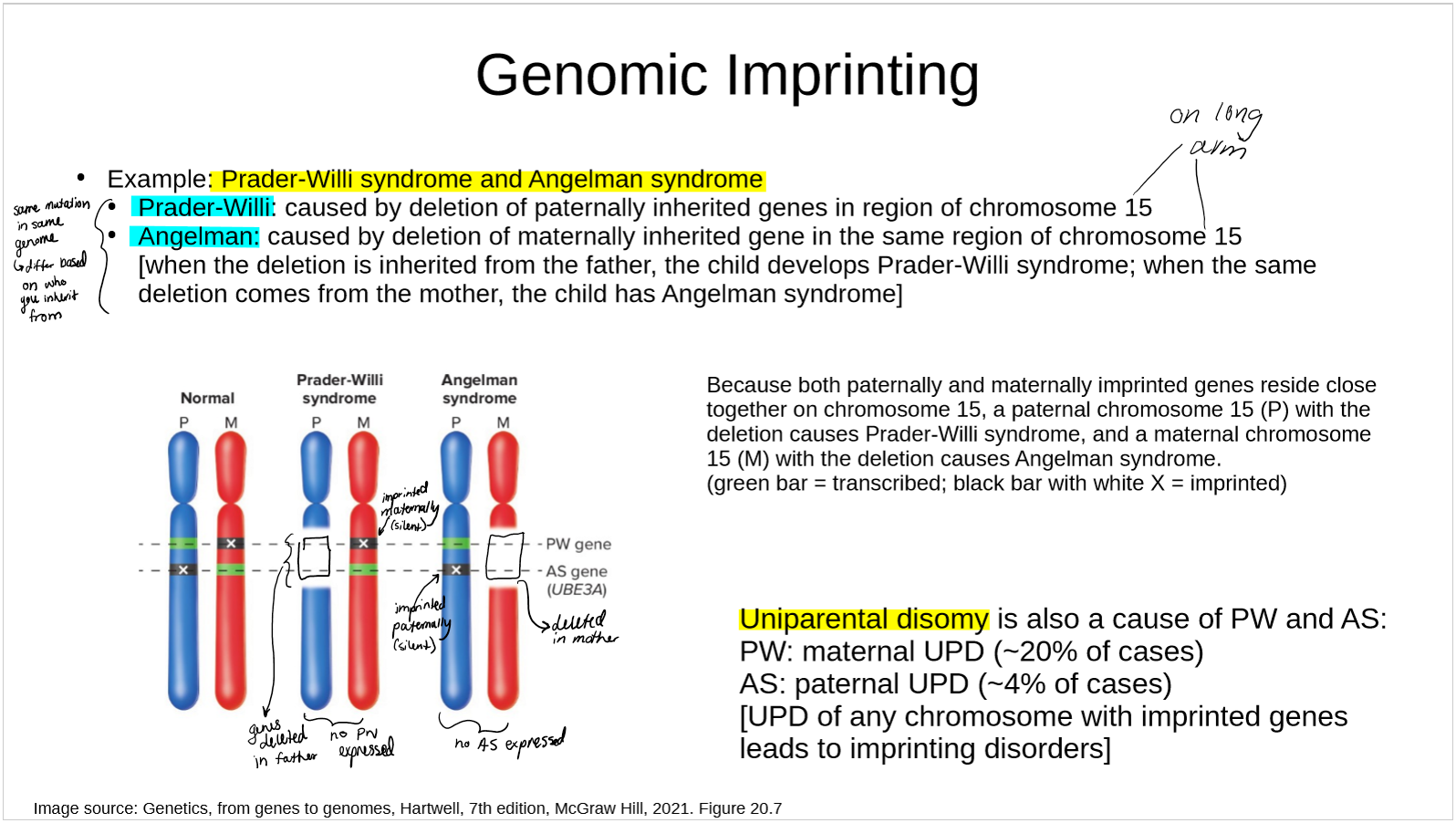
describe Prader-Willi syndrome and Angleman syndrome in terms of genomic imprinting
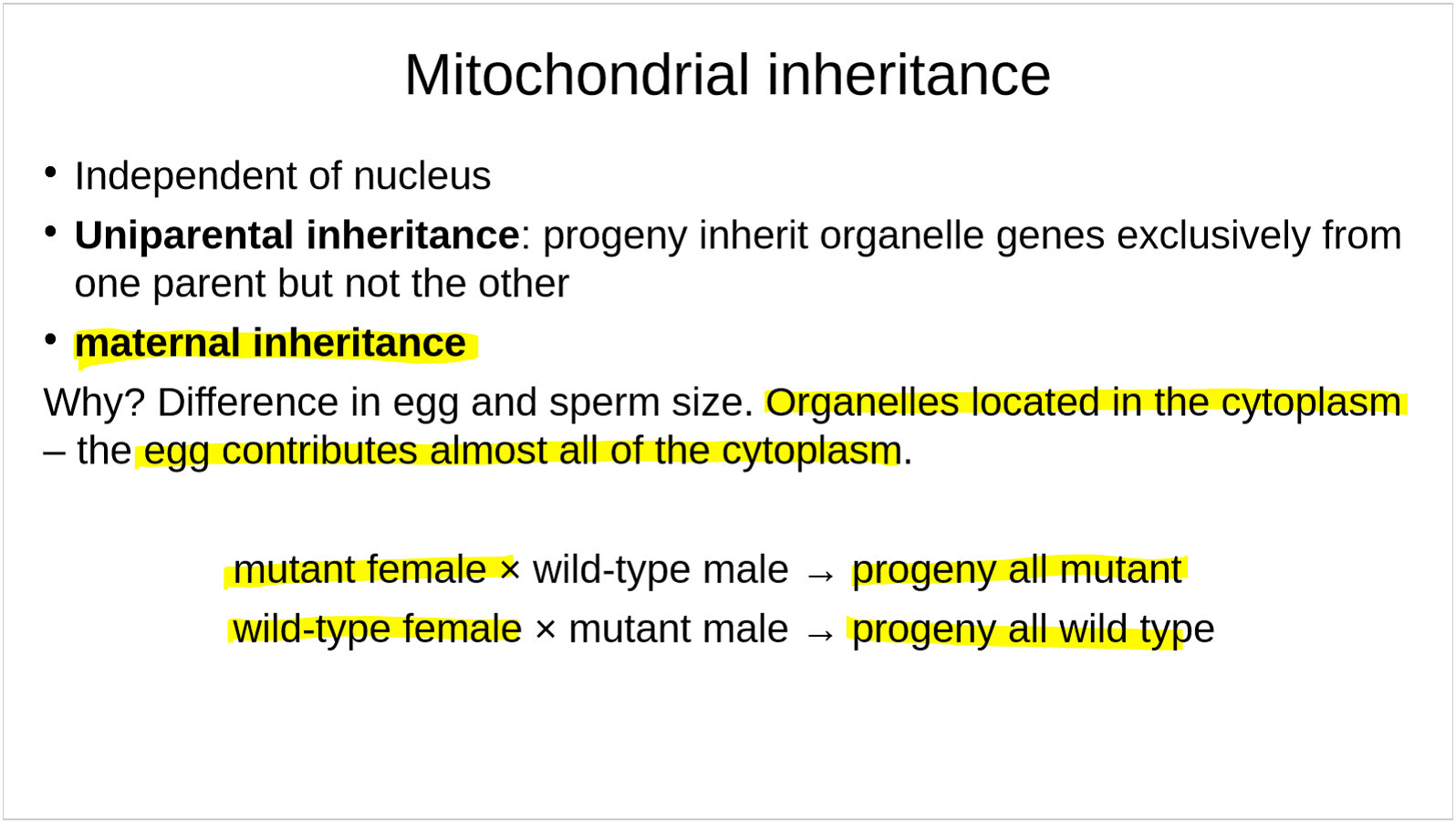
how are mitochondria inherited? why? what does this mean in terms of mitochondria phenotype?