BIO 125 | 2.1 Transcriptional Regulation
1/80
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
81 Terms
T/F: Microbes regulate gene expression at three different levels
TRUE
3 gene regulatory mechanisms utilized by microbes
Transcriptional regulation
Translation regulation
Regulation of protein activity
Explain
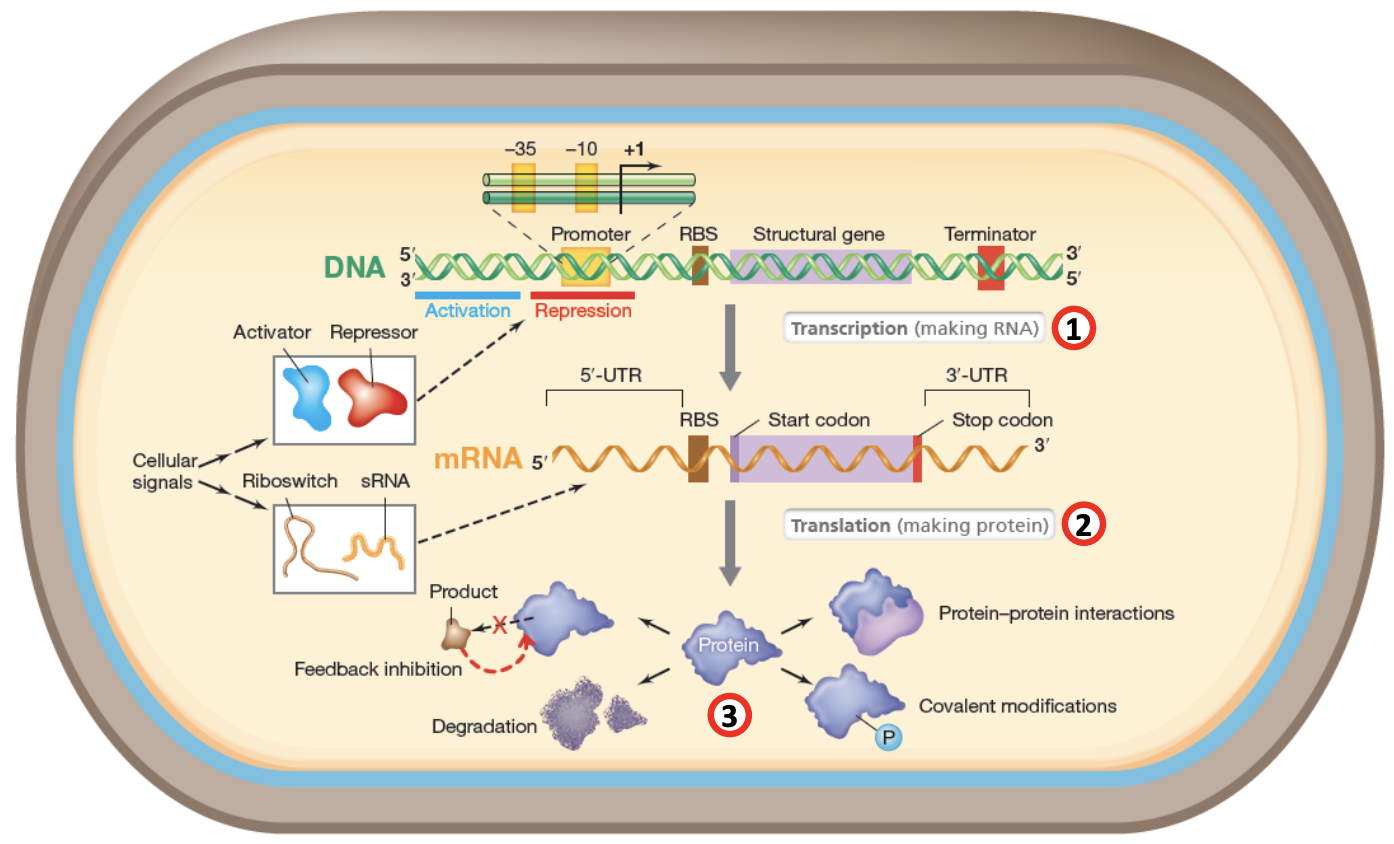
Microbes regulate gene expression at 3 levels
Transcriptional regulation
Control synthesis of RNA from DNA
DNA with promoter region (-35, -10, +1), RBS, structural gene that will be transcribed
Cellular signals
Activators promote transcription by helping RNA polymerase bind to promoter region
Repressors block transcription by preventing RNA polymerase from binding
If transcription is activated, mRNA is produced. If repressed, transcription is blocked.
Translation regulation
Occurs at initiation of translation
Cellular signals
Riboswitches: regulatory segments of mRNA that bind to specific ligands, changing their conformation and hiding RBS from ribosome in hairpin structures.
In the absence of ligands, riboswitches will remain in open conformation and won’t be able to block translation.
sRNAs: noncoding RNAs regulate translation through direct base-pairing interactions with mRNA
sRNAs sometimes bind to mRNA, overlapping with RBS and thus preventing ribosome from physically binding to it
sRNA could also promote translation if it binds to mRNA in such a way that disrupts inhibitory structures like hairpin, exposing RBS to ribosome.
Regulation of protein activity
Covalent modification, e.g., phosphorylation could activate or deactivate protein’s function
Protein-protein interactions: proteins forming complexes that are only functional when certain conditions are met
Degradation: Proteins no longer needed are marked for degradation to prevent accumulation
Feedback inhibition: Final product of pathway inhibits earlier steps of pathway to prevent overproduction of products and maintain homeostasis
_ interacts with major groove DNA to control transcription
DNA-binding proteins (DNABPs)
Part of DNA-binding protein that fits into major groove of DNA and interacts with base pairs
Domain
Parts of protein with specific structures and functions
Domain
Property of DNABPs having 2 identical subunits with various domains
Homodimerism
Where do DNABPs bind in DNA
They bind to inverted repeats in major groove DNA, which is within promoter region
Specific sequences that are identical when read in opposite directions in 2 strains; binding site for DNABPs
Inverted repeats
Why DNABPs bind to major groove DNA
Provides more access to nucleotide bases, making it a more suitable site for protein binding
Explain how DNABPs control transcription
DNABPs recognize specific sequences, often the inverted repeats, in the major groove DNA (which provides more access to nucleotide bases than minor groove).
DNA-binding domain of protein will then interact with base-pairs within recognized sequence.
The binding of DNABPs to these specific sites can influence whether a gene is turned on or off.
Specific regions of protein with specific folding structure and function
Domains
Typical site of binding, usually at inverted repeats
Major groove
Various DNABPs interact with DNA in _ and control transcription
promoter region
Specificity of DNABPs depend on
interactions between several DNABP amino acids and DNA bases
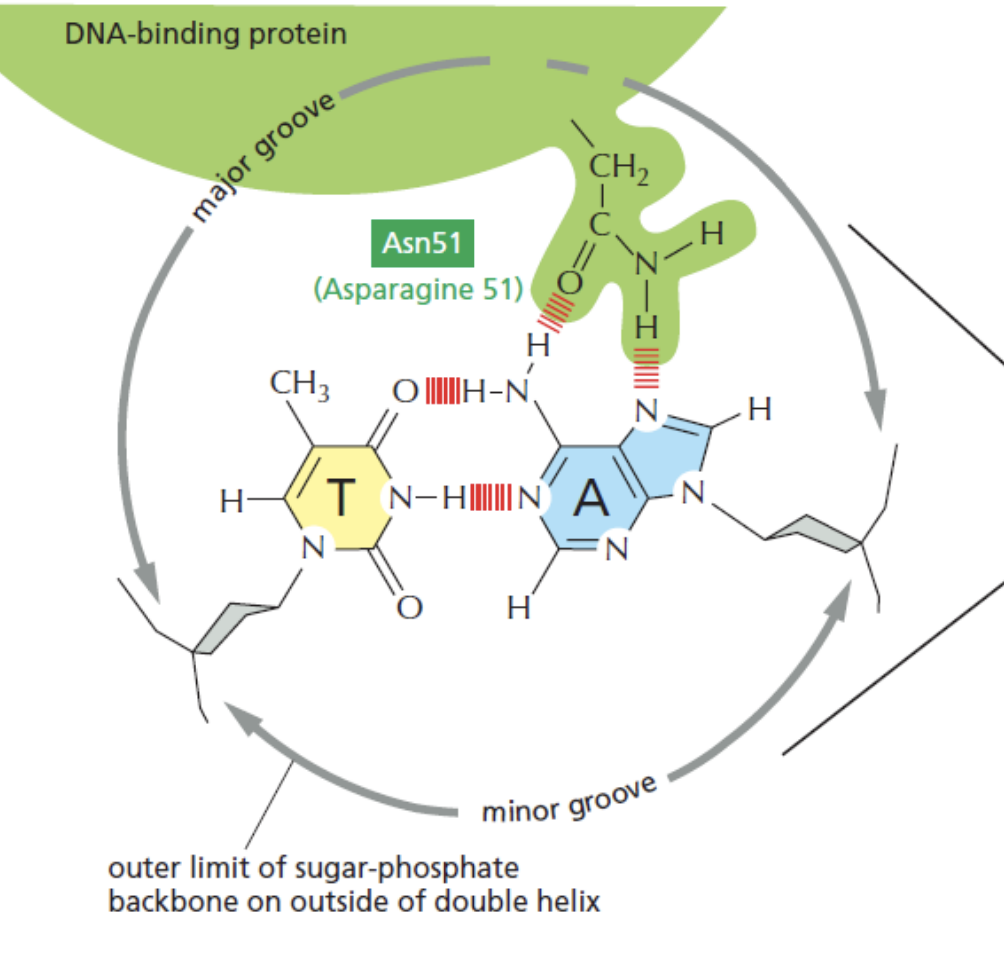
Explain
Asparagine 51 (Asn51) of DNABP interacts with Thymine-Adenine (TA) base pair on the DNA by forming hydrogen bonds with the N & O atoms of the bases.
DNABPs can read and recognize specific sequences based on the arrangement of hydrogen bond donors and acceptors in DNA.
T/F: Each DNABP amino acid interacts with a base through hydrogen bonds, hydrophobic interactions, or other forms of molecular recognition.
TRUE
T/F: DNABP specificity arises from their AA side chains fitting into major grooves of DNA and forming specific interactions with the DNA bases
TRUE
T/F: Many DNABPs interact with DNA minor groove than major groove
FALSE
DNABPs bind more often on major groove since it’s wider and the bases are more exposed, allowing for more specific interactions. Meanwhile, minor grooves are narrower and some AA are still intact, making it less accessible to DNABPs.
Common structural motifs for DNABPs
Helix turn helix proteins
Helix loop helix
Homeodomain
Zinc finger
Leucine zipper
Beta sheet recognition proteins
T/F: Helix-based motifs (HTH, HLH, Leucine zipper) primarily bind the major groove DNA
TRUE
Why is helix turn helix the most common structural motif for DNABPs?
Its recognition helix fits precisely into the major groove of DNA, allowing for specific base pair interactions.
Its stable structure, with 2 helices connected by a short turn, ensures proper orientation for effective binding.
Additionally, the simplicity and versatility of its structure make it adaptable across a wide range of DNABPs, from repressors to activators.
First helix, responsible for recognizing and binding to specific DNA sequences
Recognition helix
Second helix maintaining overall structure of motif
Stabilizing helix
Short loop connecting 2 helices in HTH motif
Turn
How to increase binding selectivity of DNABP
Dimerizing DNABPs allow for more selective binding to sequences
T/F: Cis-regulatory sequences are regions of noncoding DNA that regulates transcription of nearby genes by encoding the proteins themselves.
FALSE
Noncoding. Instead they serve as binding sites for proteins (e.g., transcription factors), which can enhance or repress transcription of associated genes.
Cis-regulatory sequences vs. Trans-regulatory
Cis-regulatory sequences are located on the same DNA molecule as the gene they regulate.
Trans-regulatory sequences are factors coming from other parts of genome and can affect gene from a distance, acting on cis-regulatory elements.
How does dimerizing a DNABP increase binding selectivity?
Dimerizing DNABPs increase their binding selectivity because it extends the length CRS necessary for binding.
With 2 recognition sites in play, the protein now has to recognize a longer and more specific sequence, making it less likely to bind to non-target DNA regions.
This higher specificity will ensure that only sequences matching the full binding site are targeted, thus reducing off-target interactions.
Explain noncooperative vs. cooperative binding
Noncooperative binding is when DNABPs already exist as dimers before binding to CRS.
Noncooperative bc each binding event is independent of others.
Exponential curve because binding is proportional to the DNABP concentration, meaning that as DNABP concentration increases, more dimers bind to CRS, leading to gradual, continuous increase of DNA occupancy.
Cooperative binding is when DNABPs exist as monomers and only dimerize upon binding to CRS.
Cooperative bc the binding of 1 monomer to CRS increases likelihood of second monomer binding.
All-or-none phenomenon leading to sigmoidal curve because cooperativity feature creates a threshold effect, where at low concentrations, binding will be slow because monomers need to form dimers.
But, once critical concentration is reached, binding will be rapid, leading to sharp increase in DNA occupancy and creating sigmoidal curve.
T/F: All activators are transcription factors
FALSE
Not all activators are transcription factors. Activators are molecules or proteins that enhance gene expression, but they don't have to directly bind to DNA. Transcription factors, on the other hand, are proteins that regulate gene expression by binding to specific DNA sequences. Some activators work indirectly, influencing transcription factors or other proteins involved in gene regulation.
Proteins that control the rate of gene transcription by binding to specific DNA sequences
Transcription factors
Proteins increasing transcription rate; bind to activator binding sites
Activator
Proteins decreasing transcription rate; bind to operator region
Repressor
Phenomenon where binding of effector molecule changes conformation of transcription factor
Allosterism
Molecules binding to TF, changing its conformation and switching transcription on/off
Effector
Effector that switches transcription on
Inducer
Effector that switches transcription off
Corepressor
Transcription factors can enhance transcription rates through _ and decrease transcription rates through _
Induction, derepression
Repression
T/F: Activities of activator and repressor proteins are complex forms of regulation that repress or induce protein production.
FALSE
Simple forms of regulation
Transcription rate control works based on availability of final product
Repression
Explain why repression usually occurs in anabolic enzymes
Because anabolic enzymes are enzymes involved in biosynthetic pathways that build complex molecules and repression ensures that unnecessary enzyme production stops once enough of the end product is produced to conserve cellular resources.
In the case of arginine biosynthesis, once the end product (arginine) accumulates, it will act as corepressor and bind to ArgR repressor, causing it to bind to the operator region and block further transcription of biosynthetic enzymes.
This prevents further synthesis of arginine when sufficient levels are already present.
What acts as corepressor to turn enzymes off in enzyme repression
Final product of biosynthetic pathway
Objective of repression
To ensure that unnecessary enzyme production stops once enough of the end product is present to conserve cellular resources.
Is arginine operon repressible or inducible
Repressible
Is lac operon inducible or repressible
Inducible
Transcription rate control where lack of substrate or presence of inducer turn enzymes on
Induction
T/F: Induction typically occurs in catabolic enzymes
TRUE
When needed substrate is present (e.g., lactose), it will trigger enzyme production
Process where repressor protein is inactivated by an inducer to allow genes to be transcribed
Derepression
When is the only time lac operon is turned on?
When bacteria does not have glucose and only have lactose
LAC OPERON ON/OFF?
+G, +L =
+G, -L =
-G, -L =
-G, +L =
Off
Off
Off
On
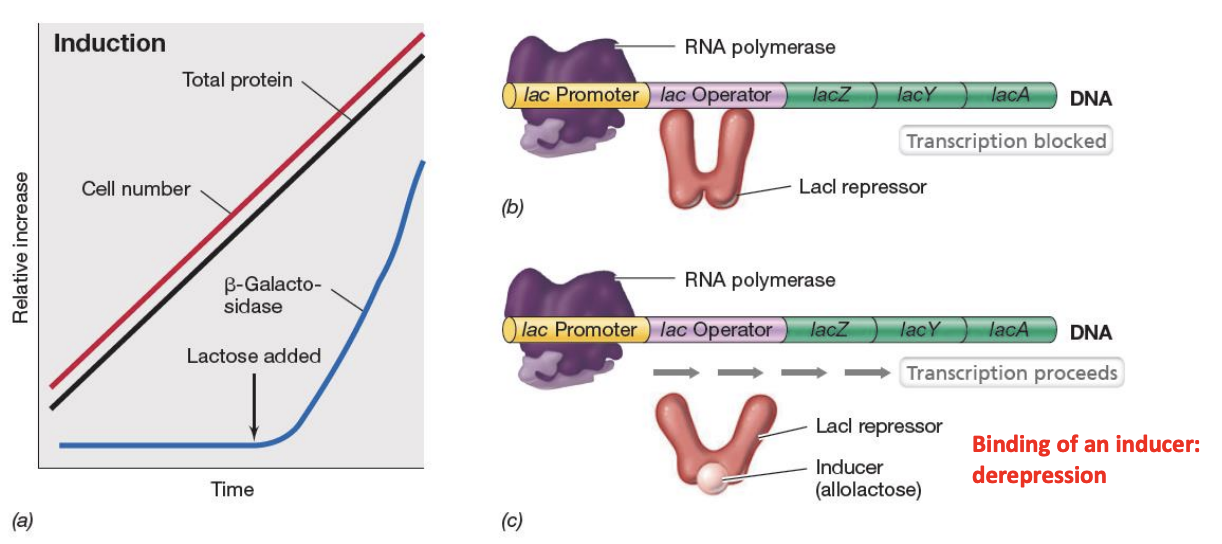
Explain induction / depression of lac operon
Lac operon will only be turned when bacterium don’t have glucose but instead have lactose.
In the absence of lactose, the LacI repressor will remain bound to lac operator region, preventing RNA polymerase from transcribing genes necessary for lactose metabolism.
In the presence of lactose, allolactose will act as inducer by binding to LacI repressor, causing it to detach from lac operator region and subsequently allowing RNA polymerase to transcribe genes for lactose metabolism.
This ensures that enzymes for metabolism of certain substrates will only be produced when substrates are actually present.
Aim of induction
To ensure that enzymes necessary for metabolism of certain substrates will only be produced when those substrates are actually present to conserve cellular resources
Form of lactose acting as inducers of LacI repressor
Allolactose
T/F: Activators are not always needed by RNA polymerase
TRUE
If promoter is sufficiently strong and RNA polymerase can easily recognize and bind to it, transcription can proceed at baseline level.
T/F: In the presence of activators, basal transcription rate is increased. In presence of repressors, basal transcription rate is decreased.
TRUE
T/F: Activator proteins can only bind to activator binding sites close to promoter.
FALSE
Activator proteins can bind to ABS both close to and far away from the promoter.
T/F: In many cases, activator protein binding is necessary because some promoters have poor matching to RNA polymerase.
TRUE
Activator protein function
To help RNA polymerase recognize the promoter and begin transcription
T/F: Looping may be necessary to activate RNA polymerases using some activators
TRUE
How is an activator binding site being hundreds of base pairs away from promoter region remedied
The DNA must loop to bring the activator binding site in close proximity to the promoter region, such that activator protein will be able to physically interact with RNA polymerase, boosting transcription rate.
Use of inducers to regulate the activators controlling RNA polymerase
Positive control
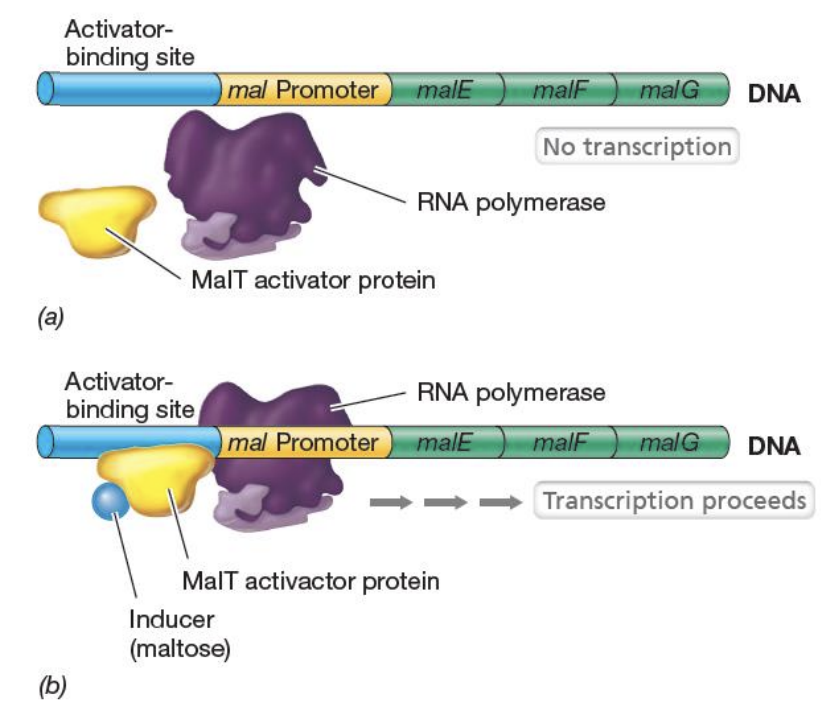
Explain this case of positive control
In the absence of maltose (inducer of malEFG operon),
neither the MalT activator protein and RNA polymerase binds to DNA
In the presence of maltose,
it will bind to the MalT activator protein, subsequently causing it to bind the activator binding site.
Binding of activator protein to ABS will allow recruitment of RNA polymerase to promoter region and begin transcription.
A group of operons under the control of a single regulatory protein
Regulon
Operon vs. Regulon
Operon
Genes grouped together physically in 1 chromosome, under the control of 1 promoter / regulatory element
e.g., LacI repressor can only bind to lac operon and regulate transcription of lactose metabolism genes located in that one position.
Regulon
Multiple operons scattered throughout genome but regulated by 1 regulatory protein
e.g., MalT regulatory protein can bind to different operons within the regulon, allowing genes for maltose utilization to be expressed in various locations of the genomes.
T/F: The genes and operons required for maltose utilization (mal) are dispersed throughout the E. coli genome and regulated by the same maltose regulatory protein (MalT)
TRUE
Advantages and disadvantages of regulon
Advantages
Coordinated gene expression across dispersed locations
Useful when the cell needs to respond quickly to a specific environmental signal, such as the presence of a nutrient or stress factor.
Also, in situations where multiple genes in different locations need to be turned on (e.g., during nutrient scarcity or in stress conditions), a single regulatory protein can trigger their expression simultaneously, making the response faster and more efficient.
Optimization of Resource Usage
By coordinating the expression of multiple genes under one regulatory signal, the cell can ensure that related genes are expressed only when needed, thus conserving energy and resources for other processes.
Disadvantages
Global Disruption in Case of Mutation
Since a single regulatory protein controls multiple operons, a mutation in the regulatory gene or its protein can lead to widespread disruption across the entire regulon. This could negatively impact several biological pathways simultaneously, potentially leading to severe consequences for the cell’s survival.
Potential for Unintended Cross-Regulation
For instance, if similar binding sequences exist elsewhere in the genome, the regulator might mistakenly activate or repress genes not intended to be part of the regulon, leading to uncoordinated gene expression.
T/F: Transcription regulation in Archaea more closely resembles that of Eukarya than Bacteria.
FALSE
The regulation of transcription in Archaea more closely resembles that of Bacteria.
Inducible or repressible?
NrpR operon
Inducible
The (+) and (-) regulation systems in Archaea controls the binding of
RNA polymerase
TBP or TFB
Explain NrpR protein as archaeal repressor in nitrogen metabolism
NrpR protein acts as archaeal repressor in nitrogen metabolism because in the presence of ammonia, it will bind to BRE, TATA in promoter, physically blocking access of RNA polymerase to DNA and preventing transcription of genes for nitrogen metabolism (since there’s no need to metabolize nitrogen in the presence of ammonia).
Meanwhile, in the shortage of ammonia, a-Ketoglutarate will not be converted into glutamate and, upon its accumulation, will bind to NrpR protein, causing the NrpR protein from being released in DNA and subsequently allowing binding of TBP, TBF to promoter, recruiting RNA polymerase and beginning transcription.
(Makes sense since in shortage of ammonia, there is a need to metabolize nitrogen and convert into ammonia)
NrpR protein functions as repressor in nitrogen metabolism of which archaeal species?
Methanococcus maripaludis
Explain how TrmBL1 can act as both repressor and activator in archaea
TrmBL1 as repressor in sugar uptake operon
Without maltose, TrmBL1 remains bound to region near promoter (BRE, TATA), preventing binding of TBP, TFB to BRE, TATA and recruitment of RNA polymerase.
No transcription of sugar uptake genes (since there is no sugar to uptake anyway)
In the presence of maltose, TrmBL1 binds to maltose, releasing itself from DNA and thus allowing binding of TBP, TFB to BRE, TATA and recruiting RNA polymerase, subsequently beginning transcription of sugar uptake genes.
There is transcriptipn of sugar uptake genes since there is sugar to uptake.
TrmBL1 as activator in glucose synthesis operon
Without maltose, TrmBL1 binds upstream of BRE, TATA, allowing binding of TFB, TBP and aiding recruitment of RNA polymerase, beginning transcription of genes for glucose synthesis.
No glucose so glucose has to be synthesized!
With maltose, TrmBL1, instead of binding upstream and aiding RNA polymerase recruitment, binds to maltose.
Release of TrmBL1 from DNA causes dissociation of components of initiation complex, including TBP, TFB, RNA polymerase.
No need to synthesize glucose since there is glucose!
T/F: Some archaeal regulators function as both repressor and activator
TRUE
_ are universally encoded transcription elongation factors in archaeal genomes
SPT4/SPT5 complexes
T/F: Some archaeal proteins interact with DNA not in promoter or activator r
TRUE
Archaeal Spt4/5 secures RNA polymerase onto the DNA template when the initiation complex transitions to the elongation complex, rendering the elongation complex more stable.
Explain how SPT4/5 functions
SPT4/5 complex does not bind at typical regulatory spots (promoters, activator regions) but instead binds to RNA polymerase during elongation phase of transcription.
Helps in stabilizing RNA polymerase by ensuring it stays attached to DNA and continues transcription efficiently.
_ regulon system of E. coli consists of 10 genes located in five operons that encode proteins involved in the uptake of maltose and short maltodextrins.
Maltose/maltodextrin regulon system
T/F Enzyme repression typically refers to catabolic enzymes that are turned off by the final product of a biosynthetic pathway.
False
Justification: Enzyme repression usually involves anabolic enzymes where the final product acts as a corepressor, turning off the enzyme.
T/F Positive control mechanisms always require the presence of inducers to enhance the activity of transcription factors.
True
Justification: Positive control mechanisms depend on inducers to activate transcription factors, thereby promoting gene expression.