RL14: PCR
1/28
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
29 Terms
What did Dr.Paabo do?
Dr. Paabo applied PCR to study genetic sequences from extinct species
He did this with thylacine (tasmanian tiger or wolf) and giant sloths
He extracted the samples from bones
Two fundamental/basic techniques in molecular biology
PCR and DNA sequencing
Other more complex techniques borrow from the same principles
Common uses of PCR (4 uses)
Sequencing
Cloning (isolating a particular gene or overexpression of proteins)
Detecting pathogens
Gene editing (ex. CRISPR)
What does PCR do?
It makes a lot of copies of a DNA molecule
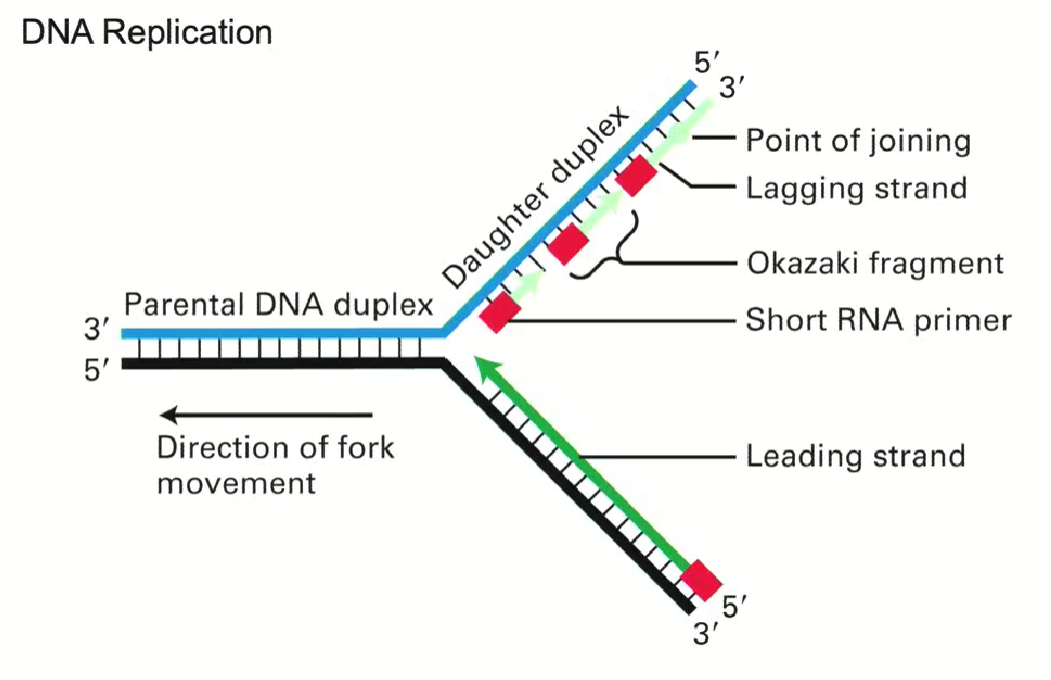
REMINDER: How does DNA replication work? (general)
Double strand unwound by the double helix
Primers allow polymerase attachment
Leading strand synthesized in same direction as replication fork movement (continuous)
Lagging strand synthesized in opposite direction as replication fork movement (discontinuous)
What do we need for Polymerase Chain Reaction (PCR)? (4 things)
The replication occurs in a tube
a DNA template (some sample that we want to amplify)
Taq DNA Polymerase (special type of DNA polymerase that comes from thermophillic bacteria - can withstand hot temperatures)
Primers (attachment point for Taq DNA Polymerase - oligonucleotides or oligos)
dNTPs (building blocks of DNA)
In PCR, how do we separate DNA strands?
Using high temperatures to denature it instead of helicase
Uses a machine called a thermal cycler
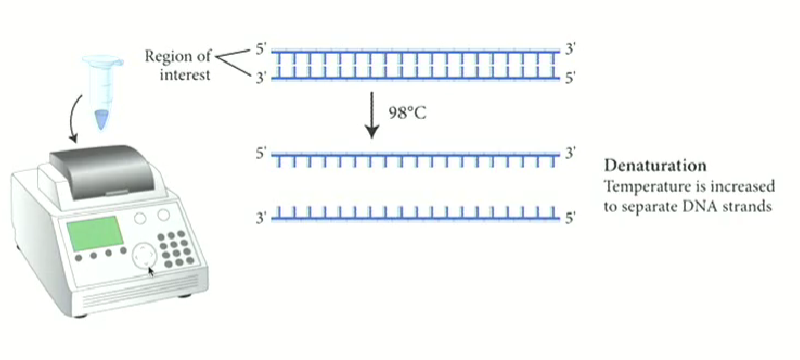
Process of DNA Denaturation
Temperature greater than 90 degrees required
As double stranded DNA denatures, it absorbs light better
the blue line shows light absorption by single stranded DNA
the pink line shows the increasing absorption of double stranded DNA as it denatures (becomes single-stranded)
This is a reversible process (strands go back together when temperature goes down)
Tm is the melting point of DNA (defined as the point at which half of the double stranded DNA denatures)
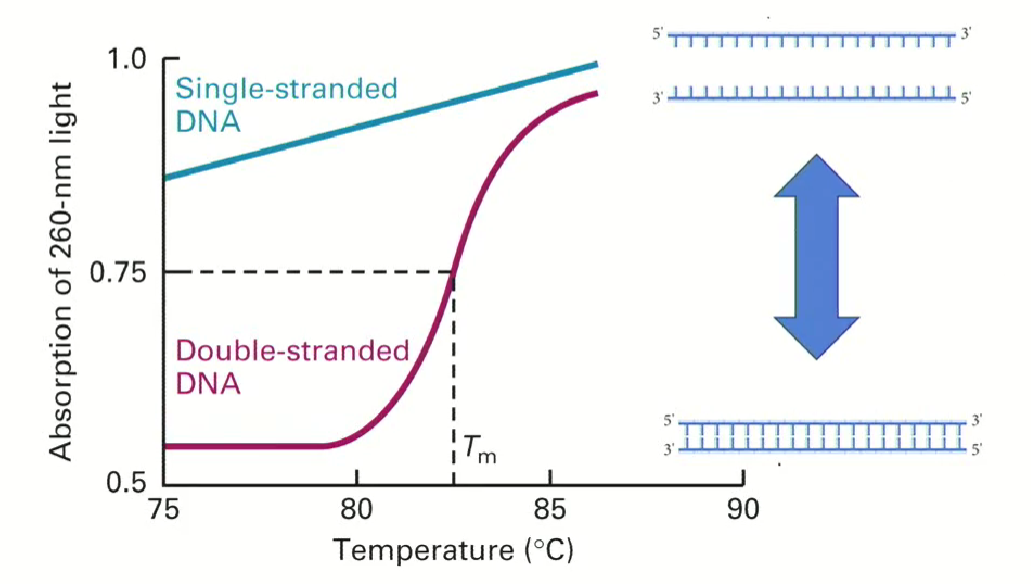
How do we know the temperature needed to separate 2 DNA strands?
It is sequence dependent (Depends on the number of hydrogen bonds between the two strands) and length dependent
The more GCs in the sequence, the higher the temperature needed (since GCs have 3 hydrogen bonds as opposed to 2)
The longer the DNA strand, the higher the temperature needed to denature.
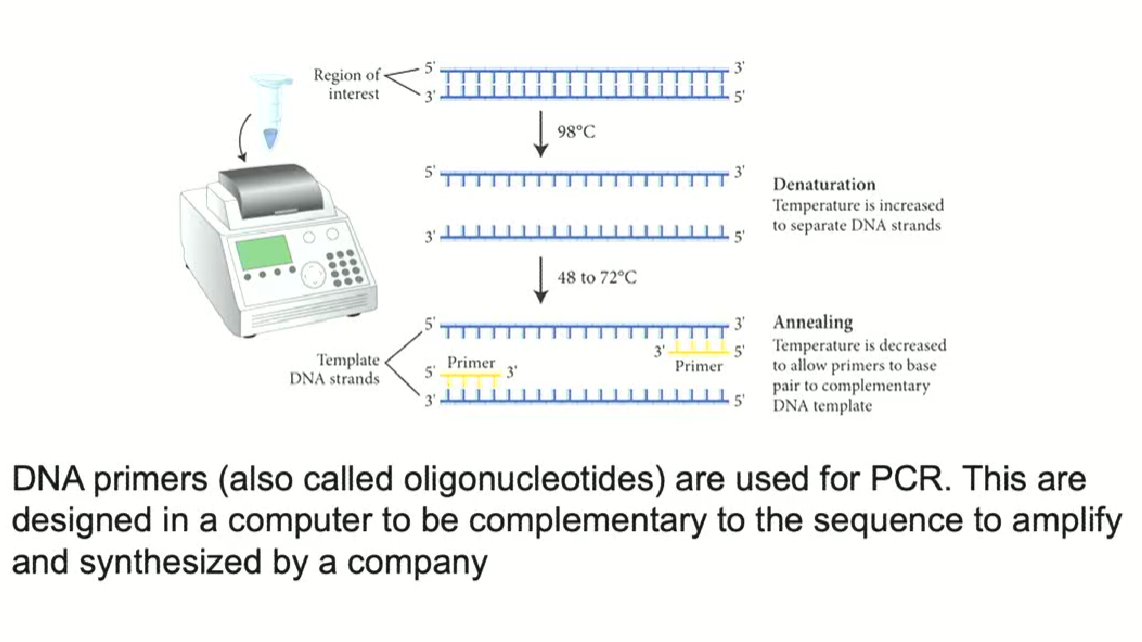
Process of PCR annealing
PCR requires primers (we use short single stranded DNA primers around 20 nucleotides long)
Primers are specific to a particular sequence
Primers are designed to be complementary on a computer, then ordered from a company
Primers have a specific temperature, which makes the pairing very specific by avoiding pairing with other parts of the chromosome
Why do we use DNA primers instead of RNA primers in PCR annealing?
RNA primers are easily degradable/ unstable due to their extra hydroxyl group
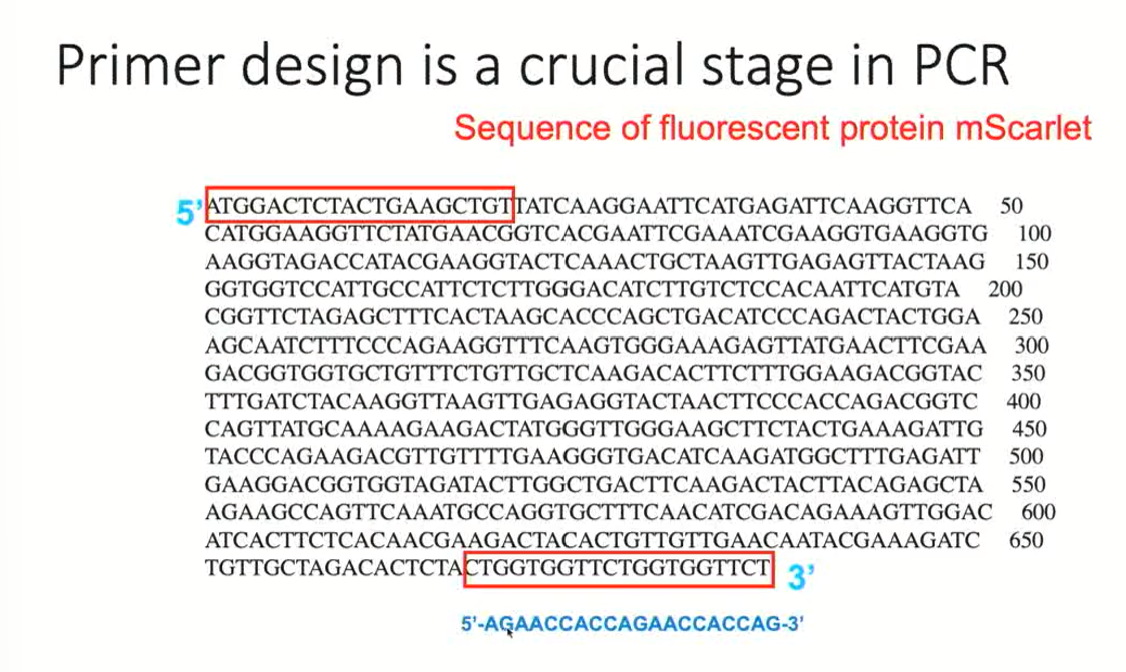
Primer Design Example: mScarlet protein
5’ end: ATG is the start codon
3’ end: TCT is the stop codon
We want to design primers to amplify the entire sequence
The first primer (at the start sequence) can be used directly on the template since it goes in the 5’-3’ direction, and the primer needs to go in this direction as well
The second primer (at the end sequence) cannot transcribe the sequence since it is going in the 3’-5’ end. Because of this, the second primer must be a reverse complement
This means that we need to take the sequence, and obtain the complementary sequence UNINVERTED
Process of PCR Extension
uses Taq DNA polymerase (comes from thermophyllic bacteria)
Taq polymerase has an optimal temperature of around 60 degrees
No proofreading exonuclease activity (error-prone)
Other enzymes with higher fidelity can also be used
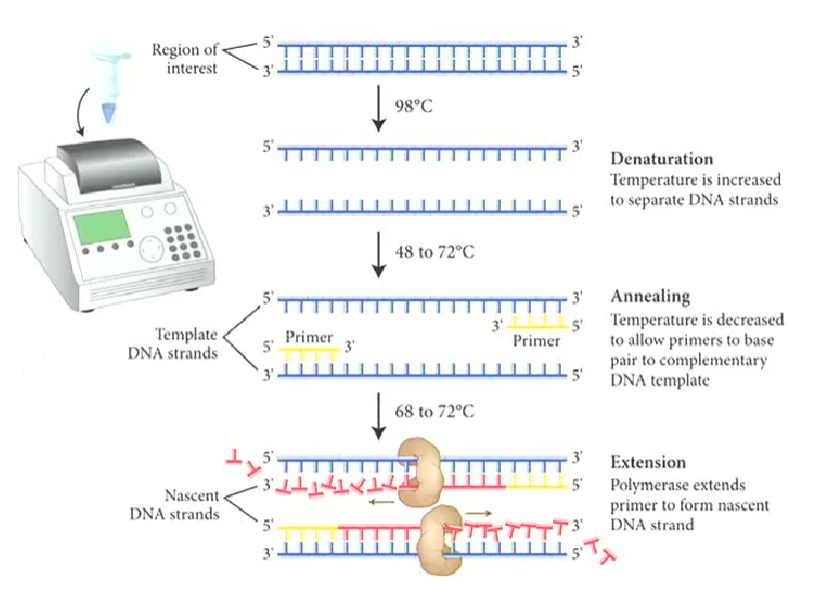
3 stages of PCR
Denaturing
Annealing
Extension
the cycle repeats many times to make many copies (20-40x)
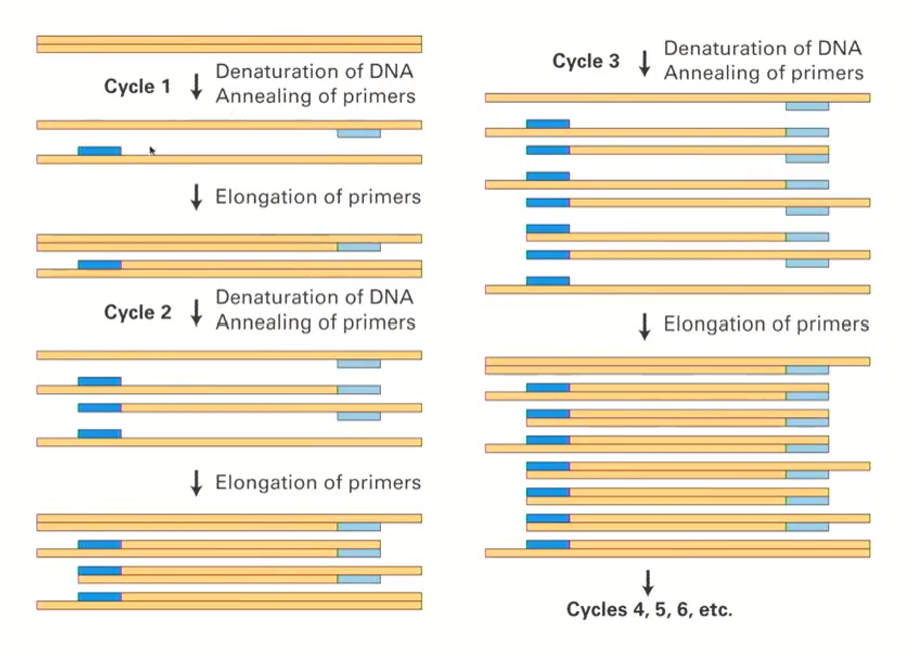
Exponential Amplification
we start with one DNA molecule (2 strands) that gets denatured, annealed, and elongated so that the DNA has the correct length (according to the primers)
We end up with 2 DNA molecules (4 strands)
Both of these DNA molecules will get denatured, annealed and elongated to become 4 DNA molecules (8 strands)
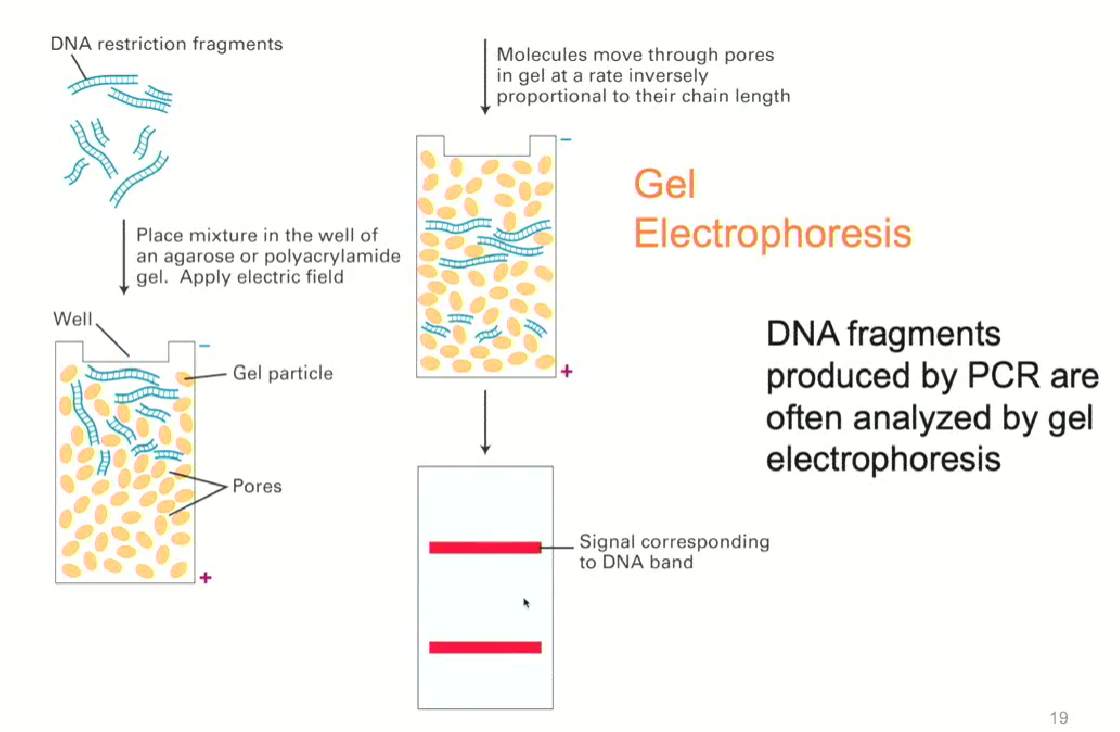
What is DNA gel electrophoresis used for?
To analyze the DNA molecules produced by PCR
DNA is easier to separate since it is negatively charged so it separates based on length
In PCR, the length of the DNA molecules is known, so the gel is used to check if the right length was obtained
DNA gel electrophoresis process
DNA molecules are loaded in a polymer called agarose gel
An electric field is applied
DNA molecules will have similar charges, so they will be separated by length
DNA molecules are stained and bands of DNA are seen
Process of using PCR for cloning
A restriction site is added at the 5’ end only as the 3’ end is needed for elongation (we leave it alone)
When PCR occurs after many cycles, we will obtain a fragment that incorporates the restriction site at both ends
We can use these restriction sites with restriction enzymes to make staggered ends and clone it into plasmids (circular DNA)
What kind of DNA did Dr. Paabo use to study thylacine and the giant sloth?
Mitochondrial DNA because it contains parts that can be used to compare organisms and know their ancestry
He compared these extinct animals’ mitochondria DNA to the mitochondria DNA of modern day animals
At that time there was no database of the DNA, so using the mitochondria DNA was helpful (this is still a limitation despite the power of PCR)
Applications of sequencing (4 options)
Phylogenetic relations between species
Human (animal) ancestry
Catching criminals (DNA fingerprinting)
Diagnosis and predisposition to disease
Sanger Sequencing
Classical technique for DNA sequencing
Helps to determine the exact order of nucleotides in a DNA sequence
What do we need for sanger sequencing?
DNA polymerase
oligonucleotide primer
DNA template
dNTPs (100mM)
In addition we need fluorescently labeled ddNTPS (4 types) so that we can differentiate
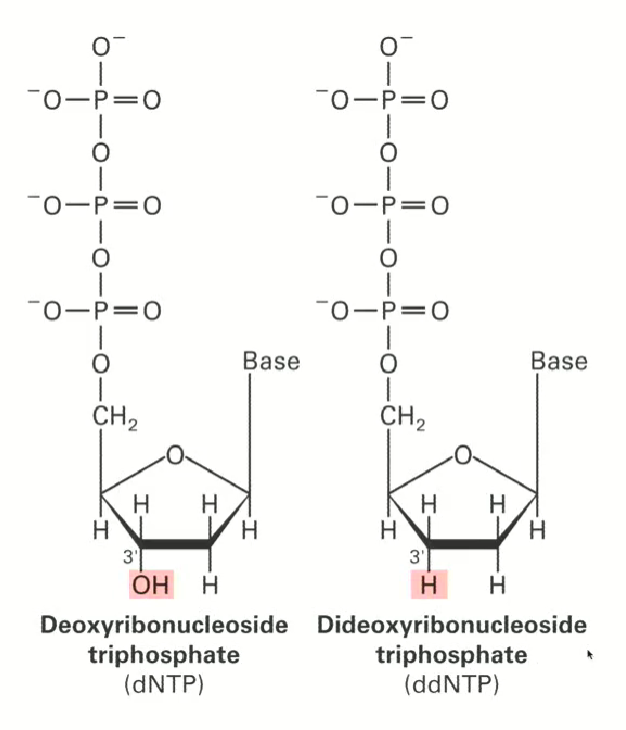
Difference between a regular dNTP (deoxyribonucleoside triphosphate) and a ddNTP (dideoxyribonucleoside triphosphate)?
ddNTP is missing the 3’ hydroxyl group (OH), it has a hydrogen instead
we need the hydroxyl group to extend the chain in the DNA sequence, so if a ddNTP, which lacks a hydroxyl, is attached, the chain will not extend further.
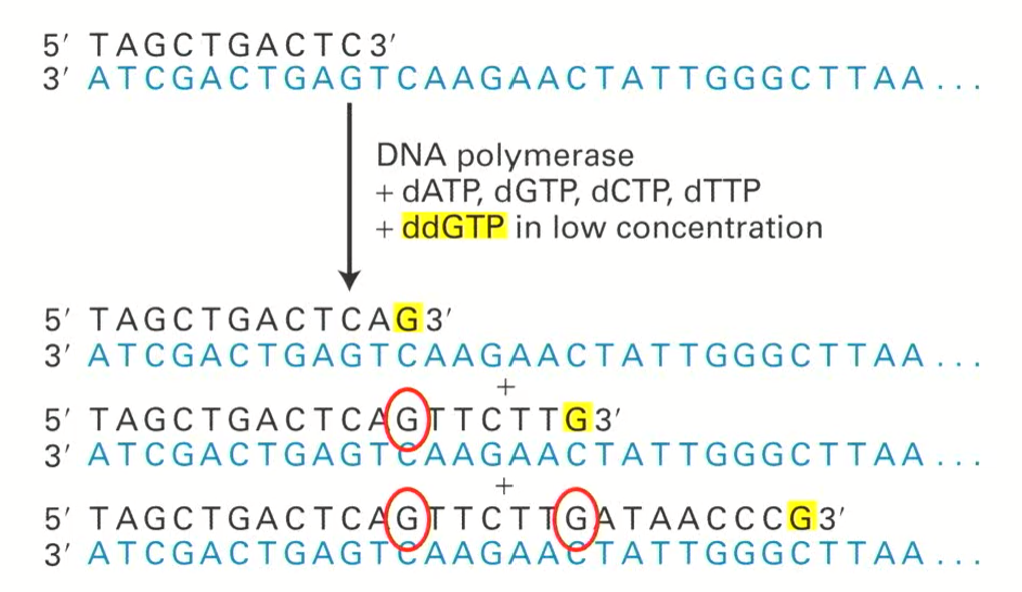
why are ddNTPs present in much lower concentrations than dNTPs? (100x less) ddGTP example included
ddNTPs are present in lower concentrations because we do not want to terminate every chain (only sometimes)
Every time a G is incorporated, the sequence MIGHT halt, but it usually continues since there is a much higher concentration of dNTPs.
Why is it helpful that the four ddNTPs are fluorescently labelled differently?
Because the ddNTPs are fluorescently different, the experiment can be done for all the ddNTPs at the same time and the ddNTP incorporated will be known by the colour
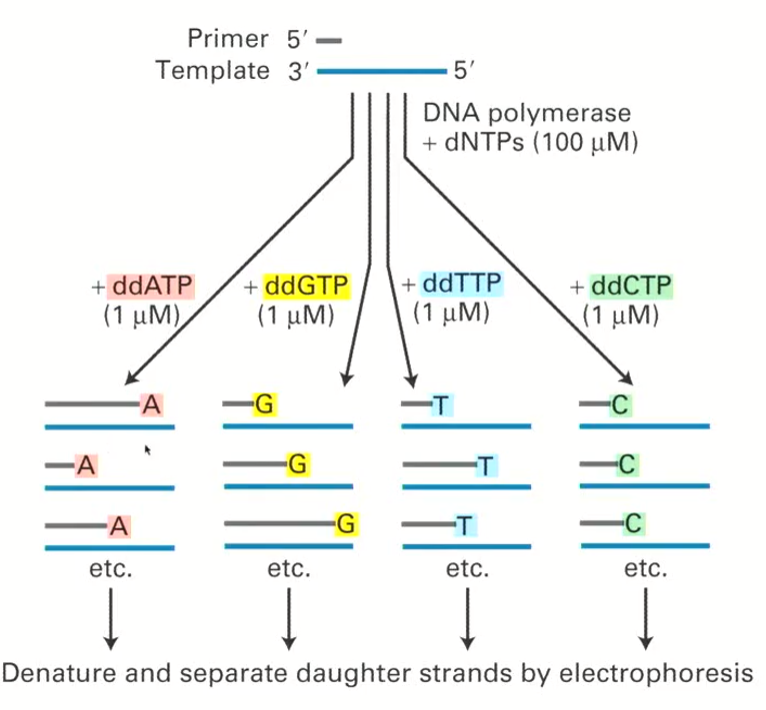
What do we do after a ddNTP is incorporated in sanger sequencing?
The daughter strands will be denatured and separated through gel electrophoresis
A gel scan will be obtained
How do you read a gel scan?
Each lane of the gel will represent a different sequence by different colours, and the length of the gel determines the number of sequences in the DNA molecule
Different colours represent different nucleotides, so we can know the sequence by converting the colours into their respective nucleotides
This is the sanger sequence
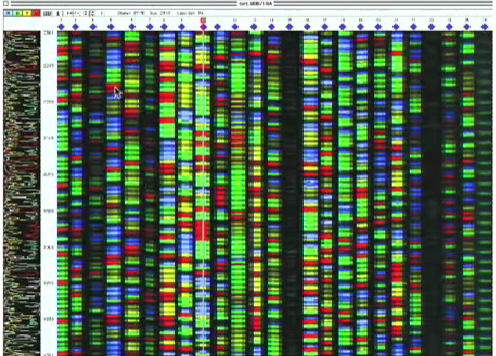
What is the number of sequences that can be obtained dependent on?
The physical capacity of the gels ( the bigger they are, the more sequences we can get)
Why do we need to do sanger sequencing a lot?
There are usually only 500-1000 nucleotides in a Sanger Sequence, but billions in the human genome. A lot of experiments should be run to get the whole genome, so this shows a limitation of Sanger sequencing.