Genetics of Microorganisms Exam 1
1/373
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
374 Terms
Friedrich Miescher
identified DNA + RNA (nucleic acids) in 1869 in waste surgical bandages
named it “nuclein” - material from nucleus of cell
later research showed DNA (and RNA) release H+ in water, making them acids
led to the rename “nucleic acids”
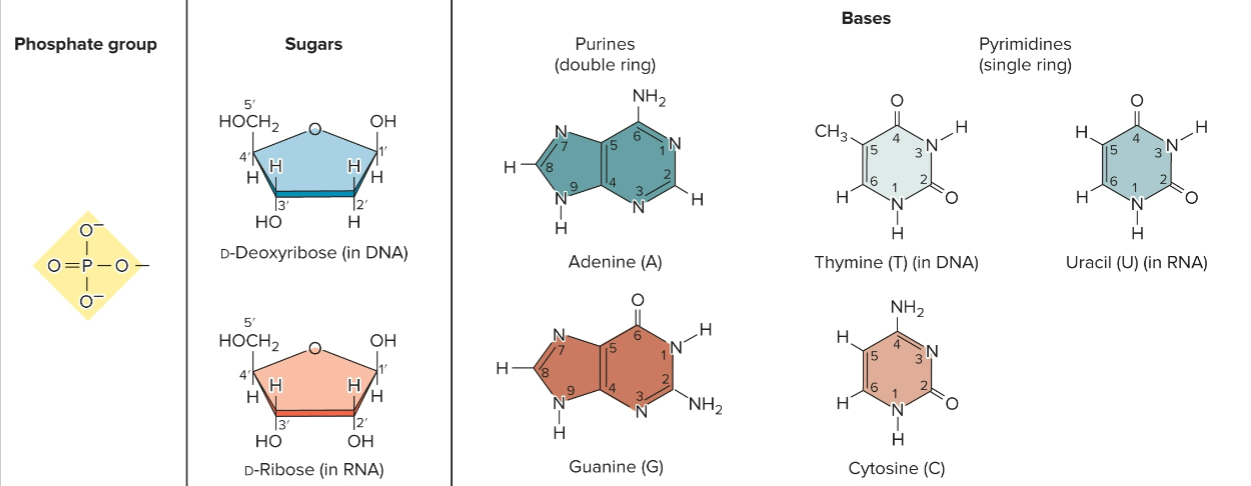
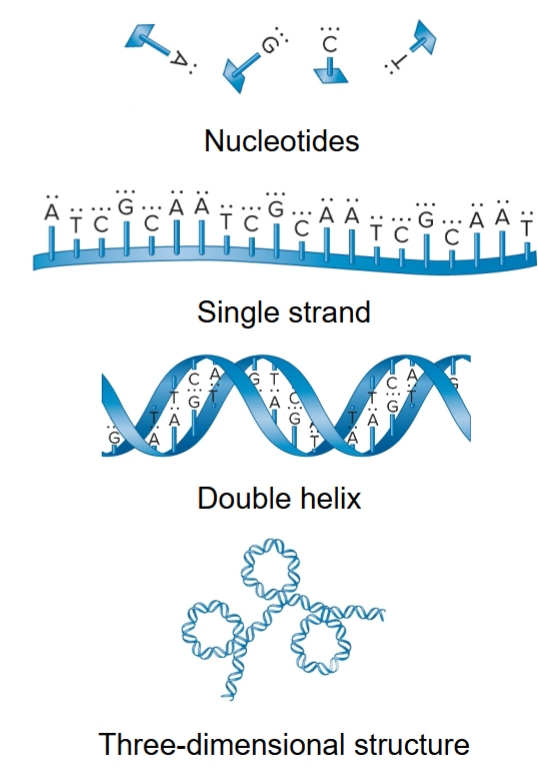
nucleotides
form repeating unit of nucleic acids, linked by ester (covalent) bonds, repeating structural unit of RNA/DNA
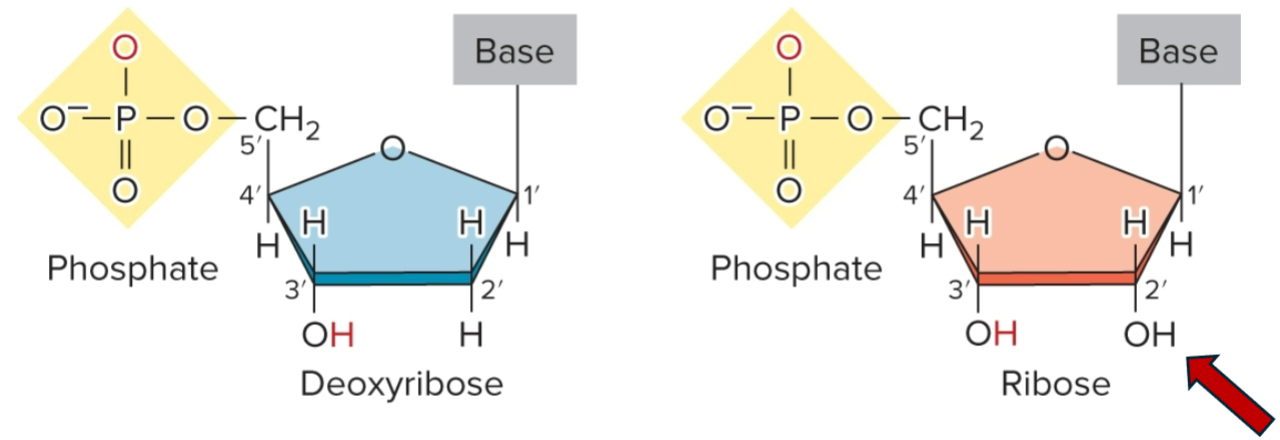
composed of phosphate group, pentose sugar (ribose/deoxyribose) and a nitrogenous base
base + sugar + phosphate; AMP, ADP, ATP
phosphodiester linkage
phosphate connects 5′ C of nucleotide to 3′ C of adjacent nucleotide
nucleoside
base + sugar
adenine + ribose = Adenosine
adenine + deoxyribose = Deoxyadenosine
DNA
3D structure from folding and bending of double helix
interaction with proteins produces chromosomes
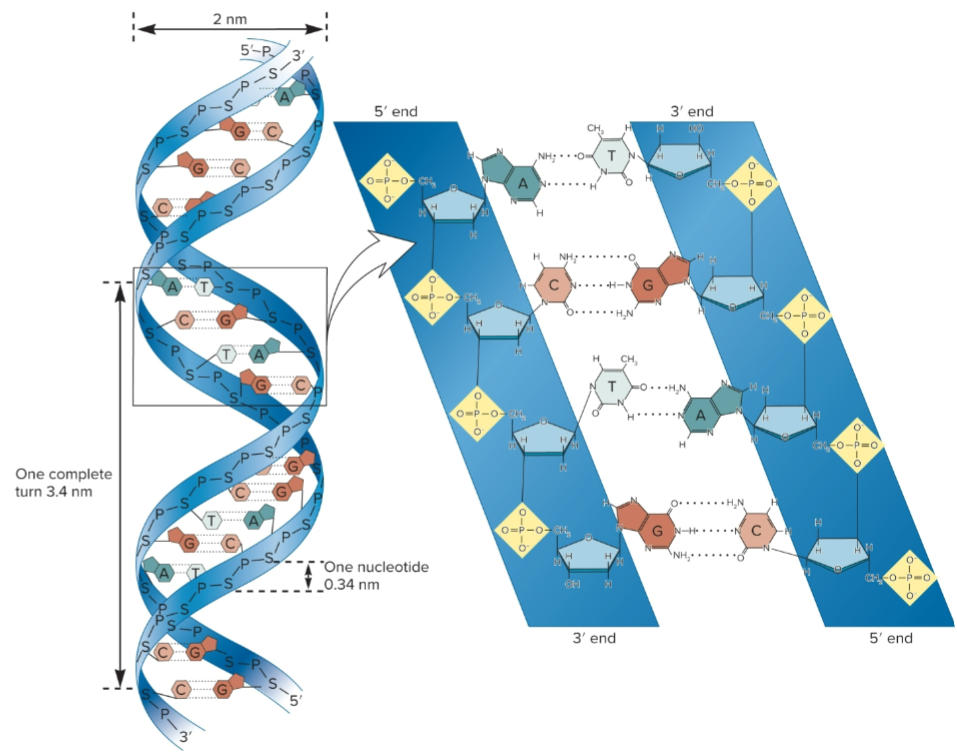
5′ to 3’, all sugar molecules oriented in same direction
phosphates and sugar form backbone of nucleic acid strand and bases project from backbone
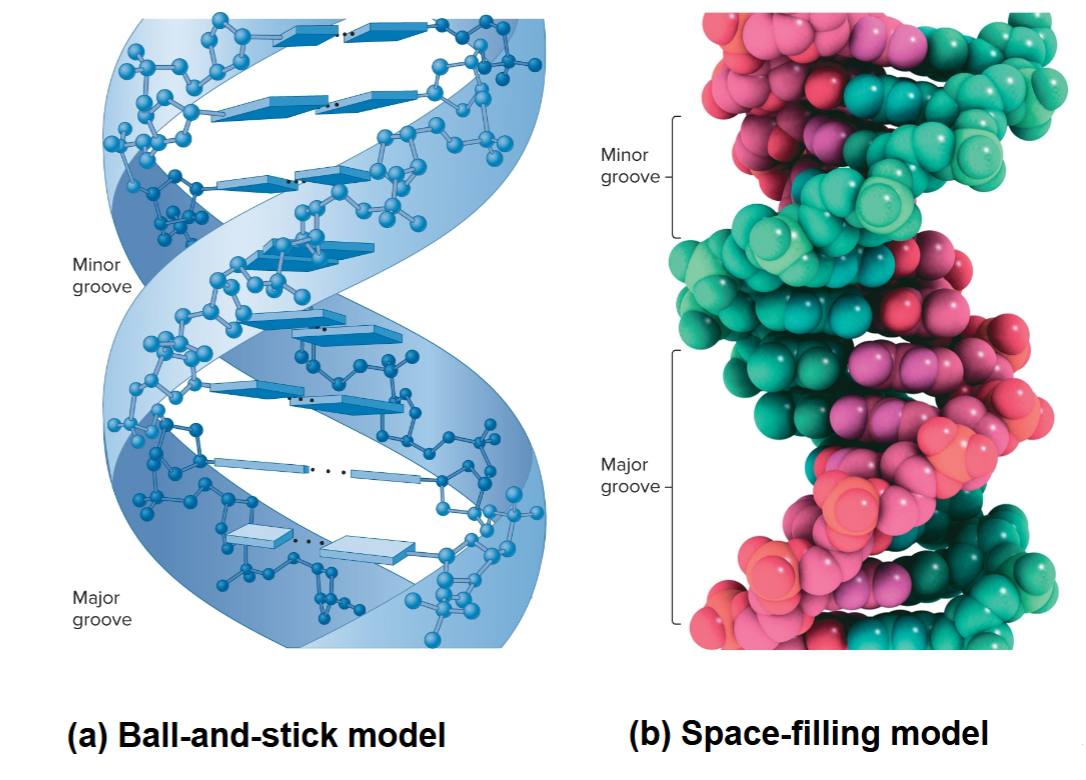
DNA structure
10 base pairs (bp) in each strand and 3.4 nm per complete turn of helix
2 strands are antiparallel, one is 5′ to 3′ and other 3′ to 5′
right-handed, as it spirals away, it turns clockwise
A-T bonded by 2 H-bonds and G-C by 3 H-bonds
Base stacking, bases oriented so flattened regions face each other
2 asymmetrical (major/minor) groves on outside that allow for protein interaction with specific sequences
Ball-and-stick vs space-filling model of DNA
structure of deoxyribonucleic acid (DNA) vs ribonucleic acid (RNA)
James Watson and Francis Crick
In 1953, they elucidated double helical structure of DNA
initially tried to build ball-and-stick models that incorporated all known experimental observations
original model had sugar-phosphate backbone on outside and bases projecting toward each other with bases forming H bonds with identical bases in the opposite strand
later realized that H bonding of A to T was structurally similar to that of C to G
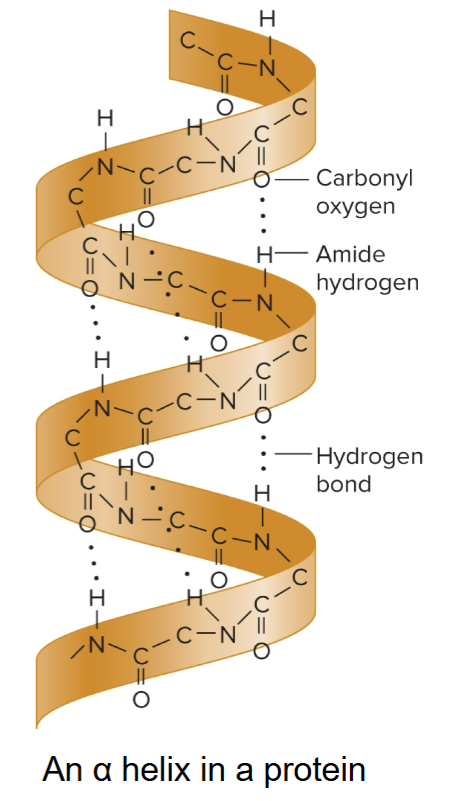
Linus Pauling
in 1950s, proposed that regions of protein can fold into a secondary structure called an α-helix
built ball-and- stick models to elucidate this structure
incorrectly proposed triple helix DNA structure
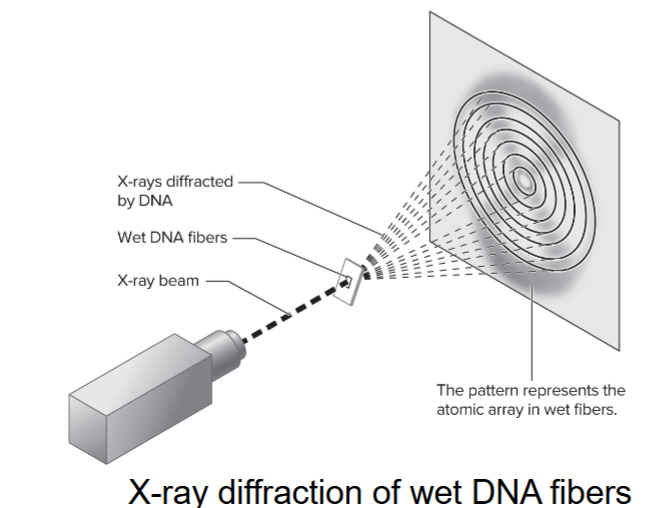
Rosalind Franklin
used X-ray diffraction to study wet fibers of DNA
diffraction pattern is interpreted (using mathematical theory) to provide info about the structure of a molecule
made advances in X-ray diffraction techniques with DNA
diffraction pattern showed DNA was helical, had more than 1 strand and 10 base pairs per complete turn
Erwin Chargaff
pioneered biochemical techniques for isolation, purification and measurement of nucleic acids from cells
analyzed base composition of DNA isolated from many different species
when base compositions from different tissues within same species were measured, there were similar results
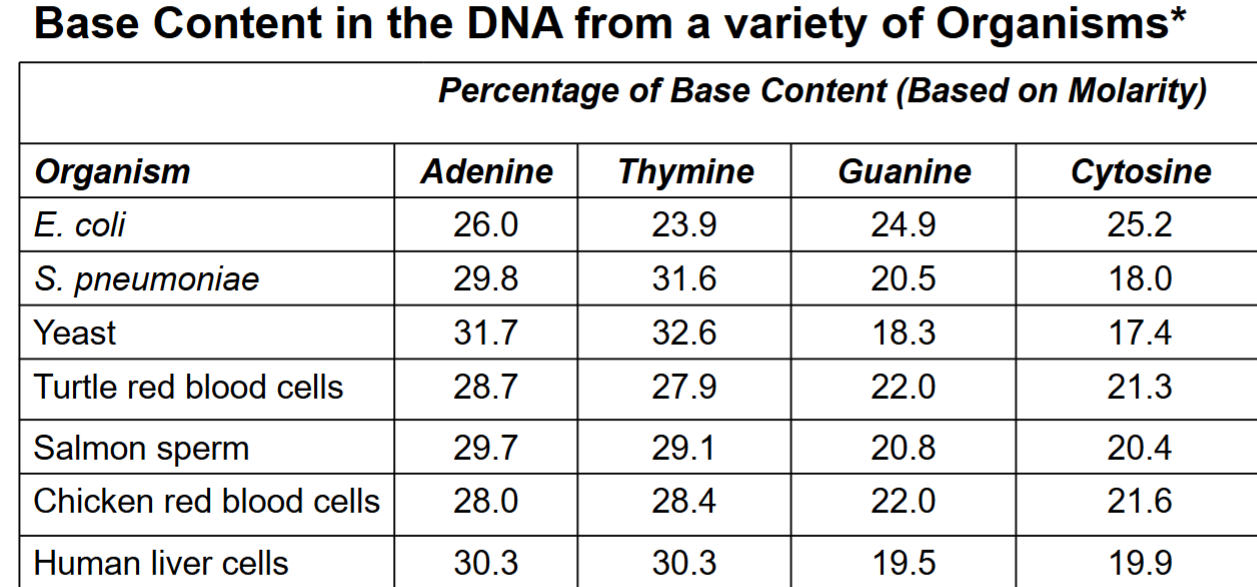
Chargaff’s rule
% adenine = % thymine
% cytosine = % guanine
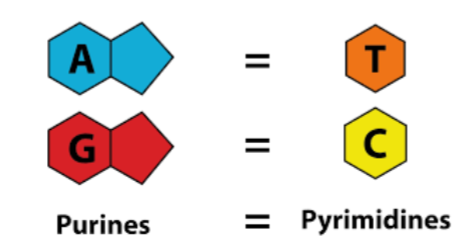
B-form DNA (DNA secondary structure)
predominant form found in living cells
bases relatively perpendicular to central axis
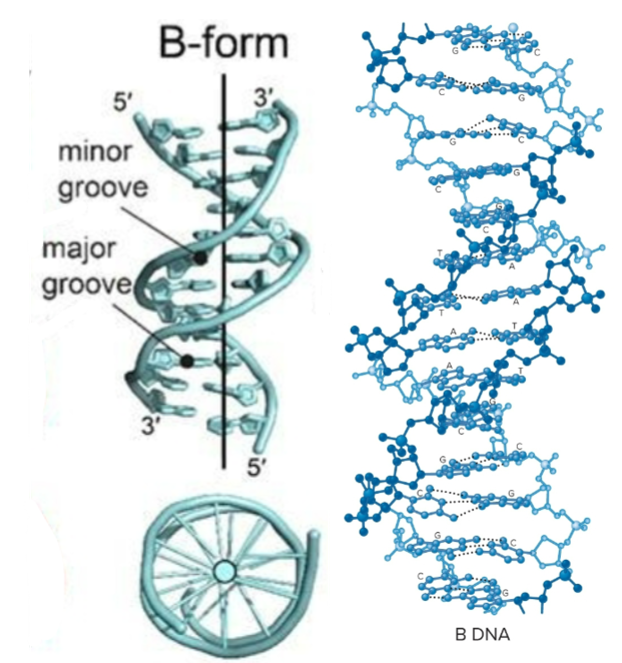
A-form DNA (DNA secondary structure)
RNA and DNA-RNA hybrids
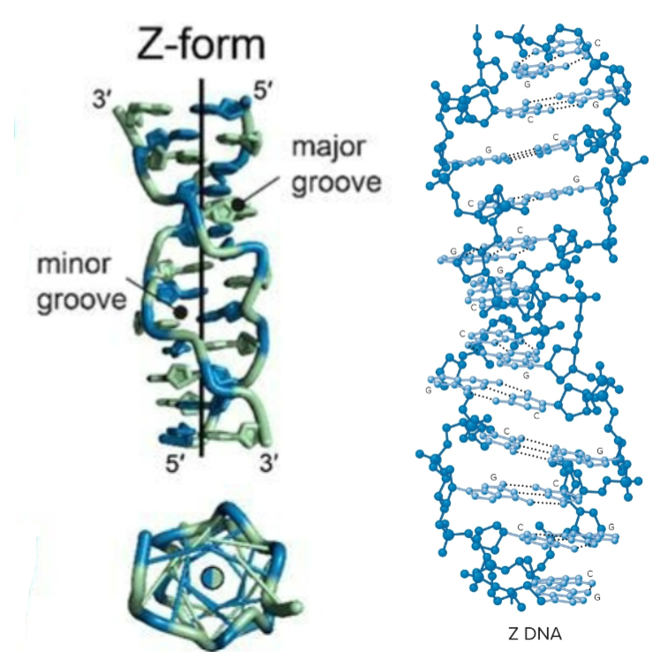
Z-form DNA (DNA secondary structure)
Left-handed helix, 12 bp per turn
formation favored by
alternating purine/pyrimidine sequences (GCGCGCGCGC), at high salt concentrations
cytosine methylation, at low salt concentrations
Negative supercoiling
may have role in transcription and chromosome structure
recognized by cellular proteins, may alter chromosome compaction
bases substantially tilted relative to central axis
sugar-phosphate backbone follows zigzag pattern
RNA
usually single-stranded, can form short double-stranded regions
double-stranded secondary structure forms due to complementary base-pairing (A/U and C/G)
double helices are right-handed and have 11-12 base pairs per turn
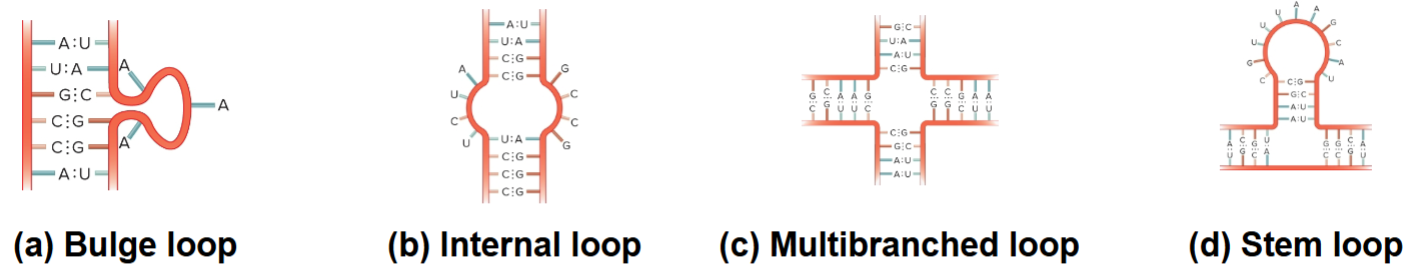
4 RNA Loop Structures
Bulge loop
Internal loop
Multibranched loop
Stem loop
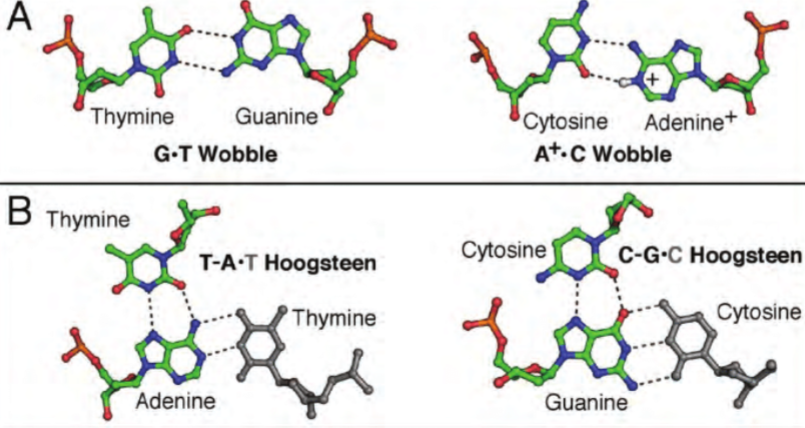
Non-canonical base pairs
Planar hydrogen bonded pairs of nucleobases
H-bonding patterns that differ from patterns observed in Watson-Crick base pairs (classic double helical DNA)
Recombinant DNA technology
use of in vitro molecular techniques to isolate and manipulate fragments of DNA
Recombinant DNA technology and gene cloning have been fundamental to our understanding of gene structure and function
recombinant DNA molecules
chimeric molecules, first constructed in 1970s by researchers at Stanford
can be introduced into living cells where they are replicated to make many identical copies, led to era of gene cloning
gene cloning
technique of isolating and making many copies of a gene; refers to use of vectors
devised during the early 1970s
includes DNA sequencing, DNA probes and expression of cloned genes
Chromosomal DNA
serves as source of DNA segment of interest
must obtain cellular tissue from organism, break open cells, and extract and purify DNA using a biochemical techniques to obtain
Vector DNA
carrier for DNA segment that is to be cloned
can replicate independently of host chromosomal DNA
vector
host cell harbors the vector
when a vector is replicated inside host cell, the DNA that it carries is also replicated
the sequence of Ori determines if a vector can replicate in a particular host cell
originally derived from two natural sources
Plasmids: have selectable markers; genes conferring antibiotic resistance to host cell
Viruses: infect living cells and propagate by taking control of host cell’s machinery
restriction endonucleases/enzymes
enzymes that cut DNA, allowing insertion of chromosomal DNA into vector
made naturally by many species of bacteria
protect bacterial cells from invasion by foreign DNA, particularly bacteriophages
bind to specific DNA sequences and cleave DNA at 2 defined locations, one in each strand
some digest DNA into fragments with “sticky ends” or generate blunt ends; NaeI cuts in middle of recognition sequence
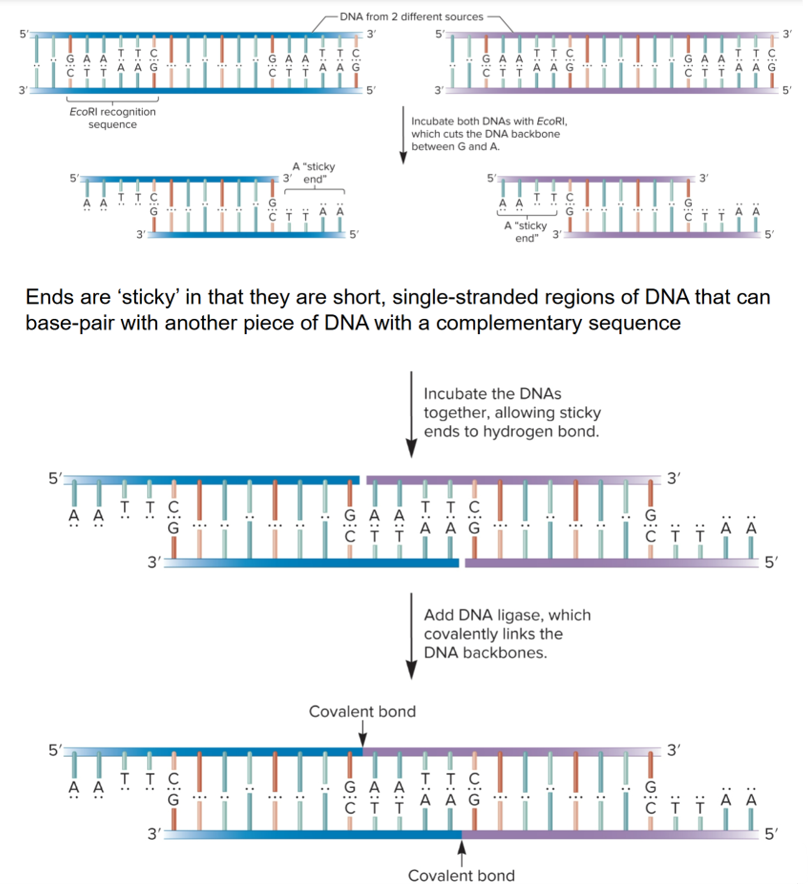
Recognition sequences
palindromic, identical when read in opposite direction in complementary strand
several hundred different restriction enzymes are available commercially
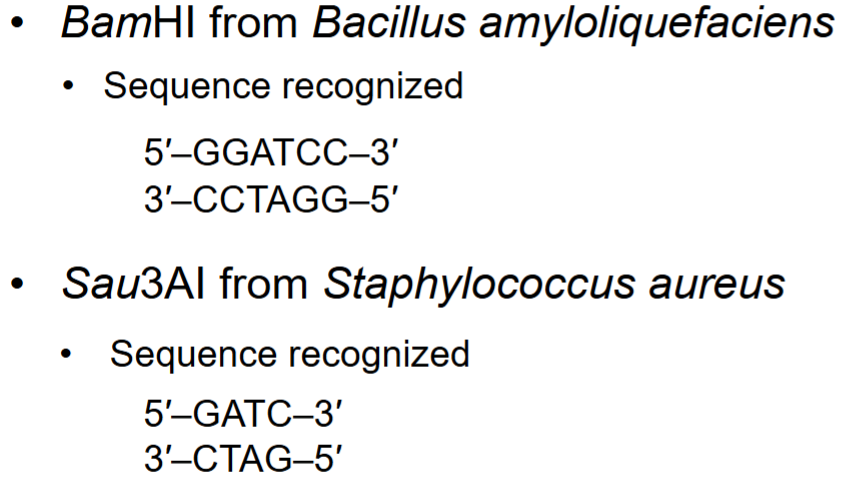
DNA ligase
covalently links sugar-phosphate backbone of DNA molecules with sticky or blunt ends in Okazaki fragments together by catalyzing formation of covalent (ester) bond
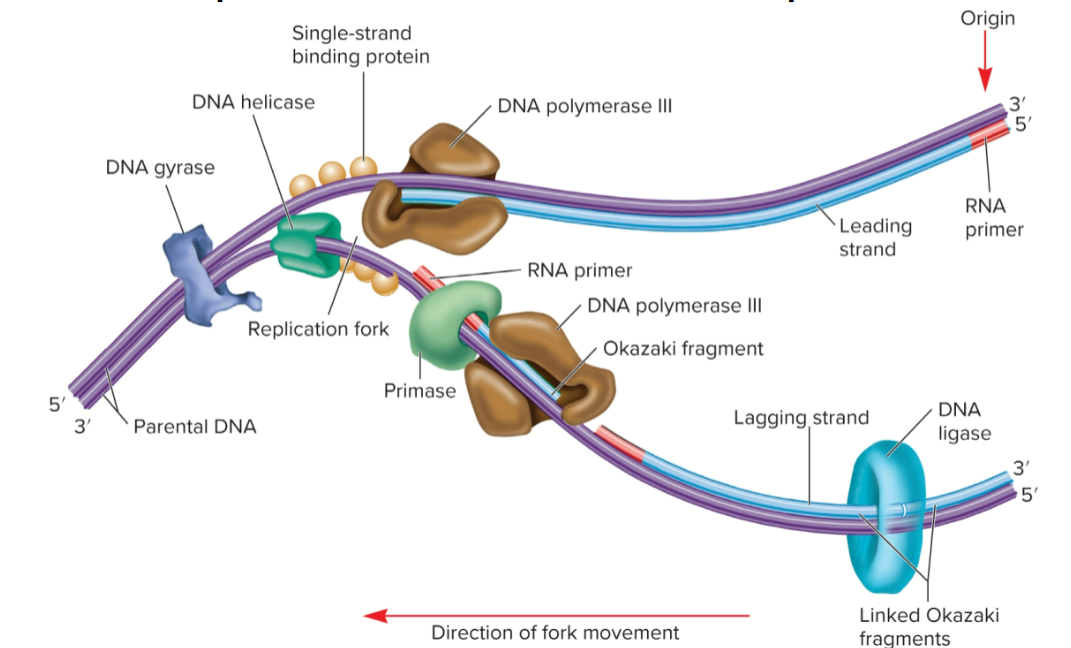
polymerase chain reaction (PCR)
can copy DNA without vectors and host cells
must know enough about gene of interest to have sequence of 2 short primers
specific DNA segment can be amplified
also used to amplify chromosomal DNA nonspecifically which uses mix of primers with many different random sequences
these will anneal randomly throughout genome and amplify most chromosomal DNA
used to amplify very small samples, ex) crime scene DNA
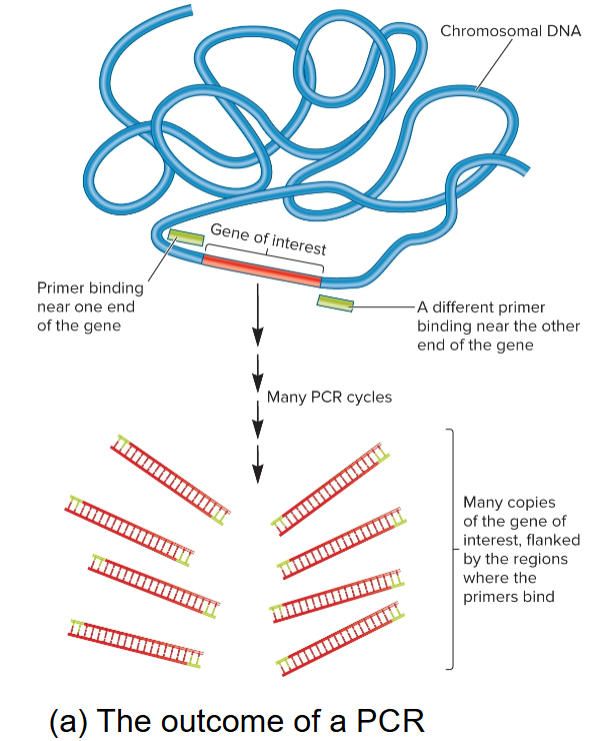
4 starting materials for PCR
Template DNA: contains region to be amplified
Oligonucleotide primers: complementary to sequences at ends of DNA fragment to be amplified, synthetic, 15-20 nucleotides long
Deoxynucleoside triphosphates (dNTPs): provides precursors for DNA synthesis
Taq polymerase (or other polymerase): DNA pol isolated from bacterium Thermus aquaticus; thermostable enzyme, used bc PCR involves heating steps that inactivates most other DNA pols
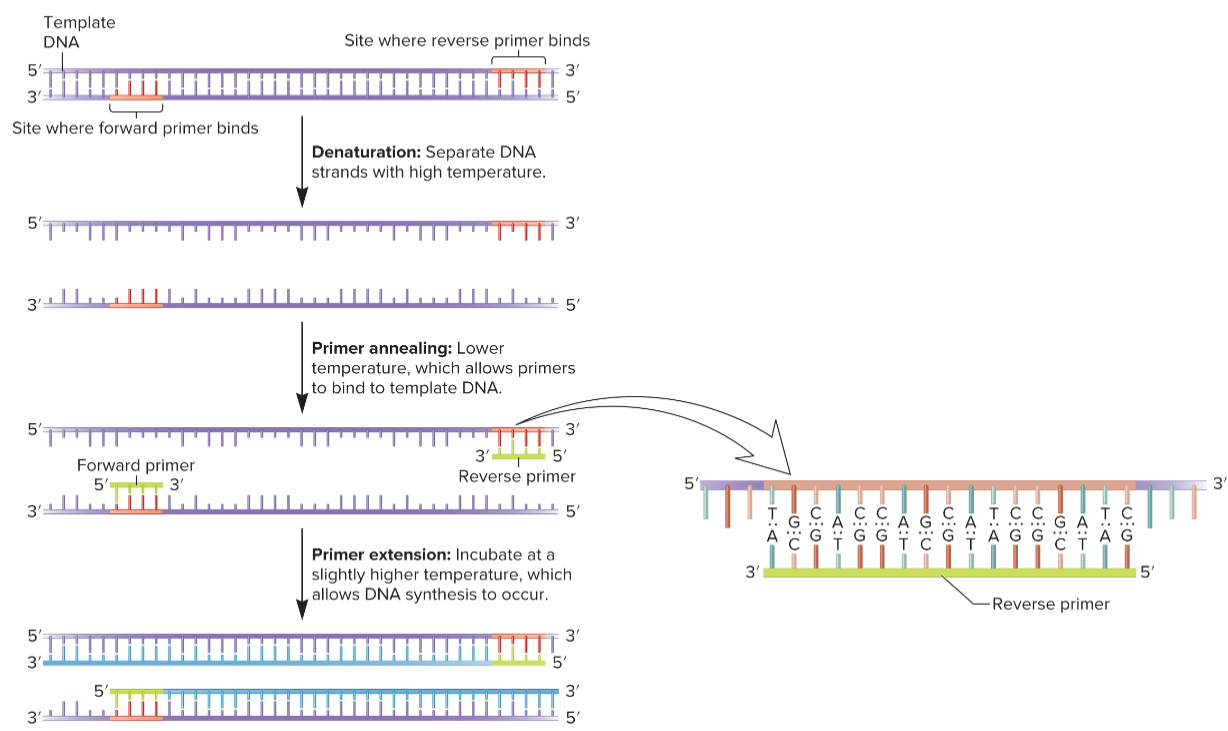
PCR steps
carried out in thermocycle, all reagents in one tube
sequential process of denaturing-annealing-synthesis repeated for many cycles, typically 20-30 cycles of replication, taking a few hours
after 20 cycles, target DNA sequence increase 220-fold (1 million-fold)
after 30 cycles, target DNA sequence increase 230-fold (1 billion-fold), assuming 100% efficiency (not real)
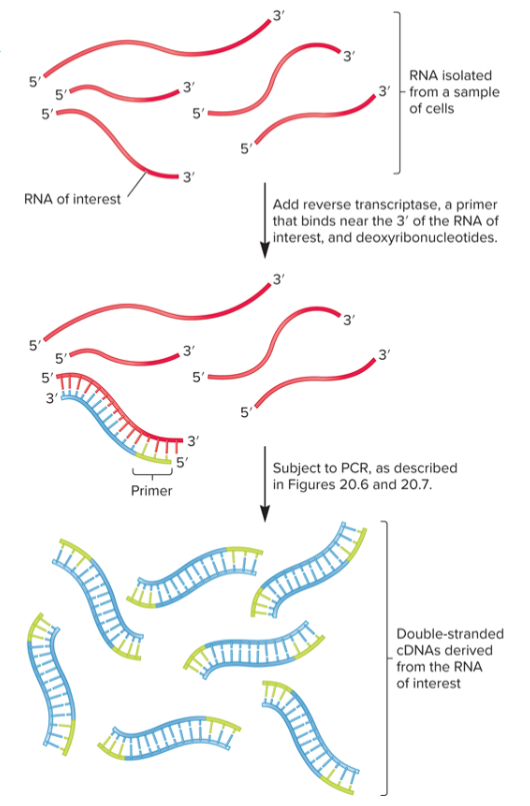
Reverse transcriptase PCR (RT-PCR)
used to detect and quantitate amount of RNA in live cells
RNA is isolated from sample then mixed with reverse transcriptase
primer will anneal to 3’ end of RNA, generating single-stranded cDNA that can be used as template DNA in PCR
extraordinarily sensitive, can detect expression of small amounts of RNA in a single cell
Quantitative PCR (qPCR)
used to quantitate amount of DNA/RNA in a specific gene or mRNA in a sample in real time
carried out in thermocycler that can measure changes in fluorescence emitted by detector molecules in the PCR reaction mix
will increase in proportion to amount of PCR product produced
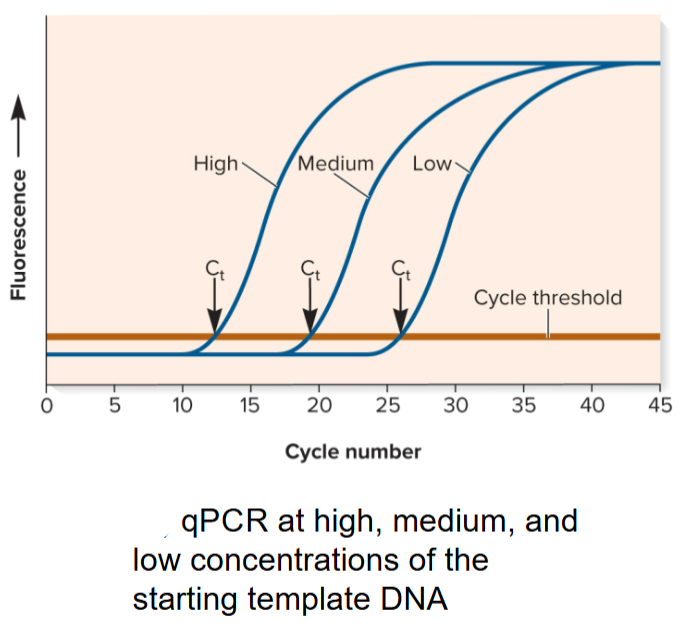
Cycle Threshold (Ct) in qPCR
reached when accumulation of fluorescence is significantly greater than background fluorescence
depends on initial concentration of template DNA
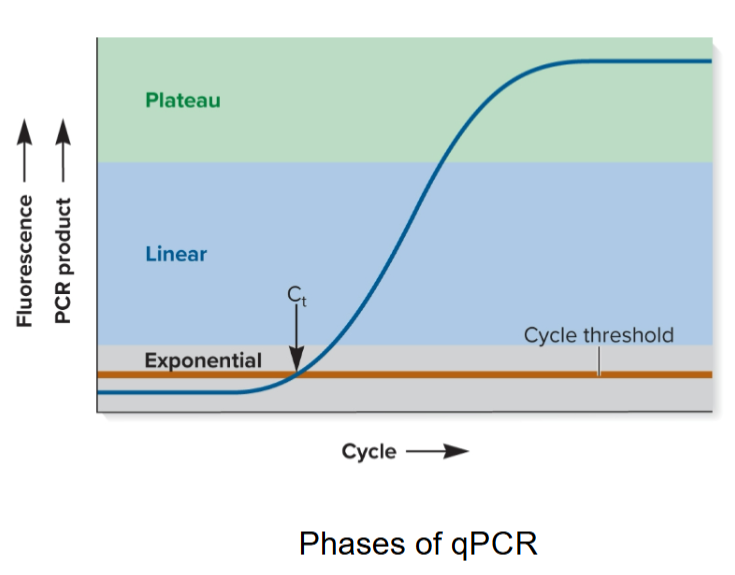
5 steps of qPCR
Initially, little product is made so no increase in fluorescence detected
When reagents are not limiting, product doubles with every cycle (exponential phase); Cycle Threshold (Ct) is reached.
Changes to linear phase when reagents are somewhat limiting
Reaction eventually plateaus when one or more reagents are used up
Concentration of unknown amount of starting DNA (or RNA) can be determined by comparing Ct with known standards by adding a known amount of DNA or amplifying another gene in sample
Chromosomes
structures that contain genetic material, complexes of DNA and proteins
genome in proks vs euks
all genetic material an organism possesses
proks: single circular chromosome, chloroplast genome
euks: one complete set of nuclear chromosomes, mitochondrial genome
4 things DNA sequences are necessary for
Synthesis of RNA and cellular proteins
Replication of chromosomes
Proper segregation of chromosomes
Compaction of chromosomes so they can fit within cells
Prokaryotic Chromosomes
most are circular w/ a few million nucleotides/bp in length
bacterial chromosome contains thousands of genes, majority of which is protein-coding genes
most contain single type of chromosome, but it may be in multiple copies
several thousand different genes and repetitive sequences interspersed throughout
at least 1 Ori to initiate DNA replication
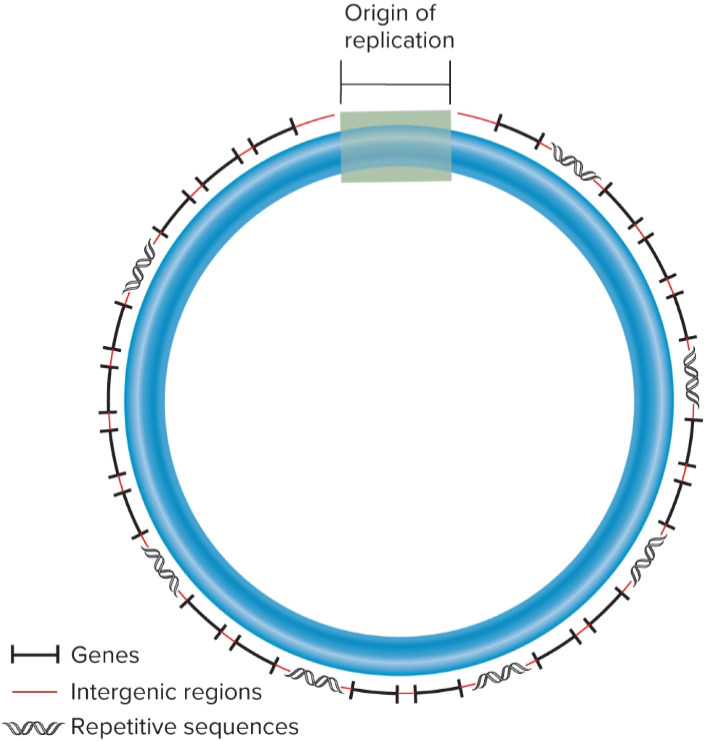
intergenic regions
non-transcribed DNA between adjacent genes
nucleoids
where prok chromosomes are found
not bounded by membrane
DNA in direct contact with cytoplasm
loop domains (microdomains)
allows chromosomal DNA to be compacted 1000-fold to fit within bacterial cell
10,000 bp
macrodomains
# of loops varies according to size of bacterial chromosome and species
E. coli has 400-500 microdomains
organizes adjacent microdomains in E.coli, 800-1000 kbp

nucleoid-associated proteins (NAPs)
DNA-binding proteins used by bacteria to form microdomains and macrodomains
facilitate chromosome compaction and organization
bend DNA or act as bridges for DNA to bind to other DNA regions
facilitate chromosome segregation
involved in gene regulation
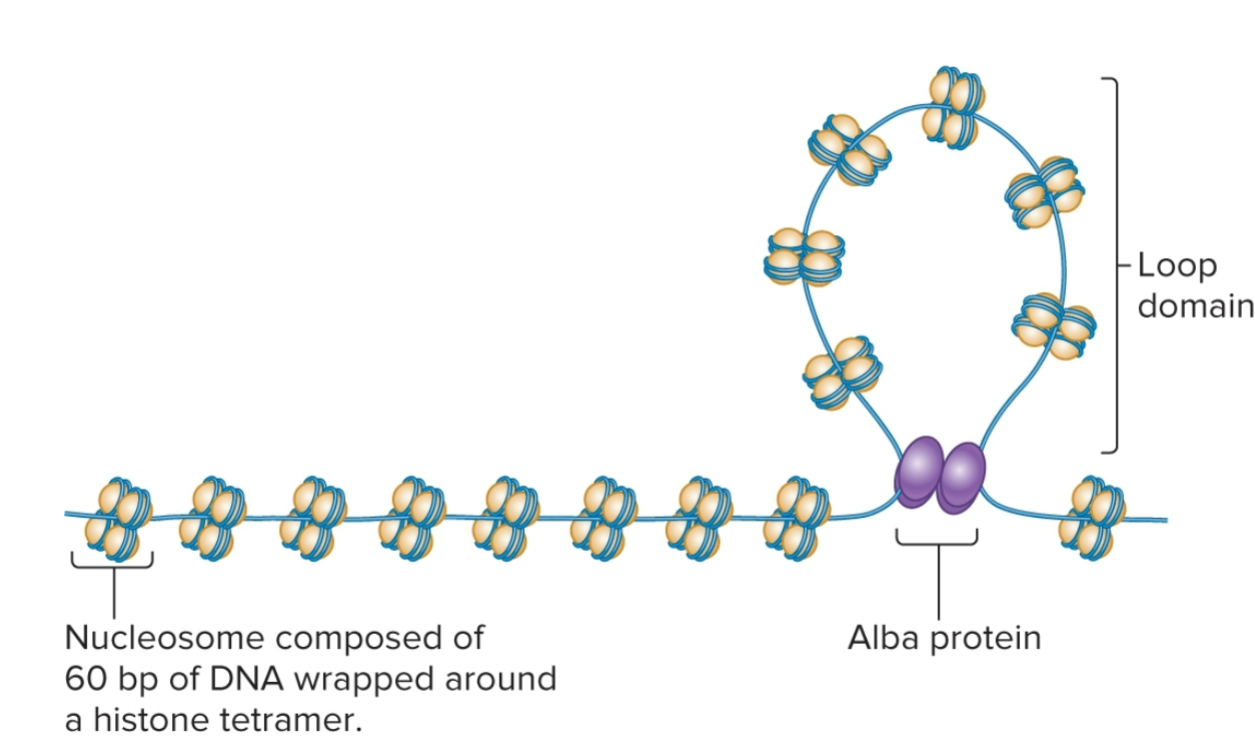
Archaeal Chromosomes
structure varies, depends on DNA-binding proteins expressed
some produce bacterial-like nucleoid-associated proteins or eukaryotic-like histone proteins
DNA wrapped around histone proteins to form nucleosomes and organized into loop domains
# of histone proteins varies among diff species, in some, structure is similar to eukaryotic chromatin
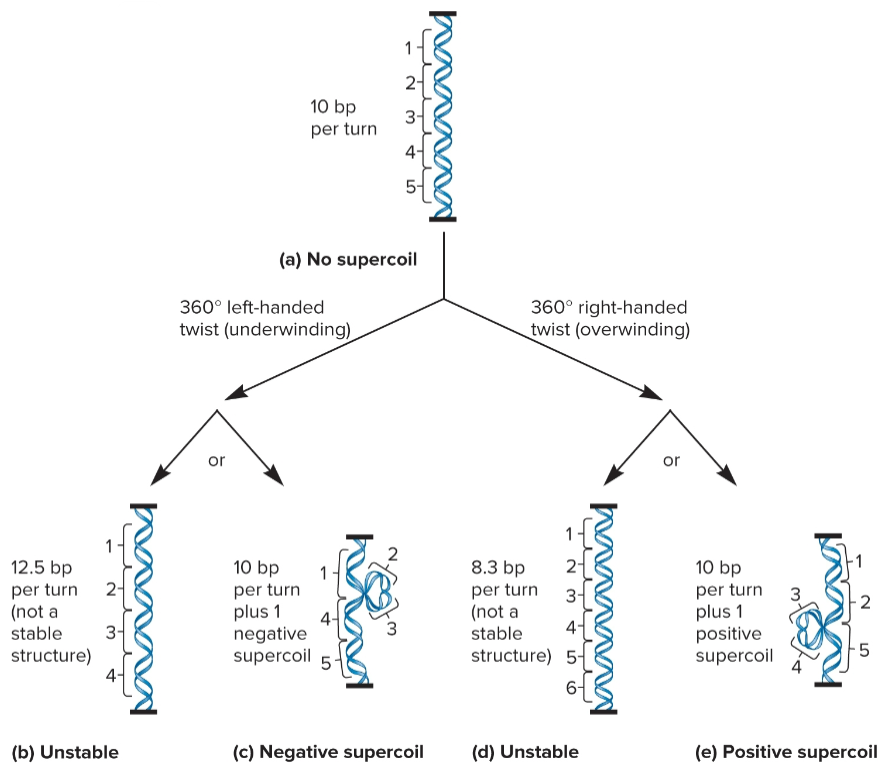
DNA supercoiling
way for prokaryotic chromosomes to become more compact
formation of additional coils due to twisting forces where the 2 strands within DNA already coil around each other
caused by both underwinding and overwinding of DNA double helix
negative DNA supercoiling
formed when DNA given a turn that unwinds helix (left)
can also cause fewer turns, topoisomers
chromosomal DNA in bacteria is negatively supercoiled
In E. coli, 1 - supercoil per 40 turns of double helix
Helps compaction of chromosome
In localized regions, creates tension that may be released by DNA strand separation which it also promotes
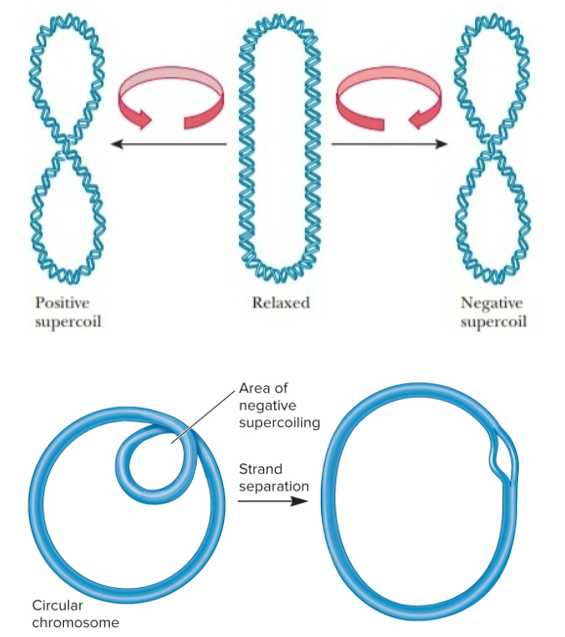
positive DNA supercoiling
formed when DNA given a turn that overwinds the helix (right)
can also cause more turns
topoisomers
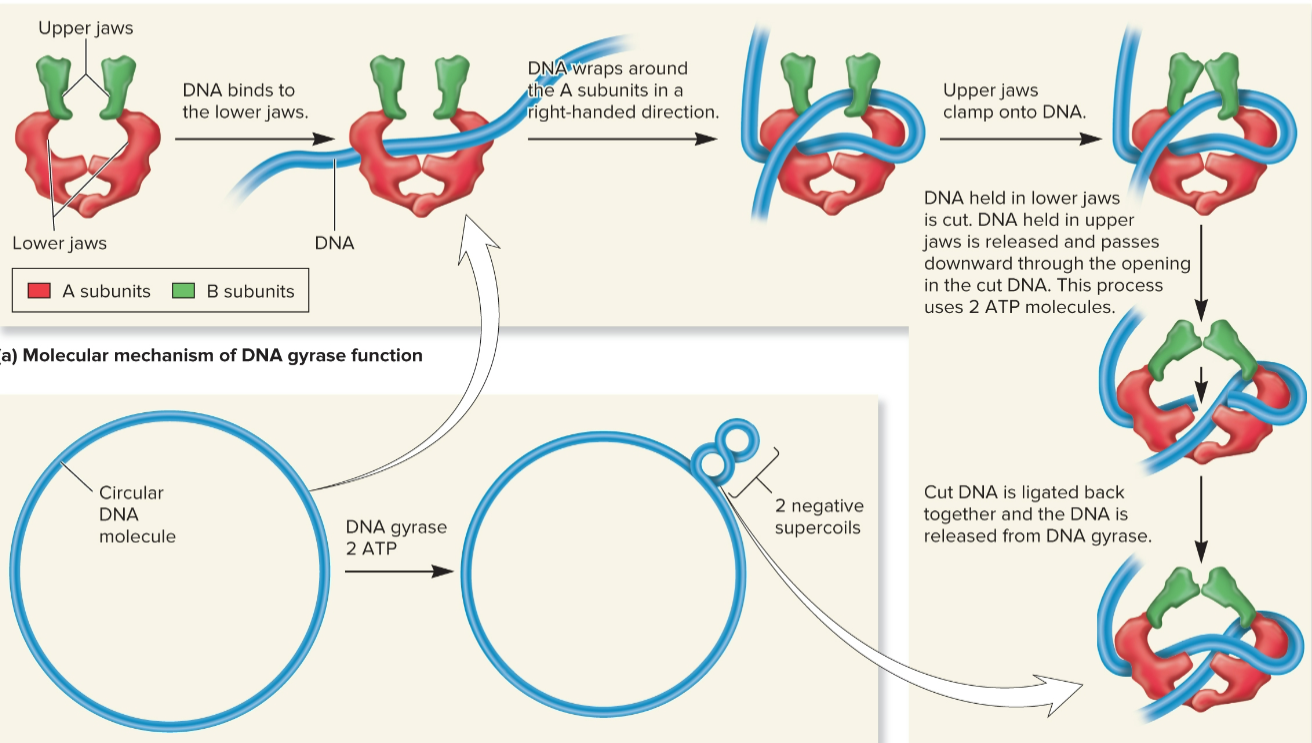
DNA gyrase (DNA topoisomerase II)
introduces - supercoils using energy from ATP
travels ahead of helicase and alleviates + supercoils
can untangle intertwined DNA molecules
crucial for bacterial survival
DNA topoisomerase I
relaxes - supercoils
2 DNA Gyrase Inhibitors
blocking gyrase can treat bacterial diseases
Quinolones: ex) Ciprofloxacin: used in treatment of anthrax, Coumarins
do not inhibit euk topoisomerases
Eukaryotic Chromosomes
contain ≥1 sets of chromosomes composed of several different linear chromosomes, many species are diploid
contains a single, linear molecule of DNA with tens of to hundreds of millions of bps and a few hundred to several thousand genes interspersed throughout
in simpler euks, genes are short (several hundred bp)
in complex euks, genes are long with many introns w/ lengths from <100 to >10,000 bp
require Ori, centromeres, and telomeres for chromosomal replication and segregation
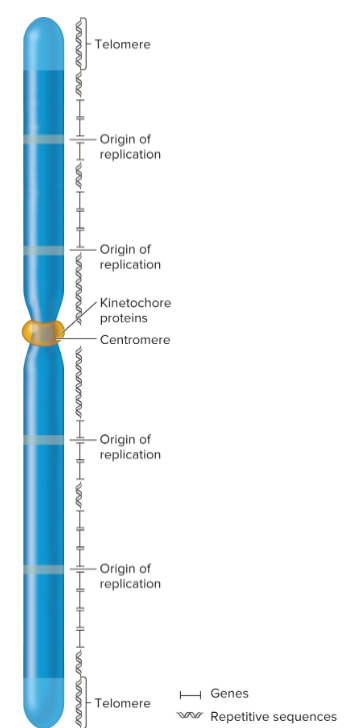
intron
noncoding intervening sequences, interrupts exons
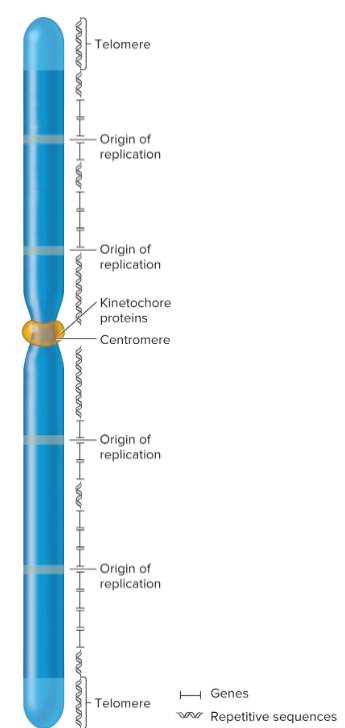
Origins of replication
chromosomal sites necessary to initiate DNA replication
eukaryotic chromosomes contain many origins
interspersed every 100,000 base pairs
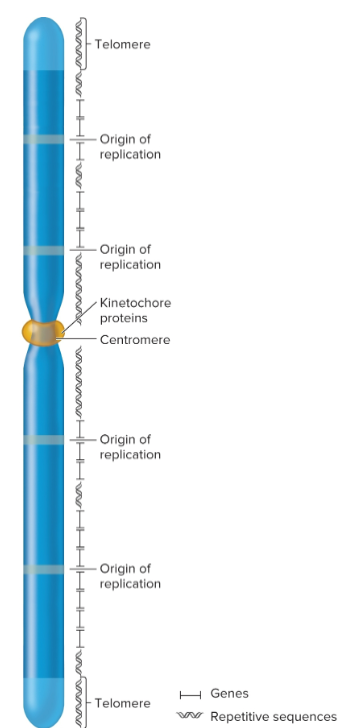
centromeres
regions that play a role in segregation of chromosomes
forms a recognition site for kinetochore proteins
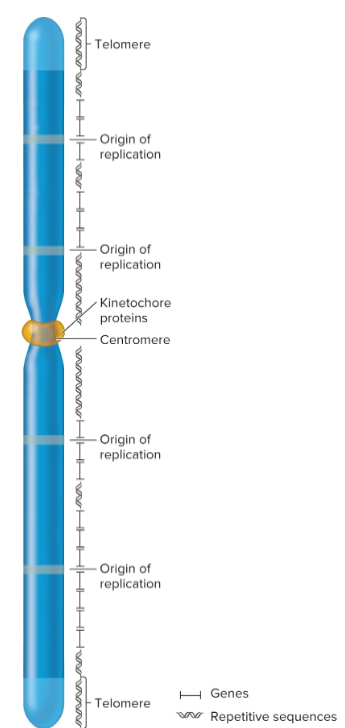
kinetochore
required for centromere linkage to spindle apparatus during mitosis and meiosis
Telomeres
specialized regions at both ends of chromosomes
important in replication and for stability
complex of telomeric DNA sequences and bound proteins
sequences have moderately repetitive tandem arrays; 3′ overhang that’s 12-16 nucleotides long, several guanine nucleotides and many thymine nucleotides
tend to shorten in actively dividing cells; 8,000 bp at birth but can shorten to 1,500 in elderly
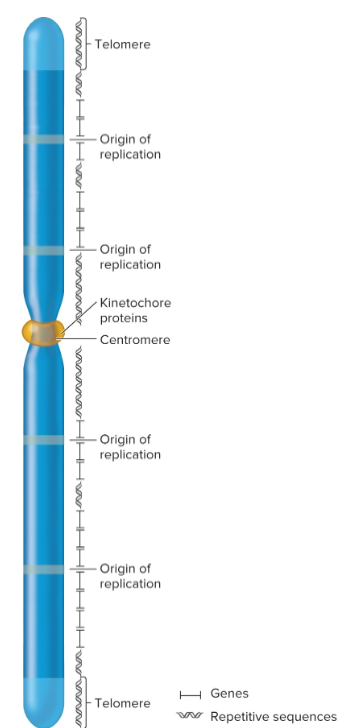
repetitive sequences
commonly found near centromeric and telomeric regions, but they may also be interspersed throughout chromosome
eukaryotic Genomes size
total amount of DNA much greater than bacterial cells
vary substantially in size, variation not related to complexity of species
difference in size due to accumulation of repetitive DNA sequences which do not code proteins
the plant Tmesipteris oblanceolata has 160 billion bps
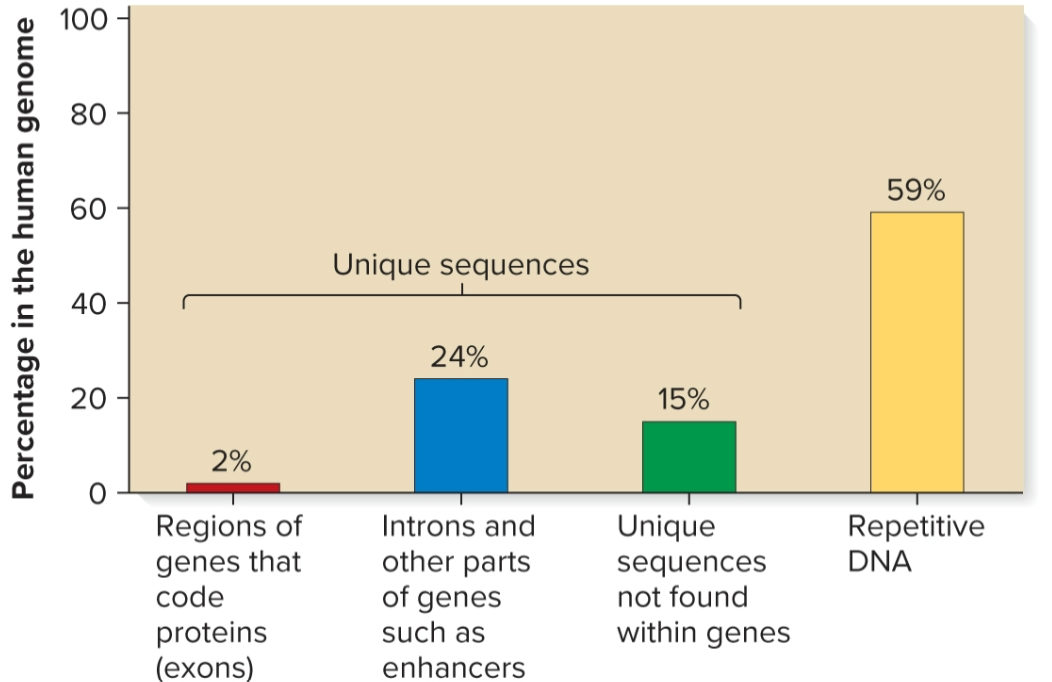
Sequence complexity
number of times a particular base sequence appears in genome
unique/non-repetitive, moderately repetitive, highly repetitive
Unique/non-repetitive sequences
found ≥1 times in genome
includes protein-encoding genes and intergenic regions
in humans, makes up 41% of genome
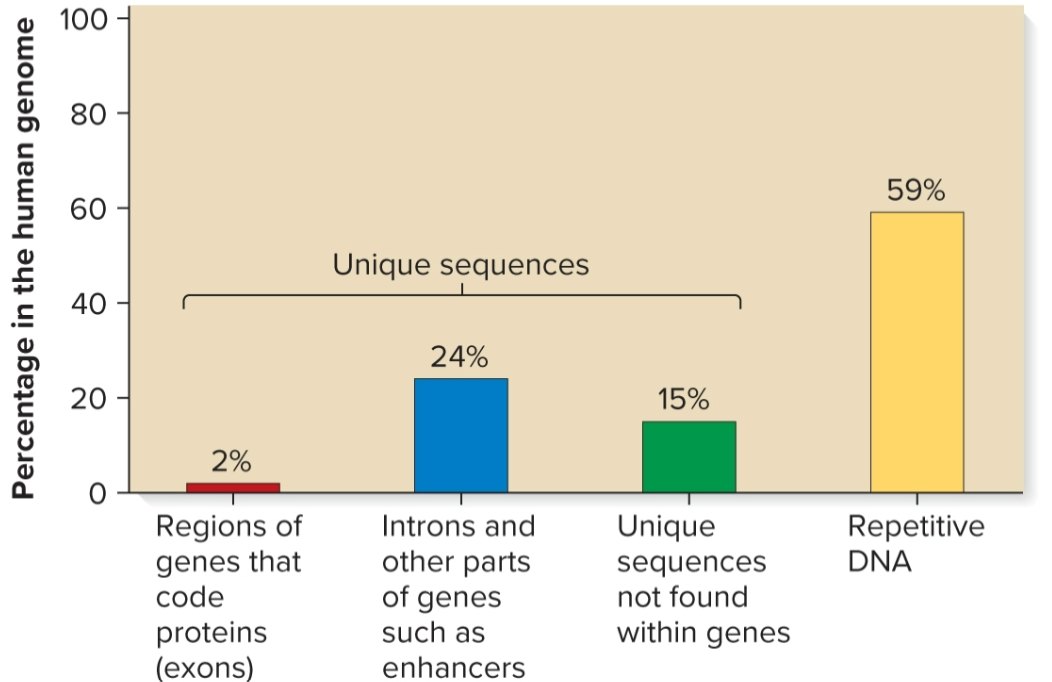
Moderately repetitive sequences
found a few hundred to several thousand times
Genes for rRNA and histones
Sequences that regulate gene expression and translation
Transposable elements
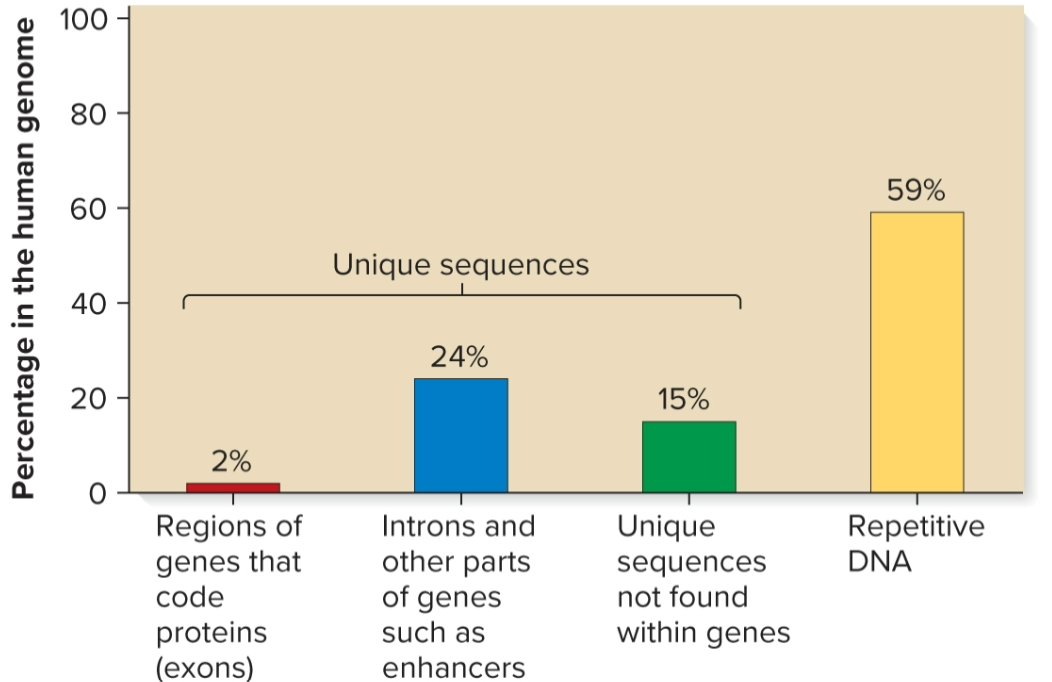
Highly repetitive sequences
found tens of thousands to millions of times
each copy is relatively short (a few nucleotides to several hundred in length)
some sequences interspersed throughout genome, ex) Alu family in humans, 300 bp, 10% of genome, found every 5000–6000 bp
other sequences clustered together in tandem arrays, ex) AATAT and AATATAT sequences in Drosophila found in centromeric regions
Transposition
integration of small segments of DNA into a new location in genome
can occur at many different locations in genome
since many outcomes are likely to be harmful, it is highly-regulated
occurs only in few individuals under certain conditions
transposable elements (TEs) or transposons
“jumping genes” or small, mobile DNA segments
move by different transposition pathways; simple transposition or retrotransposition
can enter genome of organism and proliferate quickly
ex) Drosophila melanogaster: TE called P element introduced in 1950s and expanded throughout D. melanogaster populations worldwide. The only strains without P element are in labs collected prior to 1950
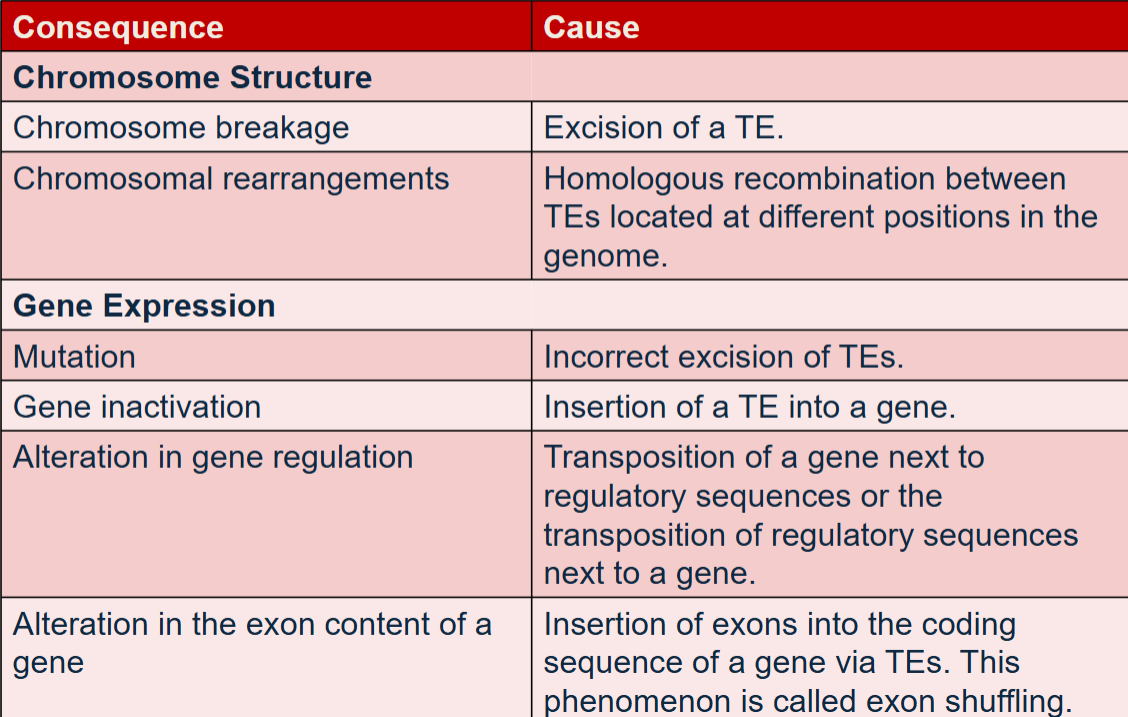
have effects on chromosome structure and gene expression
Agents such as radiation, chemical mutagens and hormones stimulate the movement
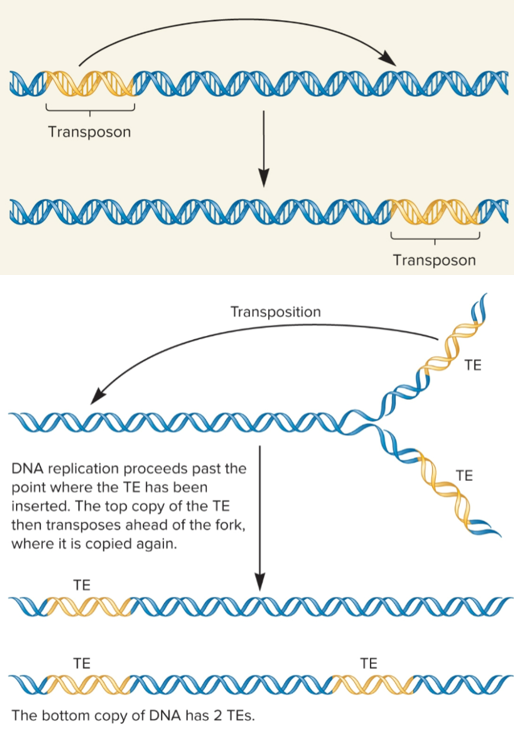
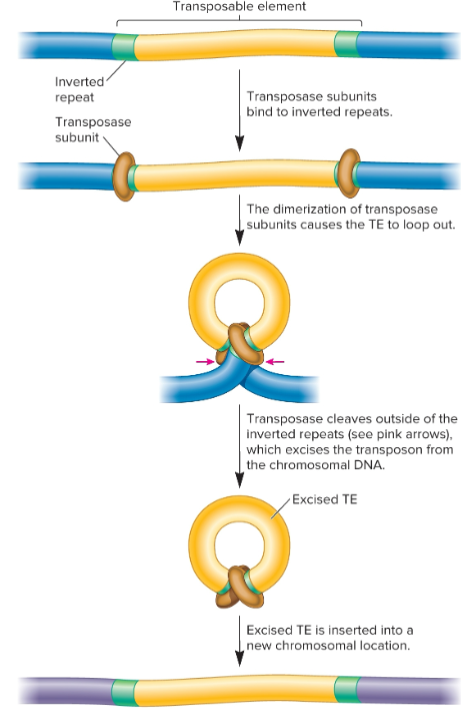
Simple transposition
used widely by transposons in bacterial and euks
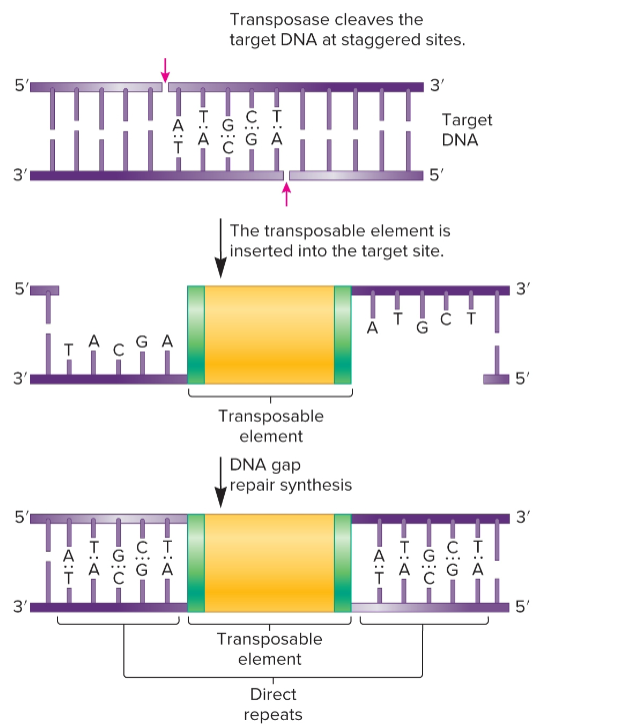
TE removed from original site and transferred to new target site, cut-and-paste mechanism
occurs after replication fork passes through TE, so there are 2 copies of TE
one TE can transpose ahead of fork where it is copied again
one chromosome will still have one TE, but the other will now have two copies
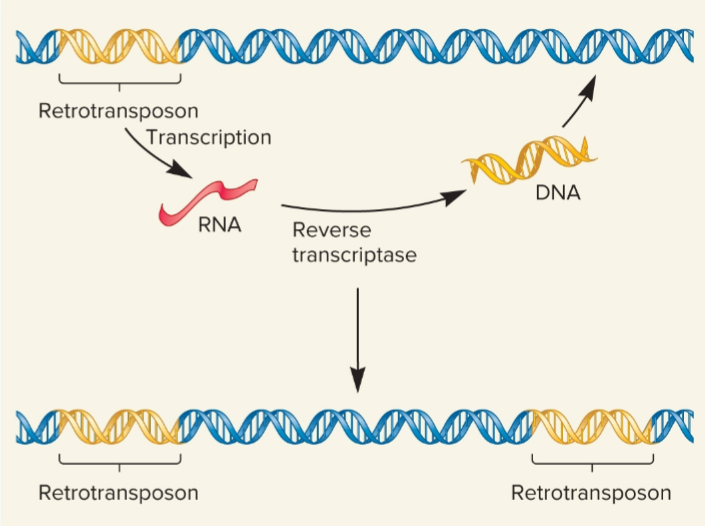
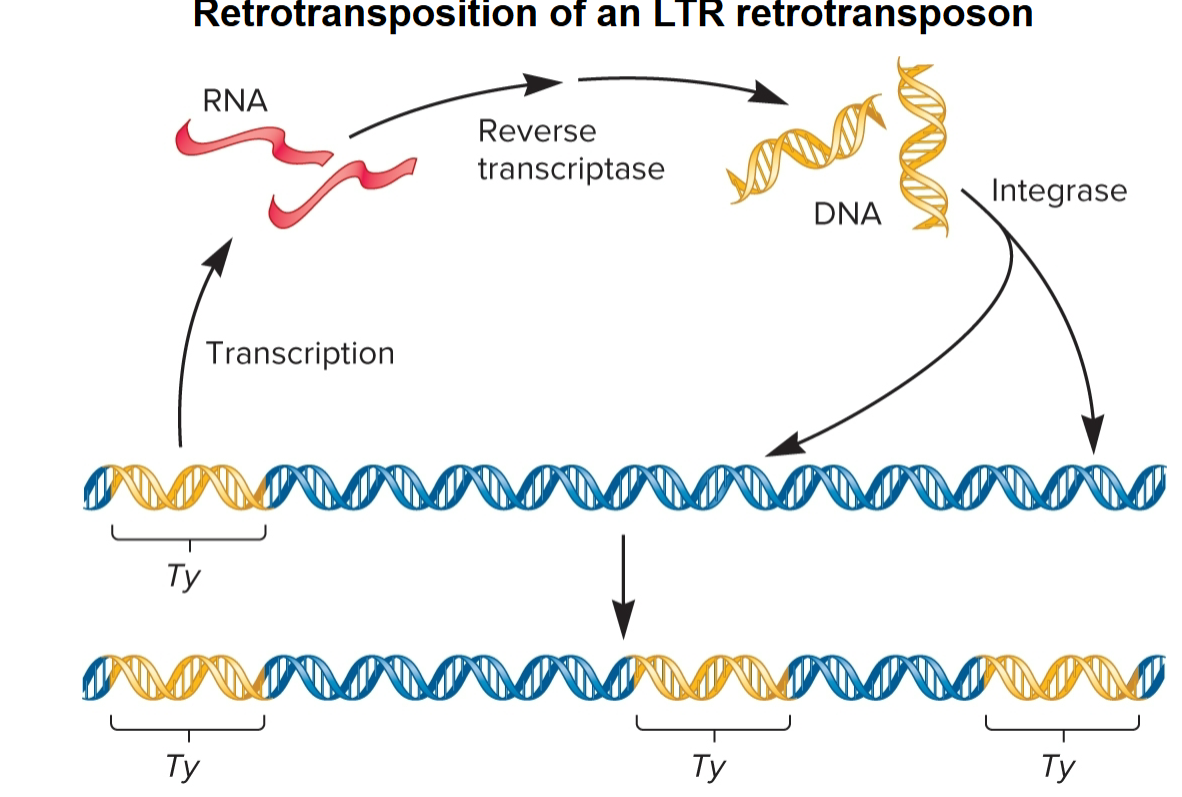
Retrotransposition (retrotransposons)
transposable elements that move via an RNA intermediate and is transcribed into RNA , found only in euks
direct repeats (DRs)
identical base sequences oriented in same direction and repeated, flank TEs
insertion element
simplest TE; flanked by inverted repeats, moves by simple transposition
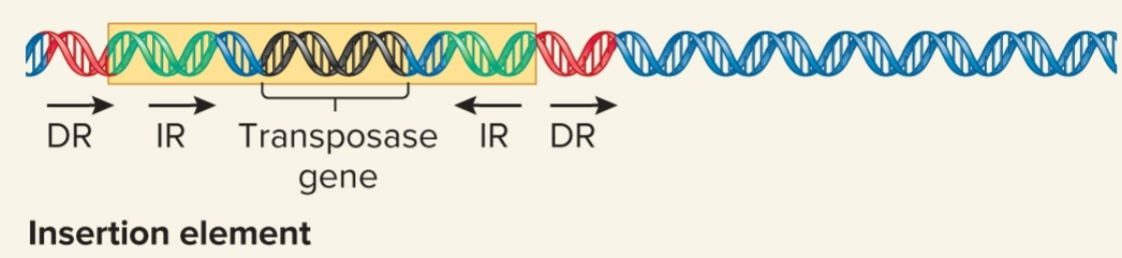
Inverted repeats
DNA sequences that are identical but run in opposite directions
9-40 bp
may contain gene for transposase
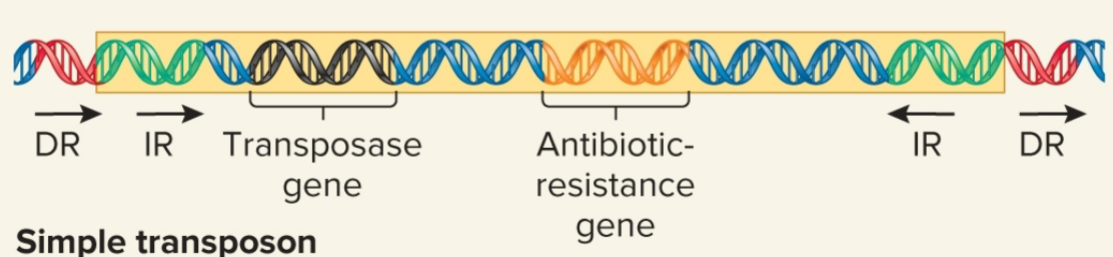
simple transposon
carries >1 genes not required for transposition, moves by simple transposition
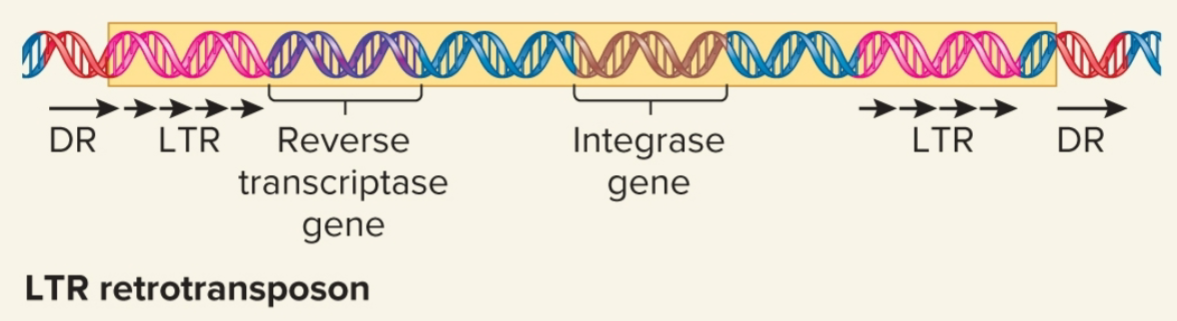
LTR Retrotransposons
evolutionarily related to known retroviruses
retain ability to move around genome but mostly do not produce mature viral particles
contain long terminal repeats (LTRs) at both ends
few hundred bps in length
code virally related proteins, reverse transcriptase and integrase, required for retrotransposition process
move by retrotransposition via RNA intermediate
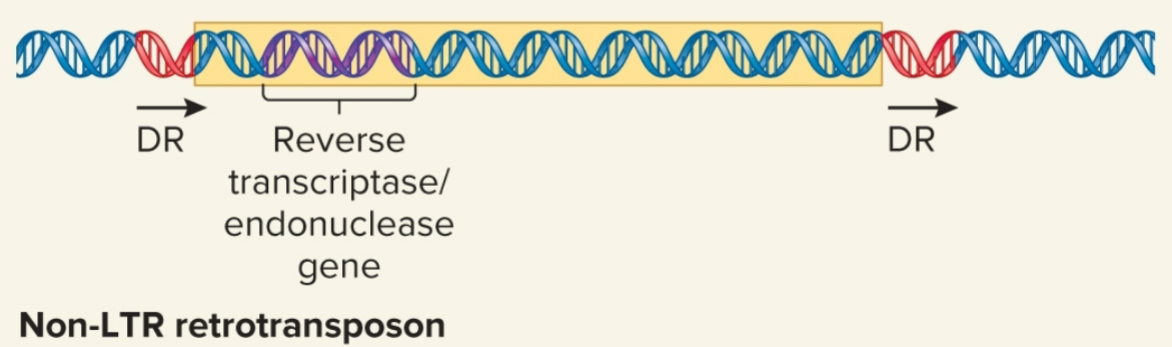
Non-LTR retrotransposons
do not resemble retroviruses in having LTR sequences
may contain gene that encodes protein that functions as both reverse transcriptase and an endonuclease
some are evolutionarily derived from normal eukaryotic genes
Alu family of repetitive sequences found in humans is derived from single ancestral gene, 7SL RNA gene which has been copied by retrotransposition many times, w/ current # of copies being 1 million
move by target-site primed reverse transcription
transposase
catalyzes transposition event, removal of a TE and its reinsertion at another location
recognizes inverted repeats at ends of TE and brings them close together
Reverse transcriptase
uses RNA as template to synthesize double-stranded DNA molecule
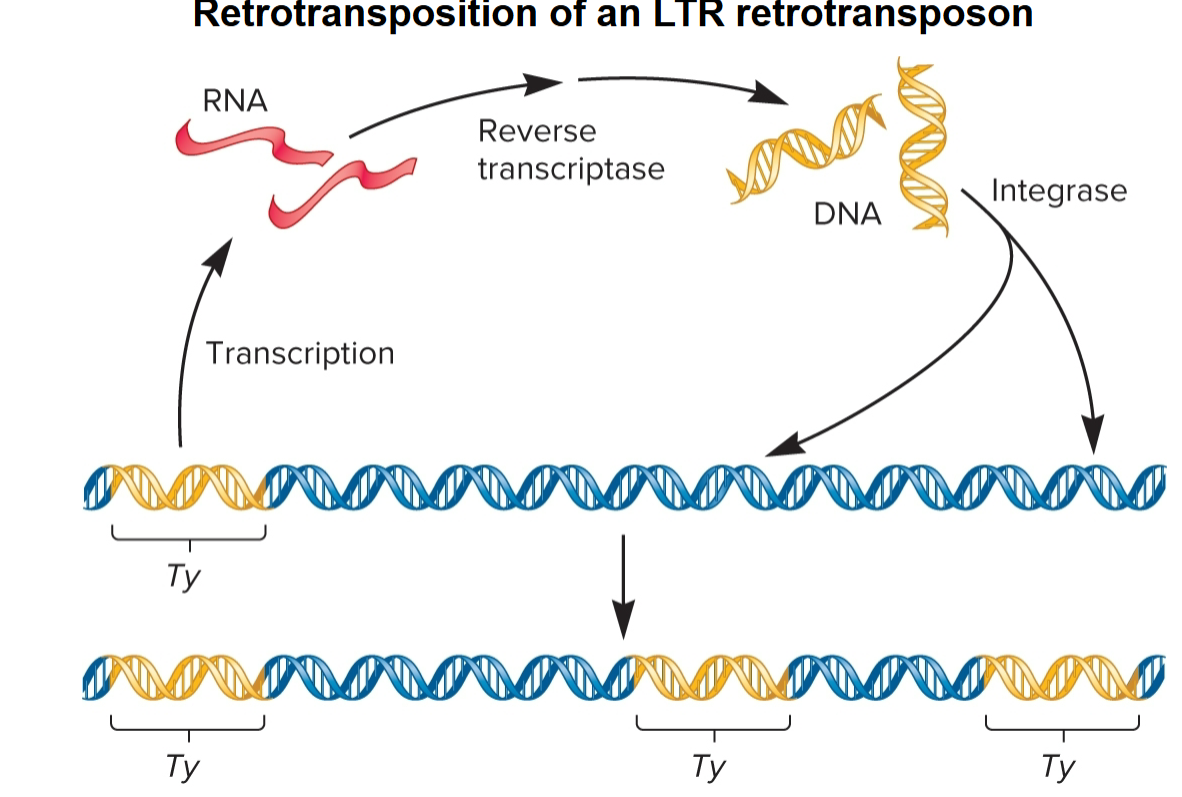
Integrase
Recognizes LTRs at ends of DNA, makes cuts at target site in host chromosome and catalyzes insertion of TE into site
target-site primed reverse transcription
allows non-LTR retrotransposons to move
retrotransposon transcribed into RNA with 3′ polyA tail
target DNA recognized by endonuclease
PolyA tail binds to nicked site
reverse transcriptase uses target DNA of primer and makes DNA copy of RNA
LINEs (Long interspersed elements)
usually 1,000-10,000 bp long
occur in 20,000-1,000,000 copies per genome
17% of human genome
SINEs (Short interspersed elements)
> 500 bp in length
ex) Alu (Arthrobacter luteus restriction endonuclease) sequence present in 1,000,000 copies in human genome (10% of genome)
2 theories of biological significance of transposons
selfish DNA theory: TEs exist because they can! They can proliferate within host as long as they don’t harm the host by disrupting survival
TEs exist because they offer some advantage. Bacterial TEs carry antibiotic-resistance genes
exon shuffling
TEs cause genetic variability through recombination
TEs cause insertion of exons into coding sequences of protein-coding genes
lead to evolution of genes with more diverse functions
nucleus
2-4 μm diameter, DNA is tightly compacted to fit within
chromatin
compaction of linear DNA in euk chromosomes involves DNA-protein complex
proteins bound to DNA subject to change during life of cell, affecting degree of chromatin compaction
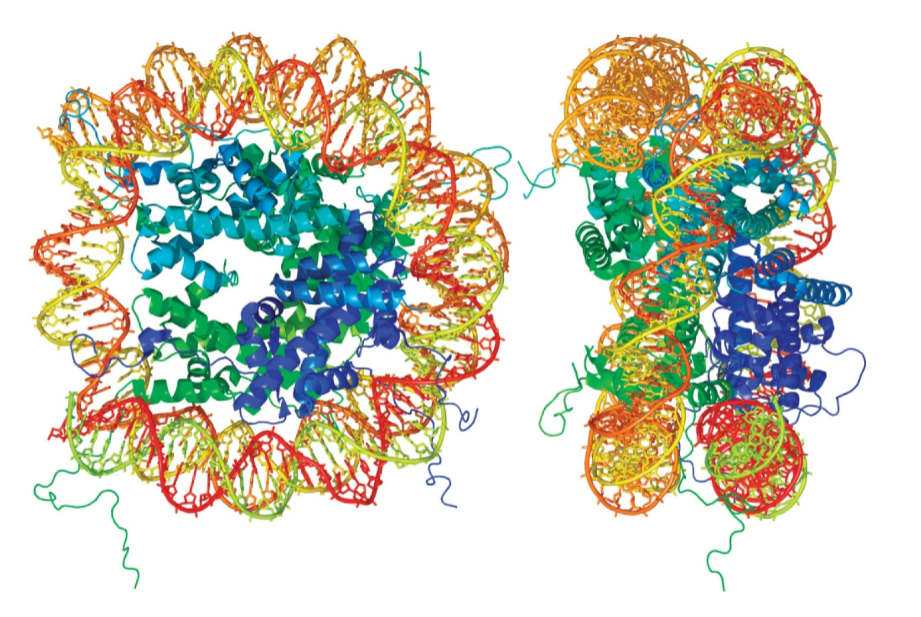
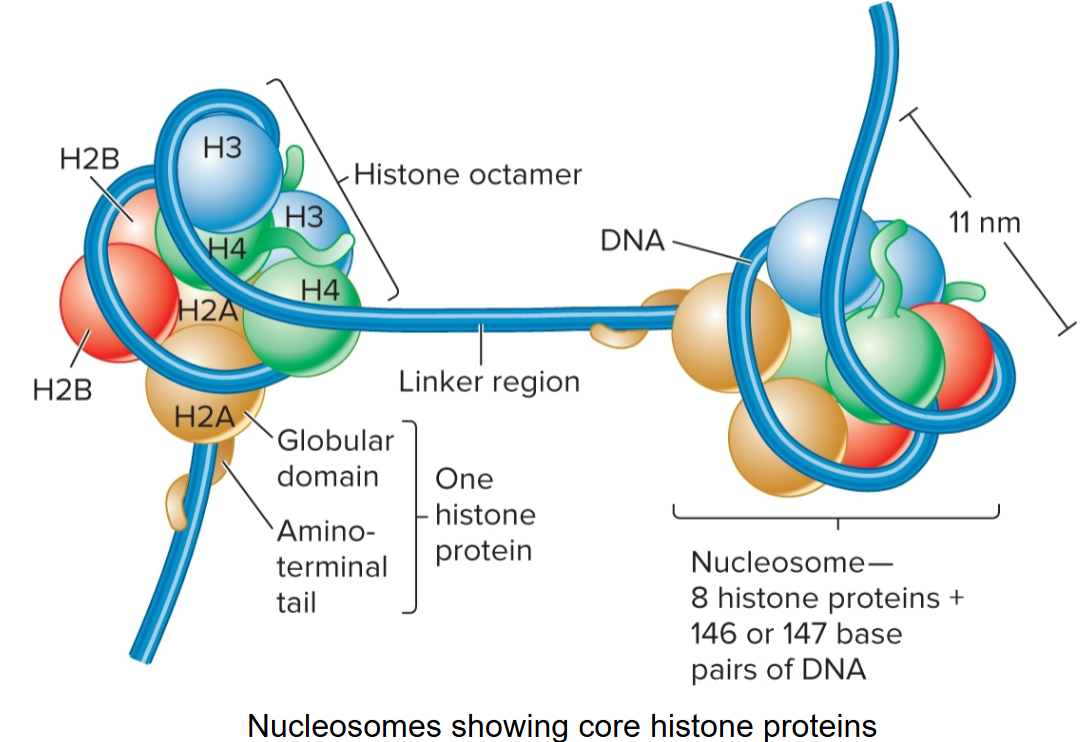
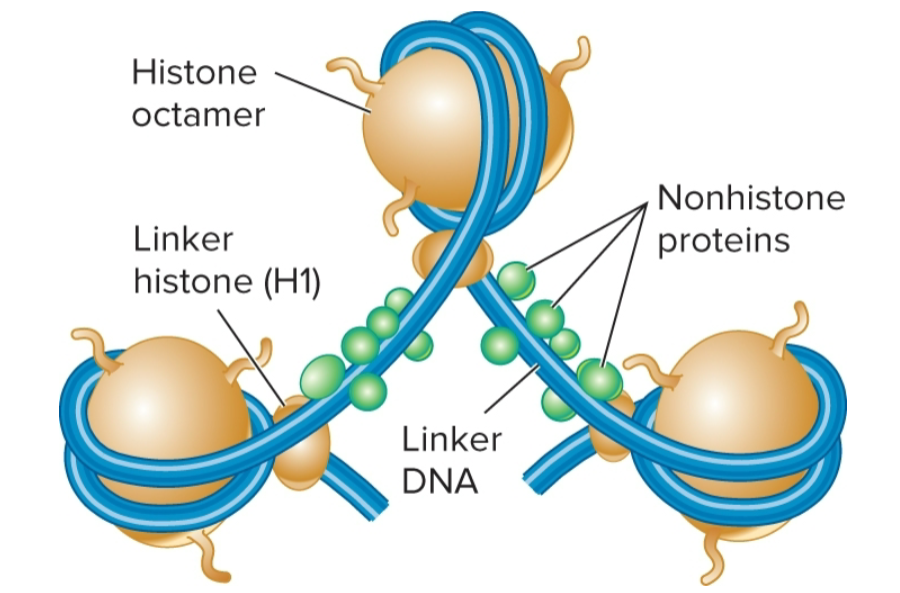
nucleosome
repeating structural unit within euk chromatin
composed of double-stranded segment of DNA wrapped around an octamer of histones
histone octamer composed of 2 copies each of 4 different histone proteins
146 bp of DNA make 1.65 negative superhelical turns around octamer
Histones
basic, contain + charged AAs; lysine and arginine
these AAs bind to - charged phosphates along DNA backbone
have globular domain and flexible, charged amino terminus or ‘tail’
core histones
H2A, H2B, H3 and H4; there are 2 of each, making up the octamer
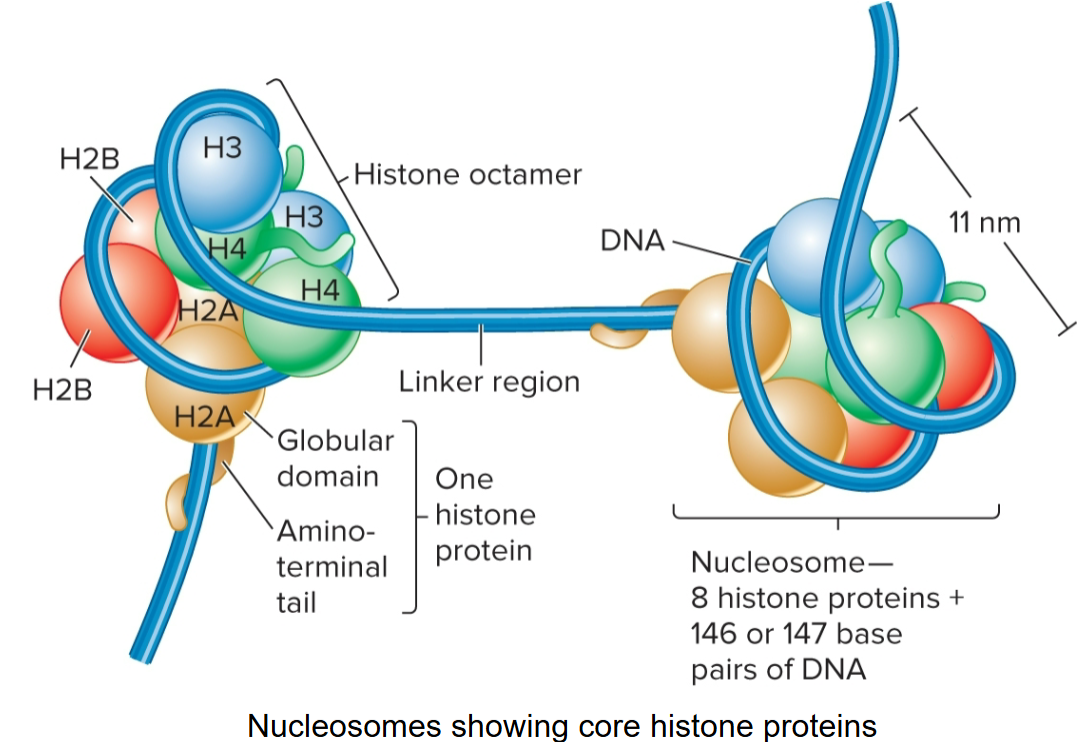
linker histone
H1, binds to DNA in linker region
less tightly bound to DNA than core histones
helps to organize adjacent nucleosomes
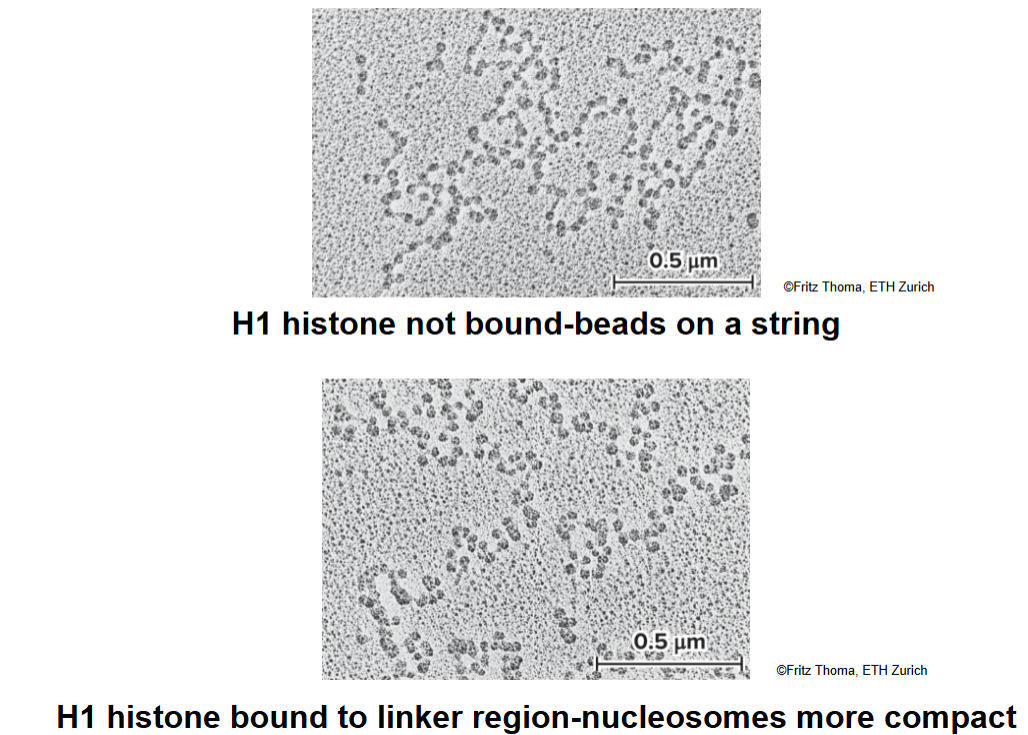
how do salt concentrations affect H1?
moderate salt concentrations: H1 is removed, classic beads-on-a-string morphology
low salt concentrations: H1 remains bound, beads associate together into a compact morphology
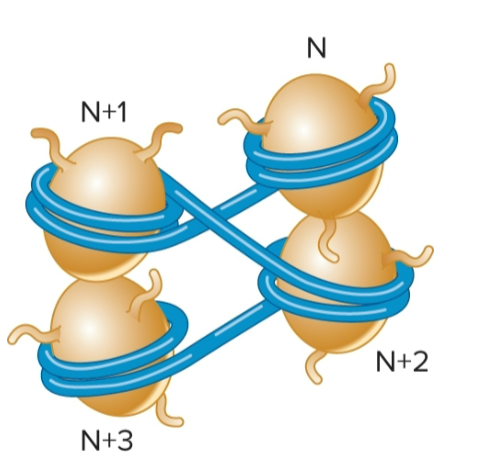
zigzag model
proposed model for interactions of nucleosomes
linker DNA is relatively straight, and nucleosomes form a zigzag arrangement
zigzag arrangement only occurs over short distances, such as 2-4 nucleosomes
a former model (30nm fiber model) depicted long-range interactions of nucleosomes to form a fiber; this model is no longer accepted
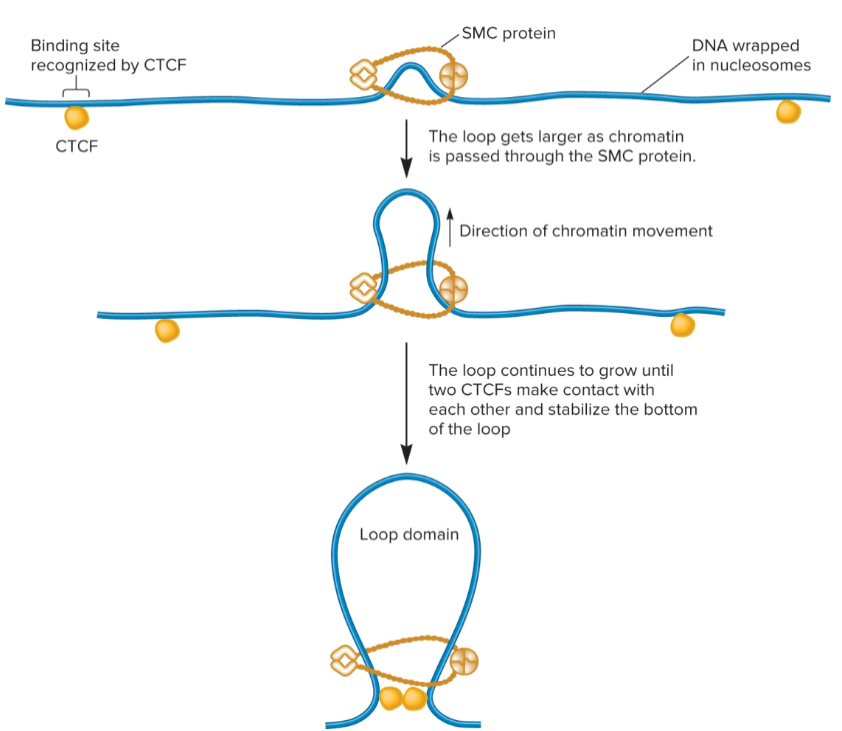
loop extrusion model
Chromatin can be further compacted by folding segments of nucleosomes into loops/loop domains
2 proteins play a role in loop formation: SMC proteins (structural maintenance of chromosomes) and CCCTC binding factor (CTCF)
SMC proteins (structural maintenance of chromosomes)
forms a dimer that can wrap itself around 2 DNA segments and promote formation of a loop
use energy from ATP to catalyze loop formation
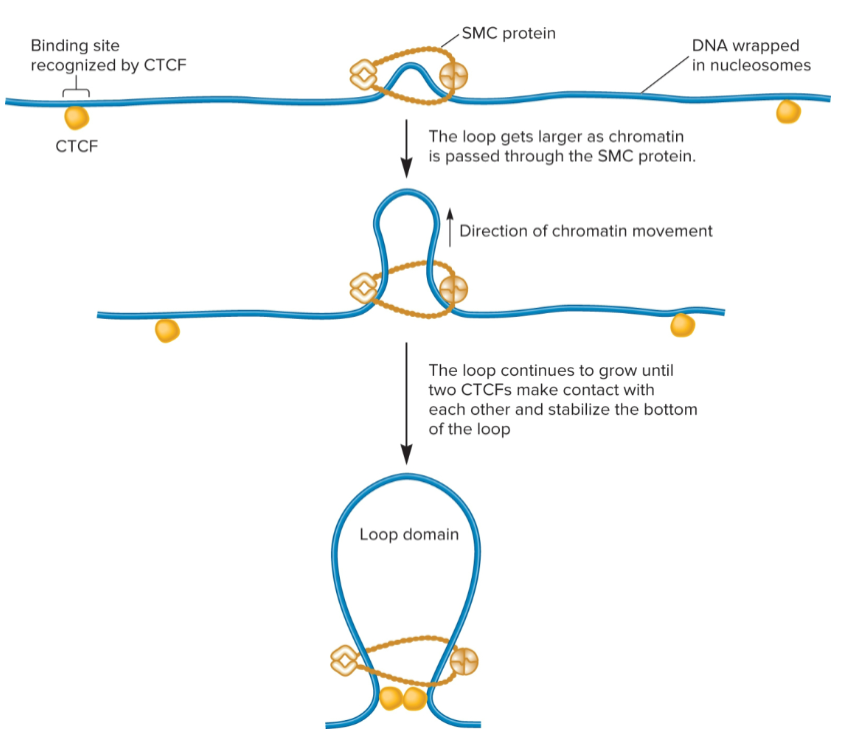
CCCTC binding factor (CTCF)
after loop has formed due to SMC proteins, 2 different CTCFs bind to DNA and then bind to each other to stabilize loop
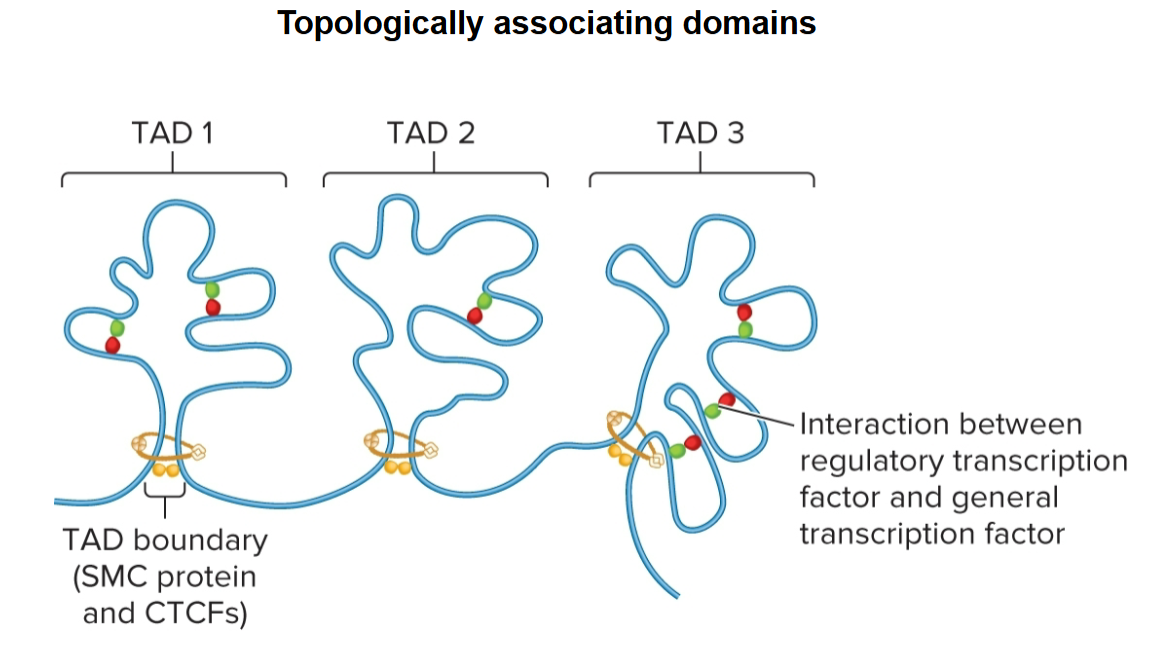
topologically associating domains (TADs)
regions that chromatin is organized into
100 kb - 1 Mb in length
segments of DNA within are more likely to interact with each other than they are with segments in other neighboring TADs
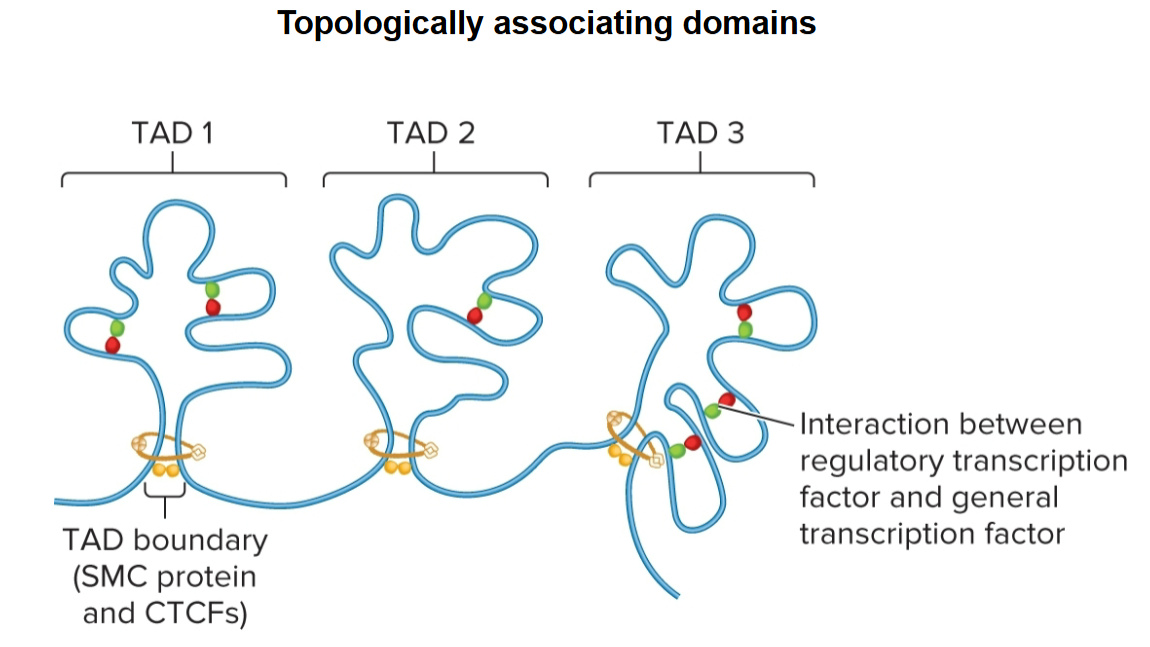
topologically associating domains (TADs) boundaries
determined by SMC proteins and CTCFs
promote interactions within
act as insulators, preventing interactions between different TADs
Heterochromatin
Tightly compacted regions of chromosomes
Transcriptionally inactive (in general)
Loop domains compacted even further
constitutive and facultative
Constitutive heterochromatin
regions that are always heterochromatic
permanently inactive with regard to transcription
usually contain highly repetitive sequences
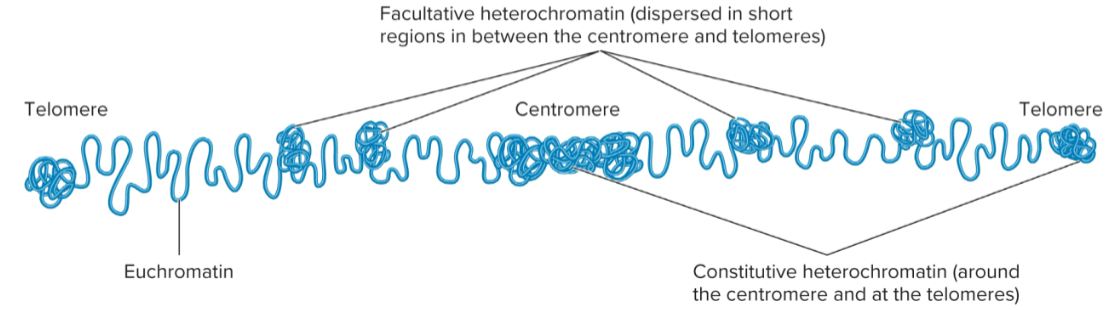
Facultative heterochromatin
Regions that can interconvert between euchromatin and heterochromatin
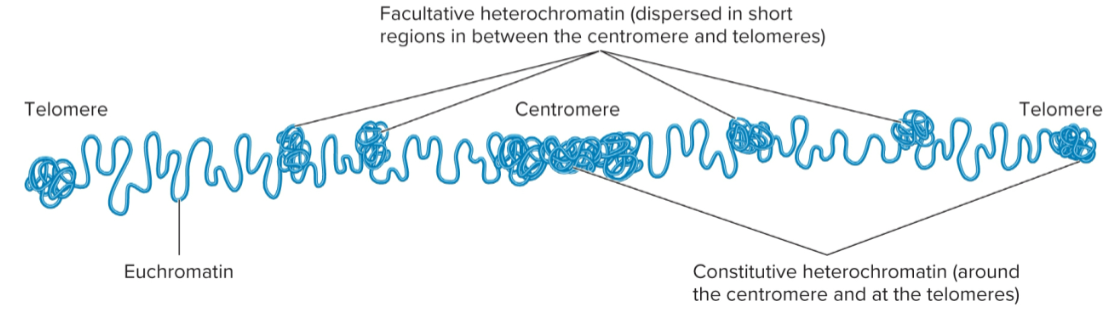
Euchromatin
Less condensed regions of chromosomes
Transcriptionally active
Loop domains are less compacted
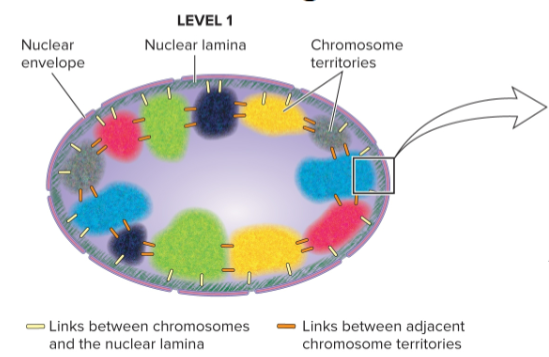
chromosome territory
each chromosome in cell nucleus is in a discrete territory
shown in studies by Thomas and Christoph Cremer and others through fluorescent staining in which each chromosome is shown in a different color
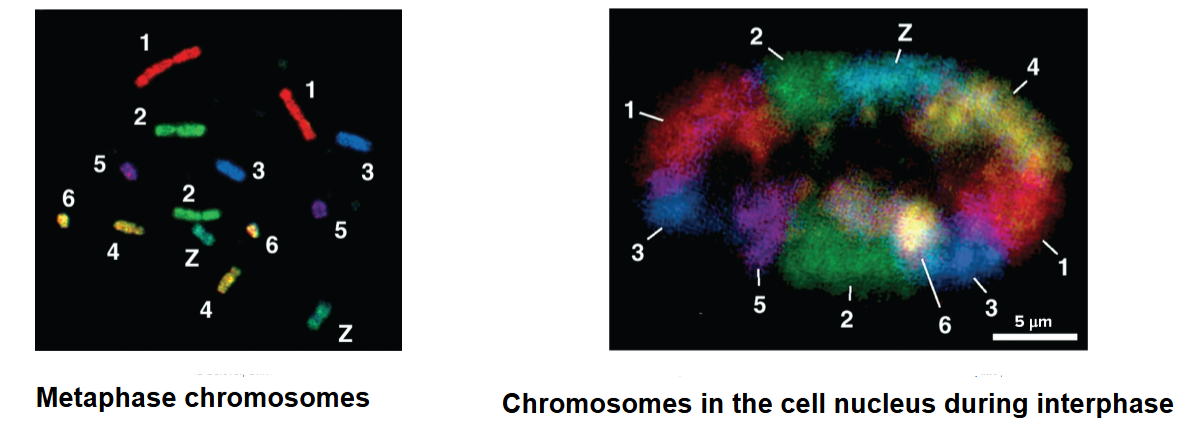
level 1 of chromosome organization
at scale of an entire nucleus
chromosomes occupy distinct territories
interchromosomal and chromosomal interactions with other nuclear structures (ex: nuclear lamina) play a role in chromosomal arrangements