Chapter 13: Translation of mRNA
1/89
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
90 Terms
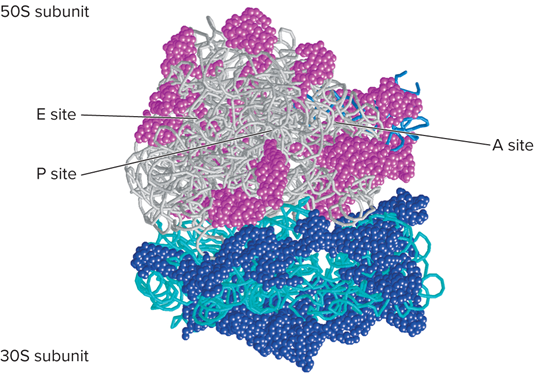
What does the molecular model for the structure of a ribosome look like?
What is translation?
Involves an interpretation of one language into another
In genetics, the nucleotide language of mRNA is translated into what?
The amino acid language of proteins
What does translation rely on?
The genetic code
What are Codons?
The genetic information is coded within mRNA in groups of three nucleotides
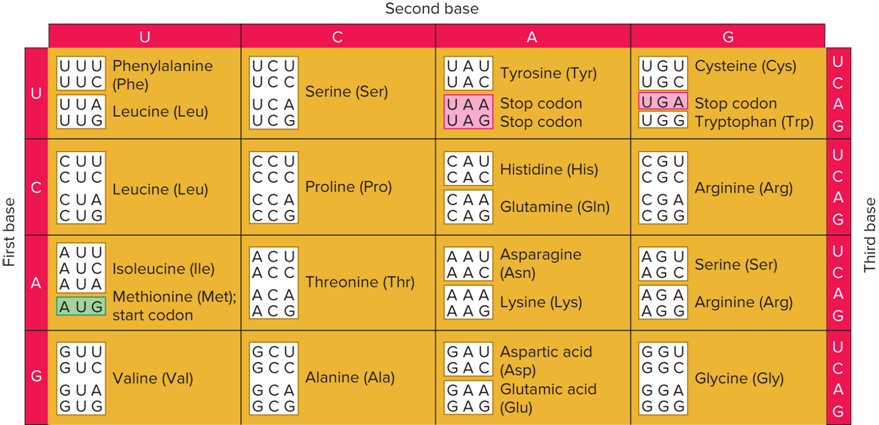
What does the Genetic Code look like?
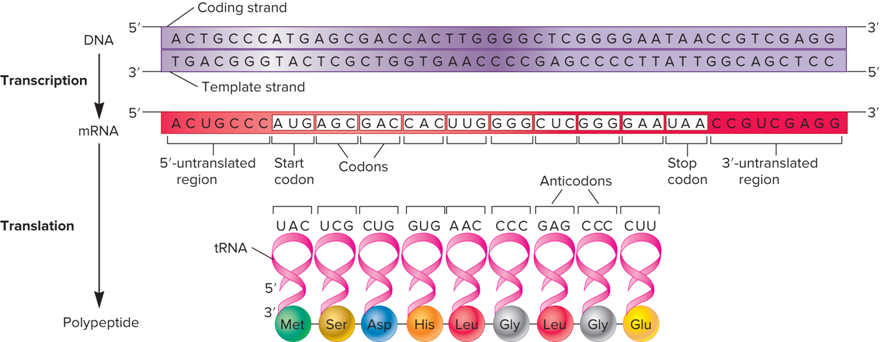
What does the Overview of Gene Expression look like?
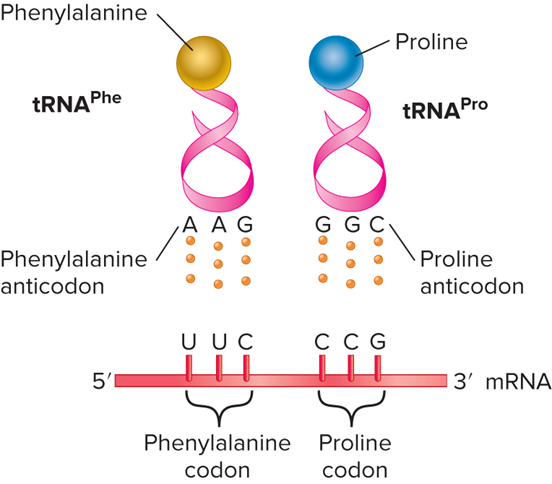
What does the Recognition between tRNA and mRNA look like?
A polypeptide chain has what?
Directionality
What does the attachment of an amino acid to a peptide chain look like?
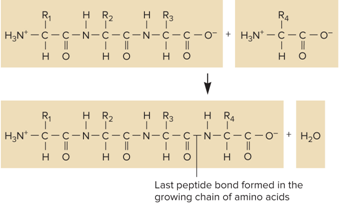
What does the Directionality in a polypeptide and mRNA look like?
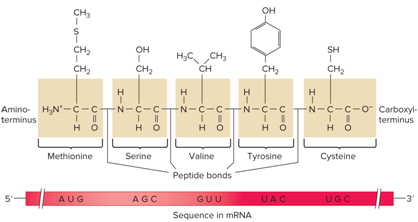
tRNAs share what?
Common Structural Features
What does the secondary structure of tRNAs exhibit?
A cloverleaf pattern
How does tRNAs exhibit a cloverleaf pattern?
Three stem-loop structures
A few variable sites
An acceptor stem with a 3’ single strand region (CCA added by enzyme at 3’ end)
The actual three-dimension (tertiary structure) involves what?
Additional folding
What do tRNAs also contain?
Have the normal A, U, G, and C nucleotides
Commonly contain modified nucleotides
How many commonly contain modified nucleotides do tRNAs have?
More than 80 of these can occur
What does the Structure of tRNA look like?
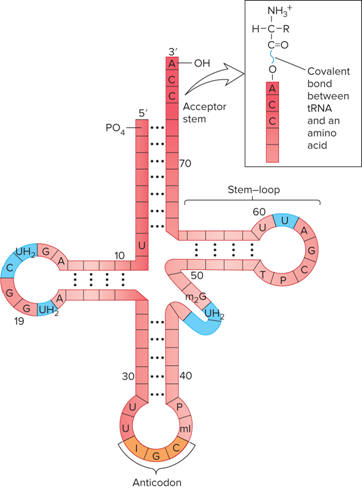
What is found in all tRNAs?
3’ ACC acceptor
What are the six modified bases of tRNAs?
I= inosine
ml = methylinosine
T = ribothymidine
UH2 = dihydouridine
m2G = dimethylguanosine
P = psuedouridine
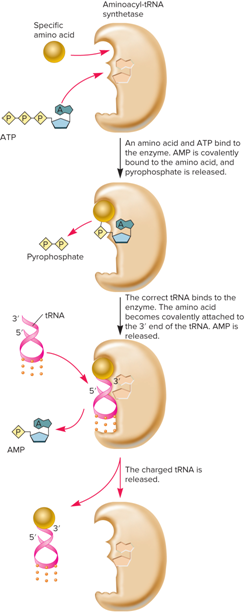
What are aminoacyl-tRNA synthetases?
The enzymes that attach amino acids to tRNAs
How many aminoacyl-tRNA synthetases are there?
20
How many aminoacyl-tRNA synthetases for each amino acid?
One
How many different molecules are involved when Aminoacyl-tRNA synthetases catalyzes a two-step reaction?
3
What are the 3 different molecules involved in the catalyzation process?
Amino acid
tRNA
ATP
What is the result of “Aminoacyl-tRNA synthetases catalyze a two-step reaction involving three different molecules: amino acid, tRNA and ATP”?
A charged tRNA
What does the charging of tRNAs look like?
What does a small subunit look like for the composition of Bacterial and Eukaryotic Ribosomes?

What does a large subunit look like for the composition of Bacterial and Eukaryotic Ribosomes?
What does a assembled ribosome look like for the composition of Bacterial and Eukaryotic Ribosomes?
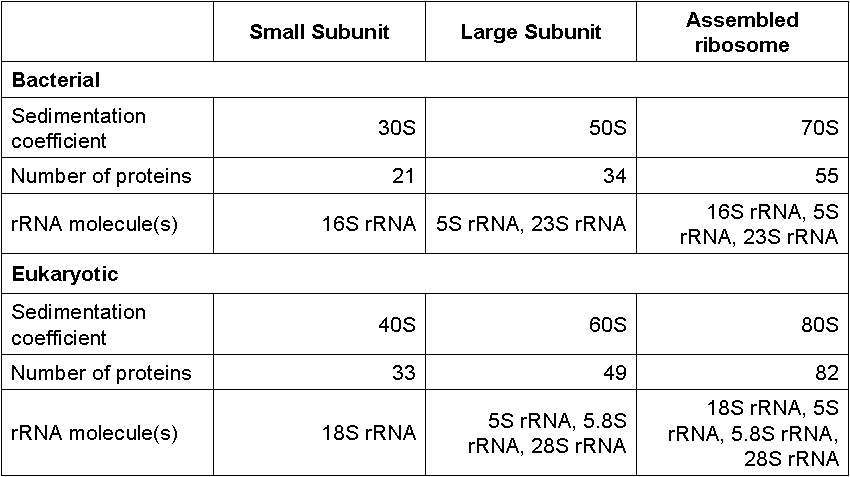
What does the composition of Bacterial and Eukaryotic Ribosomes Table look like?
For the functional sites of ribosomes, during bacterial translation, the mRNA lies on what?
On the surface of the 30s subunit
During bacterial translation, as a polypeptide is being synthesized what happens?
It exits through a channel within the 50S subunit
What are three discrete sites of Ribosomes?
Peptidyl site (P site)
Aminoacyl site (A site)
Exit site (E site)
What does the Bacterial ribosome model based on X-ray diffraction studies look like?
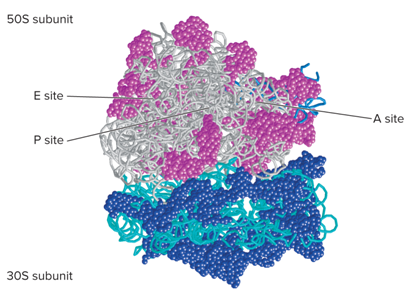
What does the model for ribosome structure look like?
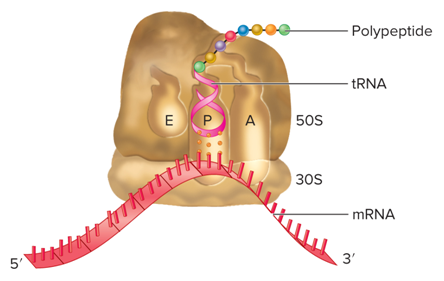
What are the three stages of translation?
Initiation
Elongation
Termination
What does the overview of Translation look like?
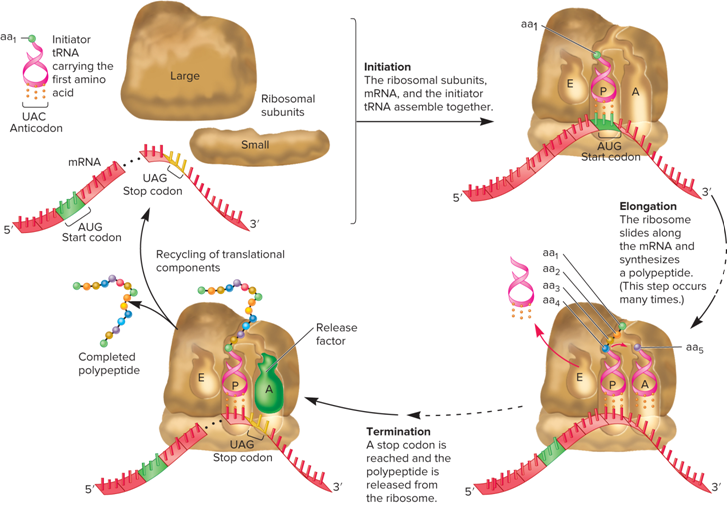
What does the first image of the Initiation stage of translation look like?
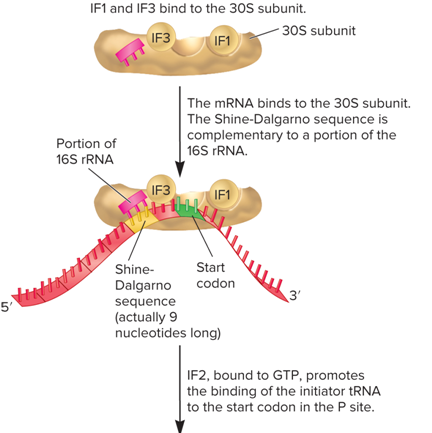
What does the fir image of the Initiation stage of translation look like?
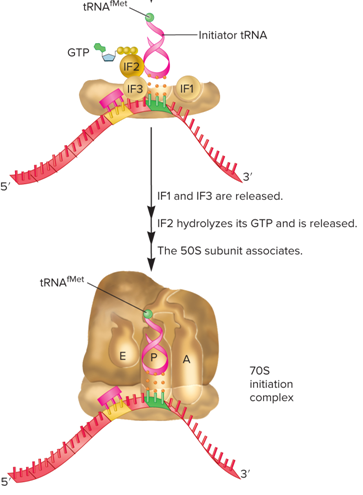
What happens during the Translation Initiation Stage?
The mRNA, initiator tRNA, and ribosomal subunits associate to form an initiation complex (this process requires three initiation factors)
The initiator tRNA recognizes the start codon in mRNA
In bacteria, this tRNA is designated tRNA^fmet (It carries a methionine that has been covalently modified to N-formylmethionine)
The start codon is AUG, but in some cases GUG or UUG (In all three cases, the first amino acid is N-formylmethionine)
In eukaryotes, the assembly of the initiation complex is similar to that in bacteria (However, additional factors are required, Note that eukaryotic Initiation Factors are denoted eIF)
The initiator tRNA is designated tRNA^met (It carries a methionine rather than a formylmethionine)
What is the start codon for eukaryotic translation?
AUG
How does the Ribosome scan the start codon?
Ribosome scans from the 5’ end of mRNA until it finds the AUG start codon (not all AUGs can act as a start)
What are Kozak’s rules?
Rules for optimal translation initiation
Where is the start codon for eukaryotic translation usually located?
Usually the first AUG after the 5’ cap
What does the start codon in eukaryotes look like?
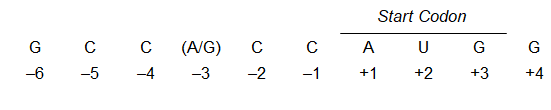
What is the process of translation initiation in Eukaryotes?
An initiation factor protein complex (eIF4) binds to the 5’ cap in mRNA
These are joined by a complex consisting of the 40S subunit, tRNA^met, and other initiation factors
The entire assembly moves along the mRNA scanning for the right start codon
Once it finds this AUG, the 40S subunit binds to it
The 60S subunit joins
This forms the 80S initiation complex
What is the Translation Elongation Stage?
During this stage, amino acids are added to the polypeptide chain, one at a time
The addition of each amino acid occurs via a series of ordered steps
This process, though complex, can occur at a remarkable rate (In bacteria → 15 to 20 amino acids per second; In eukaryotes → 2 to 6 amino acids per second)
After peptide bond formation, tRNAs at the P and A sites move into the E and P sites
The 23S rRNA (a component of the large subunit) is the actual peptidyl transferase (Thus, the ribosome is a ribozyme!)
What does the elongation stage of translation look like?
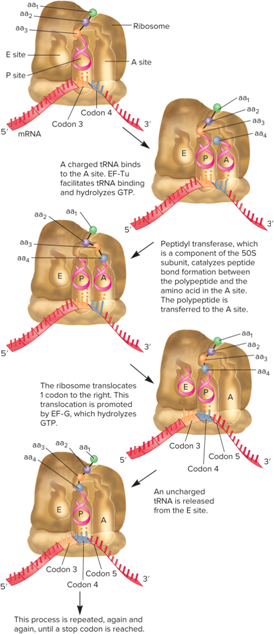
What occurs during the translation termination stage?
The final stage occurs when a stop codon is reached in the mRNA
In most species there are three stop or nonsense codons (UAG, UAA, UGA)
These codons are not recognized by tRNAs, but by proteins called release factors (The 3-D structure of release factors mimics that of tRNAs)
Bacteria have three release factors (RF1 recognizes UAA and UAG; RF2 recognizes UAA and UGA; RF3 does not recognize any of the three codons - required for the termination process)
Eukaryotes only have two release factors (eRF1 recognizes all three stop codons; eRF3 required for termination process)
What does the termination stage of translation look like?
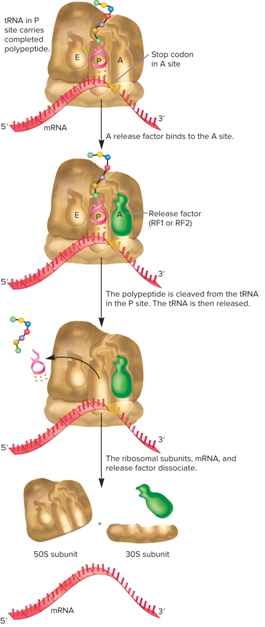
Can bacterial translation begin before transcription is completed?
Yes
Bacteria lack what?
A nucleus
What occurs because bacteria lack a nucleus?
Both transcription and translation occur in the cytoplasm
What happens as soon as mRNA strand is long enough?
A ribosome will attach to its 5’ end
Why does translation begin before transcription ends?
Because as soon an mRNA strand is long enough, a ribosome will attach to its 5’ end
What are characteristics of mRNA being long enough such that a ribosome attaches to its 5’ end?
So translation begins before transcription ends
This phenomenon is termed coupling
Does not occur in eukaryotes (Transcription is in nucleus; Translation is in cytosol)
Concept: Which genetic disorder occurs when homogentisic acid oxidase is defective?
Alkaptonuria
Concept: What enzymatic function is missing in the strain 2 mutants in Figure 13.2: An example of an experiment that supported Beadle and Tatum’s one-gene/one-enzyme hypothesis?
The strain 2 mutants are unable to convert O-acetylhomoserine into cystathionine
Textbook 13.1: An inborn error of metabolism is caused by?
A mutation in a gene that causes an enzyme to be inactive
Textbook 13.1: The reason why Beadle and Tatum observed four different categories of mutants that could not grow on media without methionine is because?
Four different enzymes are involved in a pathway for methionine biosynthesis
Concept: Describe the role of DNA in the synthesis of a polypeptide
The role of DNA in a polypeptide is storage. It stores the information that specifies the amino acid sequence of a polypeptide
Concept: Which two amino acids do you think are the least soluble in water?
Tryptophan
Phenylalanine
Concept: What type of bonding is primarily responsible for the formation of the two types of secondary structures?
Hydrogen bonding
Textbook 13.2: What is the genetic code?
The relationship between a three-base codon sequence and an amino acid or the end of translation
Textbook 13.2: The reading frame begins at a ______ and is read _____
start codon, in groups of three bases
Textbook 13.2: The fourth codon in an mRNA is GGG, which specifies glycine. Assuming that no amino acids are removed from the polypeptide, which of the following statements is correct?
The fourth amino acid from the N-terminus is glycine
Textbook 13.2: A type of secondary structure found in proteins is?
An a helix (alpha)
A B sheet (beta)
Concept: Explain how the use of radiolabeled amino acids in the experiment by Nierenberg and Leder helped to reveal the genetic code
Only one radiolabeled amino acid was found in each sample. If radioactivity was trapped on the filter, this meant that the codon triplet specified that particular amino acid.
Textbook 13.3: Lets suppose a researcher mixed together nucleotides with the following percentages of bases: 30% G, 30% C, and 40% A. If RNA was made via polynucleotide phosporylase what percentage of the codons would be 5’-GGC-3’?
2.7%
Textbook 13.3: In the triplet-binding assay carried out by Nirenberg and Leder, an RNA triplet composed of three bases was able to cause the?
Binding of a tRNA carrying the appropirate amino acid
Concept: What are the two key functional sites of a tRNA molecule?
Has an anticodon that recognizes a codon in the mRNA
Has an acceptor stem where the correct amino acid binds
Concept: what is the difference between a charged tRNA and an uncharged tRNA?
A charged tRNA has an amino acid attached to it
Concept: how do the wobble rules affect the total number of different tRNAs that are needed to carry out translation?
The wobble rules allow a smaller number of tRNAs to recognize all of the possible mRNA codons
Textbook 13.4: If a tRNA has an anticodon with the sequence 3’-GAC-5’, which amino acid does it carry?
Leucine
Textbook 13.4: The anticodon of a tRNA is located in the?
loop of the 2nd stem-loop
Textbook 13.4: An enzyme known as _____ attaches an amino acid to the ___ of a tRNA, thereby producing _____
Aminoacyl-tRNA synthetase, 3’ single-stranded region of the acceptor stem, a charged tRNA
Textbook 13.5: Each ribosomal subunit is composed of?
Multiple proteins
rRNA
Textbook 13.5: The site(s) on a ribosome where tRNA molecules may be located include?
A site
P site
E site
Concept: Describe the roles that mRNA plays in the three stages of translation
A site in an mRNA promotes the binding of the mRNA to the ribosome during initiation. The codons in an mRNA specify the polypeptide sequence during elongation. The stop codon terminates translation.
Concept: Why does bacterial mRNA bind specifically to the small ribosomal subunit?
The Shine-Dalgarno sequence in bacterial mRNA is complementary to a region in the 16S rRNA within the small ribosomal subunit. These complementary sequences hydrogen bond with each other.
Concept: what is the role of peptidyl transferase during the elongation stage?
Catalyzes the formation of a peptide bond between the amino acid at the A site and the last amino acid in the growing polypeptide
Concept: Explain why release factors are called “molecular mimics”
The three-dimensional structures of release factors, which are proteins, resemble those of tRNAs
Textbook 13.6: During the initiation stage of translation in bacteria, which of the following events occur?
IF1 and IF3 bind to the 30S subunit
The mRNA binds to the 30S subunit, and tRNA^fMet binds to the start codon in the mRNA
IF2 hydrolyzes its GTP and is released; the 50S subunit binds to the 30S subunit
Textbook 13.6: Kozak’s rules determine
The choice of the start codon in complex eukaryotes
Textbook 13.6: During the peptidyl transfer reaction, the polypeptide, which is attached to a tRNA in the _____, becomes bound via _____ to an amino acid attached to a tRNA in the ____
P site, a peptide bond, A site
Textbook 13.6: A release factor is referred to as a “molecular mimic” because its structure is similar to that of
a tRNA
Concept: If a mutation prevented IRP from binding to the IRE in the ferritin mRNA, how would that mutation affect the regulation of ferritin synthesis? Do you think there would be too much or too little ferritin in the cell?
The mutation would cause the overproduction of ferritin, because ferritin synthesis would occur even if iron levels were low
Textbook 13.7: The binding of iron regulatory protein (IRP) to the iron response element (IRE) in the 5’-UTR of the ferritin mRNA causes?
The inhibition of translation of the ferritin mRNA
Textbook 13.8: Which of the following is not similar between archae and eukaryotes?
Initial binding of mRNA to the ribosome