lecture 6 - transcription (general info and specifically in prokaryotes)
1/43
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
44 Terms
in prokaryotes, where does transcription begin? end?
begins at promoter region of the gene
ends at termination signal
is transcription specific? what DNA participates?
yes specific
only genes participate
what DNA strand is read by RNA pol? what does the mRNA resemble?
template/non-coding strand and makes a strand complementary to it (identical to the coding strand/non-template strand)
name the required components for transcription
Enzyme - RNA pol
ATP, GTP, CTP, UTP
Mg++
DNA template
explain the experiment that described how RNA pol works. what were the conclusions?
Experiment:
Used DNA with alternating AT
sequence and following the
incorporation of different dNTPsIsolated new RNA molecule and
analyzed for content
conclusions
RNA pol copies DNA
the product RNA molecule only had As paired with Us
RNA pol used complementary base pair rule
full name for RNA pol
DNA dependent RNA polymerase
describe the chemical mechanism of RNA synthesis
Requires Mg++
Adds nucleotides in 5’ to 3’ direction one by one
only one strand on DNA serves as a template in 3’ to 5’ direction but it could be either strand
describe the structure of bacterial RNA pol
polymerase core
2 copies of α subunit, one of each β, β’ and ω
α2ββ’ω (MW = 390,000 Da)
sigma factor
One subunit of σ (many different σ subunits, σ70 is most common)
σ70 (MW = 70,000 Da)
holoenzyme = core + sigma factor
function of sigma factor
decides which promoter to bring to RNA pol core to begin transcription
how does eukaryotic RNA pol differ from bacterial?
eukaryotic DNA pol has no sigma
uses other transcription factors
compare the 5 subunits of prokaryotic RNA pol
Prokaryote) RNA polymerases have five types of
subunits: α, β, βʹ, and ω have rather constant sizes in different bacterial species, but σ varies more widely
σ gives specificity
how are promoter sequences recognized in prokaryotes vs eukaryotes?
Recognized by sigma factor of bacterial RNA polymerase
Recognized by initiation factors eukaryotic RNA polymerase
describe the promoter sequences sections
-35 is RNA pol recognition site
-10 is RNA pol binding site (TATA box aka Pribnow box)
transcription start: at +1 site also -10 from TATA box
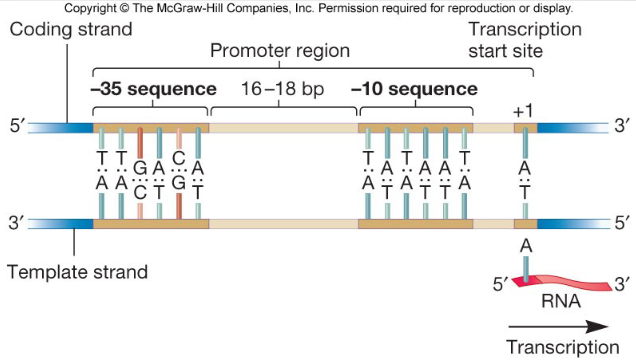
define transcription unit
a sequence of DNA transcribed into a single RNA starting at the promoter and ending at the terminator (the genes transcribed)
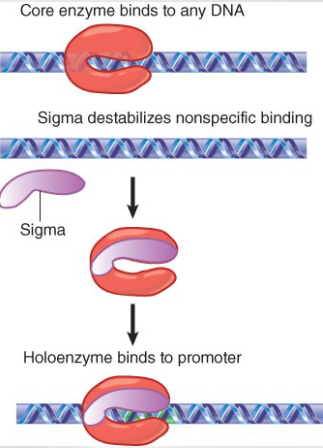
explain how the RNA pol binds to DNA (role of the core enzyme and sigma factor)
Core enzyme binds indiscriminately to any
DNA.Sigma factor reduces the affinity for
sequence-independent binding and confers specificity for certain promotersi.e., core enzyme binds any DNA, sigma factor will reduce stability of non-specific/random binding and increase affinity of holoenzyme to specific promoters
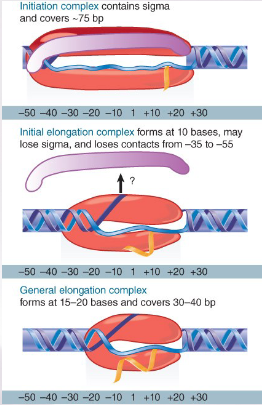
explain how the general elongation complex is formed
RNA polymerase initially contacts the region from −55 to +20.
When sigma dissociates, the core enzyme contracts to −30; when the enzyme moves a
few base pairs, it becomes more compactly organized into the general elongation complex
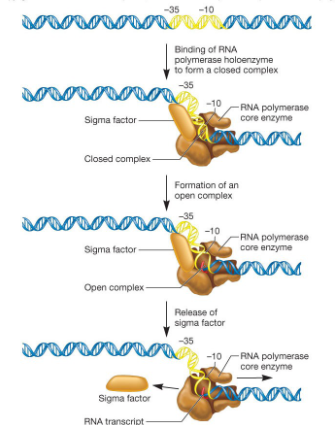
describe sigma cycle
binding of RNA pol holoenzyme to form closed complex (RNA pol binds to ds helix)
formation of open complex (open helix - form replication bubble)
sigma factor is released now that initiation is over because it is only needed to give specificity
RNA pol core enzyme transcribes
elongation
generally describe the process
describe how RNA pol moves during elongation
A continuous process until termination
Basically, elongation is the stage when the RNA strand gets longer, thanks to the addition of new nucleotides.
During elongation, RNA polymerase "walks" along one strand of DNA, known as the template strand, in the 3' to 5' direction. For each nucleotide in the template, RNA polymerase adds a matching (complementary) RNA nucleotide to the 3' end of the RNA strand.
how does RNA pol ensure the transcript is correct?
kinetic proofreading
nucleotide proofreading
both done by RNA pol
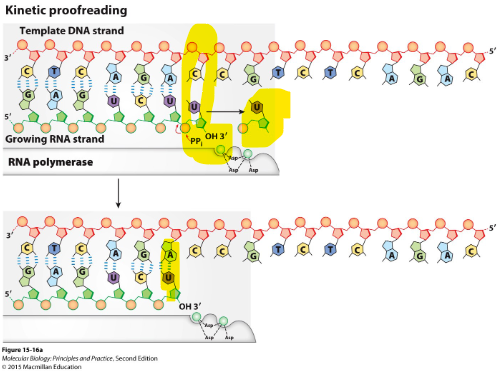
describe kinetic proofreading
when nucleotides are mis-matched they tend to dissociate more rapidly then when correctly matched
Nucleotide is removed by pyrophosphorolysis
then another nucleotide is added again (try again)
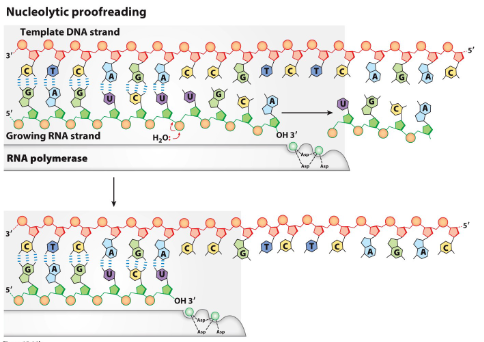
describe nucleotide proofreading
RNA polymerase reverses direction to remove mismatched bases (usually create a bulge)
RNA pol goes forward again once done proofreading
describe generally how termination occurs
what are the two main types in bacteria?
The DNA sequences required for
termination are located upstream of
the terminator sequence. Formation
of a hairpin (ds) in the RNA may be
necessary.hairpin tells RNA pol to stop/disassemble
2 main types in bacteria:
Rho-independent
rho-dependent
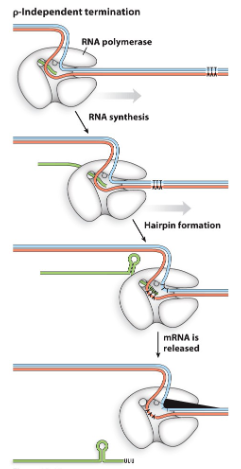
what are the two features of the Rho-independent termination pathway?
indirect repeats containing lot’s of G-C pairs which forms stem+loop (hairpin)
sequence of As in DNA to cause a string of Us in the RNA
these cause the hairpin configuration which causes RNA to dissociate
describe the Rho-independent termination pathway
once RNA synthesis is done
hairpin forms
regions of As and Ts in DNA cause RNA to be released when RNA pol reaches this segment
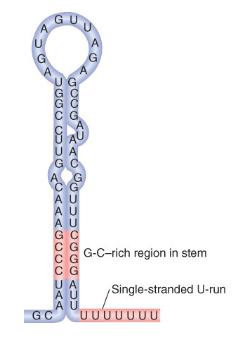
describe the hairpin structure
has double stranded G-C rich region in the stem which allows for strong binding
has a single stranded U-run which will have weaker binding with DNA’s As so is good for releasing the RNA
the hairpin does not need to be perfectly complementary
what are intrinsic terminators?
palindromic regions that form hairpins
(stem and loop) varying in length from 7 to 20 bp
describe Rho’s structure
Rho monomer has 2 domains
Rho has an N-terminal, RNA- binding domain
C-terminal ATPase domain
A hexamer in the form of a gapped ring can form and bind RNA along the exterior of the N-terminal domains
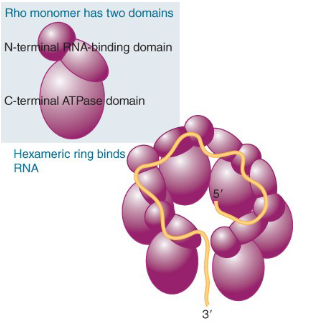
describe the Rho-dependent termination pathway
Rho factor attaches to RNA’s rut site (rho utilization site)
rho translocates along RNA until it reaches
the RNA–DNA hybrid in RNA polymerase,
where it releases the RNA from the DNA.rho which has ATPase activity causes release of RNA polymerase (rho helicase)
what makes the RNA more likely to undergo Rho-dependent termination?
no GC rich regions
lack of sequence repeated A residues (therefore no run of Us added to RNA)
how did they find promoter sequences?
did DNA footprint analysis → looks to see if there are parts of the DNA protected from enzymes/DNAase (which covered by sigma)
sequences regions protected by RNA pol
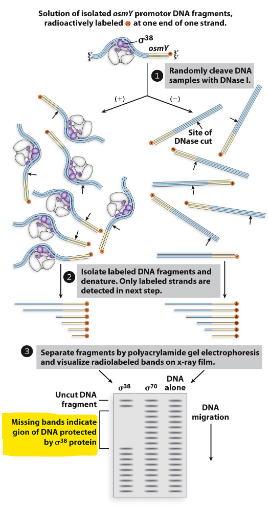
how does the location of the promoter mutation effect transcription?
mutations that effect transcription occur in -35 region and -10 region
mutations in between the -35 and -10 region do not effect transcription
can mutations increase transcription/enzyme activity
yes if they make the sequence more similar to the consensus sequence (mutations that make the sequence less like the consensus sequence have the opposite effect)
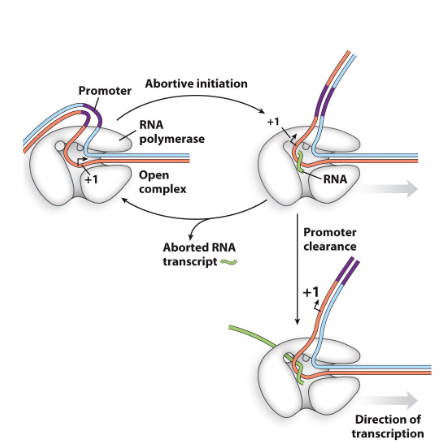
describe abortive initiation
RNA pol falls off promoter and tries again
happens if RNA pol binds to the wrong promoter, binds wrong or not enough RNA has been made yet to stabilize binding
Happens in the first 8- 10 nucleotides, after
that the association becomes more stable
describe sigma 70
function of the genes it activated
consensus sequence
“house-keeping” genes expressed in all growing cells
-35: TTGACA
spacer is 16-18 bp
-10: TATAAT (perfect -10 sequence)
what sigma factor is responsible for transcribing osmotic shock gene -osmY
how was this discovered?
sigma 38 is responsible
able to figure it out because with increasing osmotic shock, more sigma 38 genes were transcribed and less sigma70 genes were transcribed
describe sigma factor’s mechanism (for all sigma’s except sigma 54)
sigma of holoenzyme binds to DNA
sigma opens up DNA strand
abortive initiation may occur
initiation begins (for real this time)
promoter is cleared
elongation occurs
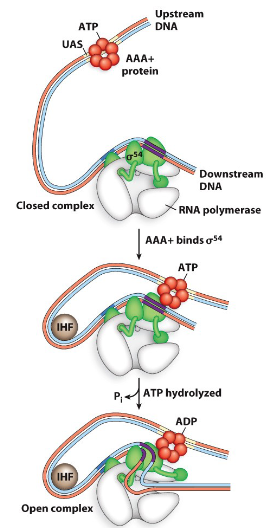
describe sigma factor 54’s mechanism
responsible for nitrogen take up
requires an IHF protein (DNA binding and bending protein)
requires AAA+ protein that hydrolyses a ATP
STEPS
AAA+ binds upstream DNA and then sigma 54
IHF binds the DNA and bends it
AAA+ hydrolyzes an ATP on the DNA
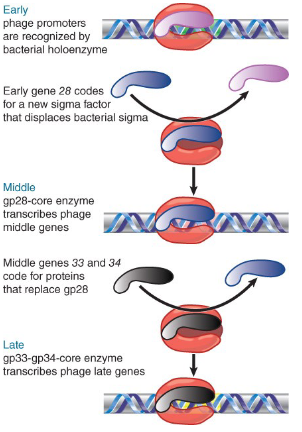
explain what happens when early promoters are transcribed by bacteriophage T4 (cascade effect)
early transcription is performed by host RNA pol, promoter are recognized by host sigma factor (sigma 20)
several genes are transcribed, one makes a phage specific sigma (comes from phage)
explain what happens when middle promoters are transcribed by bacteriophage T4 (cascade effect)
Middle transcription is performed by host RNA polymerase with phage sigma factor made during the early transcription phase
genes in this region code for another new sigma factor
explain what happens when late promoters are transcribed by bacteriophage T4 (cascade effect)
Late transcription if performed by host RNA
polymerase and phage sigma factor made
during the middle transcription phase
explain what happens when late promoters are transcribed by bacteriophage T4 (cascade effect)
Each new factor affinity for new promoters and therefore transcription from those genes starts, the order of transcription cannot be changed as each step is dependent on what happened at the previous step
explain cascade transcription in sporulation
Transcription of phage SPO1 genes is controlled
by two successive substitutions of the sigma
factor that change the initiation specificity.
what are antiterminators?
regions that contain a terminator that no longer
read as terminators usually because there has been a modification to RNA polymerase so it no longer recognizes the terminator sequence
control transcription
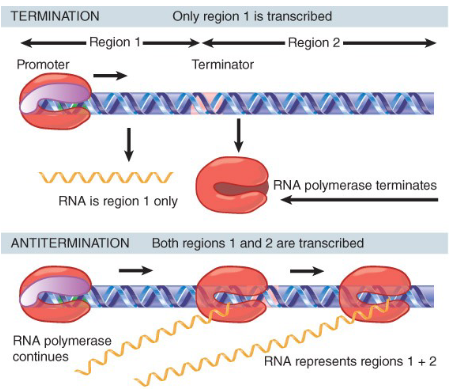
how do antiterminators affect RNA pol?
An anti-termination protein can act on RNA polymerase to enable it to read through a specific terminator (helps it ignore the terminator)
keeps RNA pol bound to DNA
allows termination to occur at the 2nd termination signal (bypasses the first)