Lesson 4: Protein synthesis and degradation: ribosomes and proteasomes
1/24
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
25 Terms
Ribosomes
Non-membranous organelles where proteins are synthesised
Ribosome Classification
Free ribosomes in the cytoplasm: bound to cytoskelten, synthesize cytoplasmic proteins
Bound to RER: external layer RER membrane, synthesize protein: membrane proteins or secreted proteins
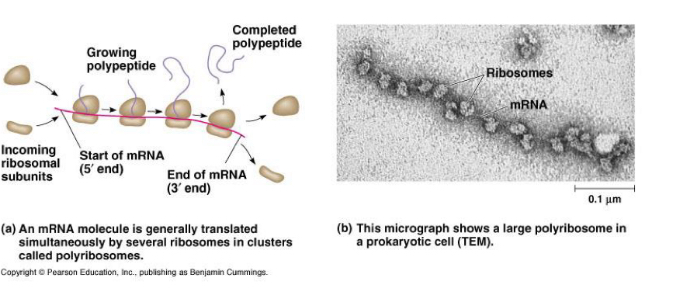
Polyribosomes (in cluster): Cytoplasmic assemblies made up of several ribosomes (5-20 ribosomes) spaced along a single mRNA molecule.
Ribosome Structure
Consist of 2 Subunits which are separated in cytosol but bind for protein synthesis: Small and big Subunit
In prokaryotic cells and Mitochchondria: 70S
In Eukaryotic cells: 80S (60S + 40S+)
Ribosome composition
rRNA (Ribosonal RNA) 60%:
-transcribed from DNA, named nuclear organizer, in nucleolus
-Eukaryotic ribosomes contain 4 rRNA molecules
-Large subunit: 28S
-Small subunit: 18S
Ribosomal proteins 40%:
-translated from their own mRNAin other already existing ribosomes (In cytoplasm)
-Large subunit: -50 proteins
-Small subunit: -30 proteins
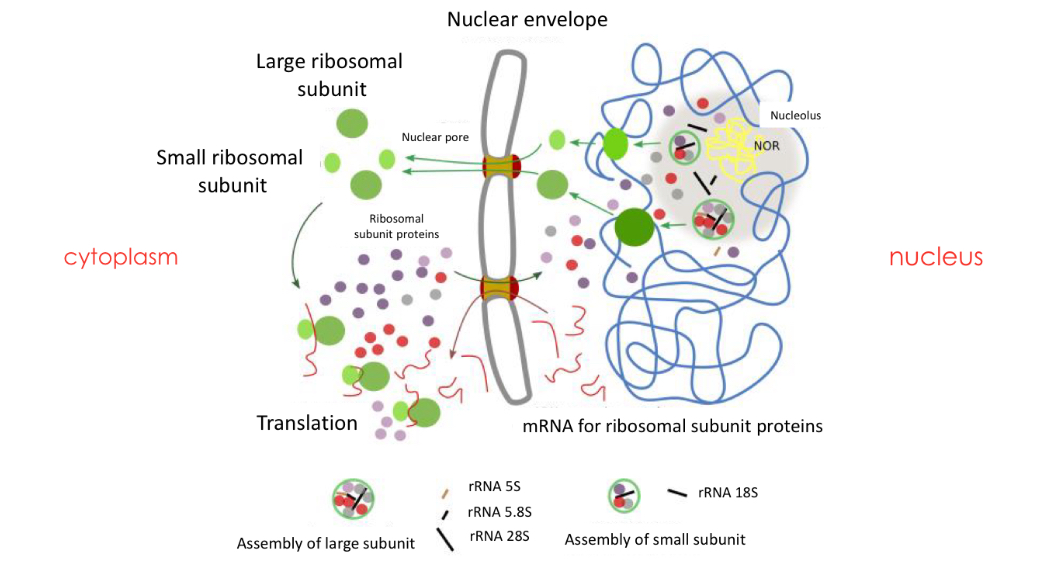
Formation of eukaryotic ribosomes
Nucleolus responsible for: rRNA formation, assembly of ribosomal formation
-Inside Nucleolus, Nucleolar organiser region (NOR) contains genes from different regions of one or more chromosomes, that encode for rRNA:
Nucleolar organizer DNA is transcribed into rRNA.
Which genes does NOR contain
•genes clustered on the short arms of 5 chromosomes: 13, 14, 15, 21 and 22.
• Once transcribed, these rRNAs and imported ribosomal
proteins will be assembled to form the different
ribosomal subunits in the nucleolus.
Formation steps
Synthesis of rRNA and proteins: separately
rRNA in in the NOR in nucleolus
Synthesised in cytoplasmic ribosomes → enter nucleus then nucleolus
Formation of 2 subunits: in the nucleolus
The proteins that have migrated to the nucleolus, assemble with the rRNA to form the two subunits (the large subunit and the small subunit) separately.
Assembly of both subunits: Exported to the cytoplasm
The two subunits leave the nucleus to the cytoplasm. They remain separate. Both subunits bind only during protein synthesis.
DNA - Desoxyribonucleic acid
-double helix nucleic acid, which contains the genetic
instructions for synthesising other components of the cell (RNA molecules and proteins).
-It is also responsible for hereditary transmission.
Nucleotides: A,T, C, G
RNA – ribonucleic acid Types
single-stranded nucleic acid, may be:
• mRNA – messenger RNA: contains the genetic Information to synthesize the proteins
• rRNA – ribosomal RNA: structure of ribosomes Ribosomes are large complexes of RNA and protein.
• tRNA – transfer RNA: identifies the appropriate amino acids for protein synthesis and brings them to the ribosome
Nucleotides: A, U, C, G
Proteins
Amino acid polymers (polypeptides) bound by peptide bonds.
Primary structure = amino acid chain
Gene
Specific part of DNA that has specific function
Genetic code
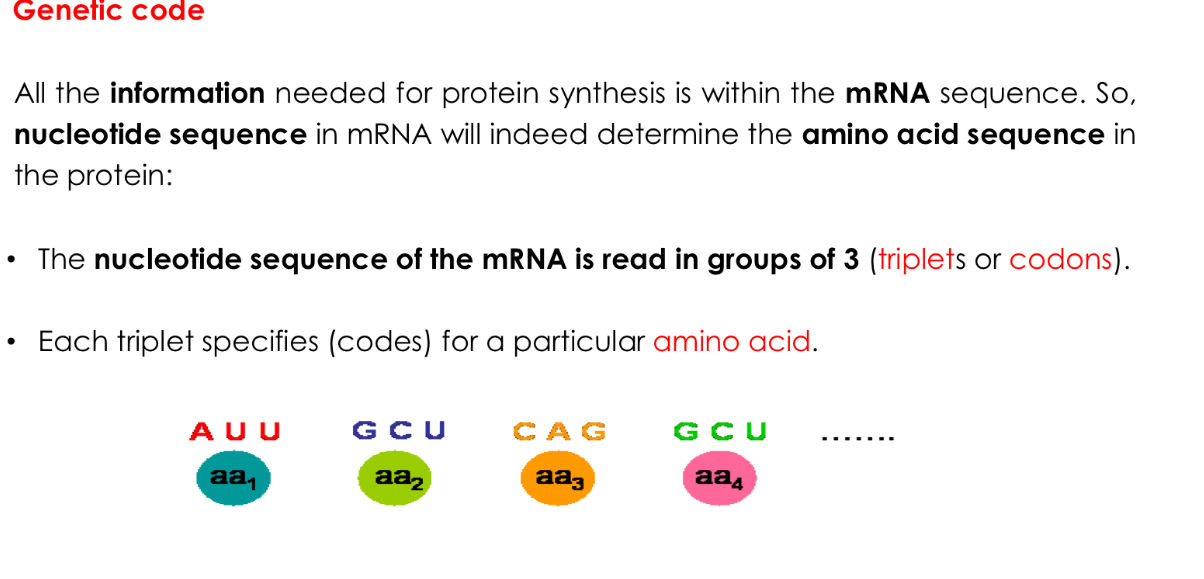
From DNA to proteins: Translation machinery
Ribosome: organelle where translation takes place.
rRNA: part of the Ribosome that:
- enables recognition with mRNA.
- catalyses the reactions of protein synthesis.
– mRNA: indicates the amino acid sequence in the protein being synthesised
– tRNA: transports amino acids to the ribosome depending on the mRNA sequence. It recognise the codon of that amino acid: it serves as a link between mRNA and amino acids.
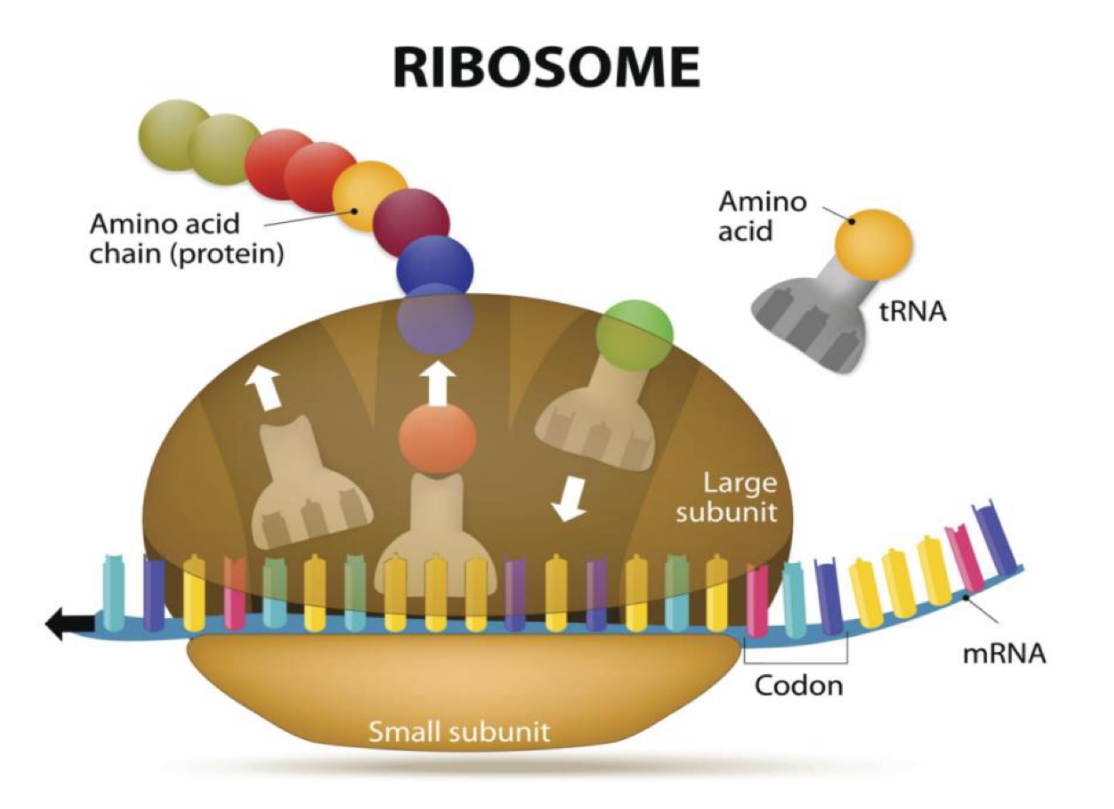
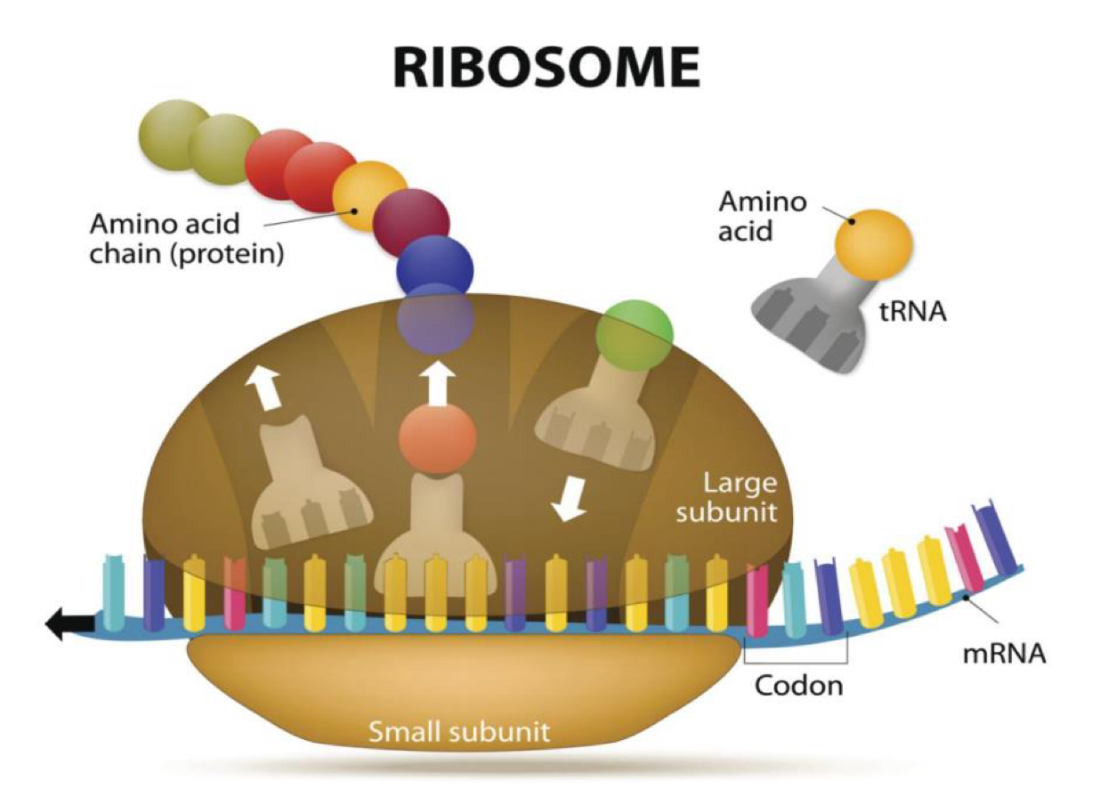
Ribosome structure
ribosomes contain four binding sites for RNA molecules:
▪ 1 binding site for the mRNA in the small subunit
▪ 3 binding sites for tRNAs in the large subunit:
– A site (aminoacyl-tRNA): where the tRNA with the corresponding amino acid enters
– P site (peptidyl-tRNA): where the peptide is placed (amino acid chain in synthesis)
– E site (exit): where the RNA that has already transferred its amino acid is placed
Step: Activation of amino acids
- Recognition between tRNA and amino acid
- Binding of both molecules to form an aminoacyl-tRNA
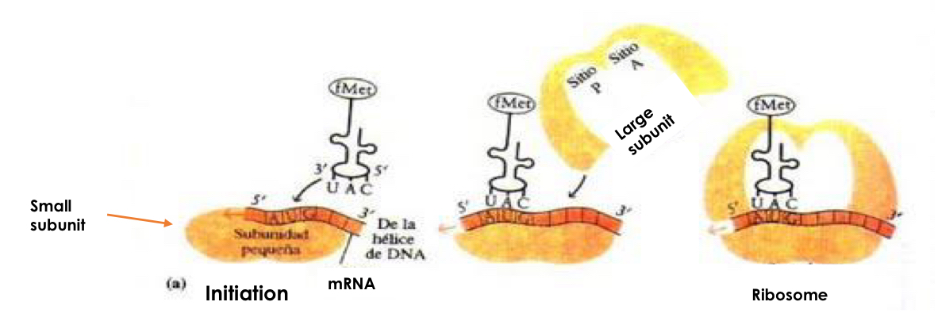
Step: Initiation
-mRNA binds to small subunit of ribosome
-first aminoacyl-tRNA binds small subunit P-site
-Initial codon is AUG and first acid acid is methionine
-large subunit binds to complex
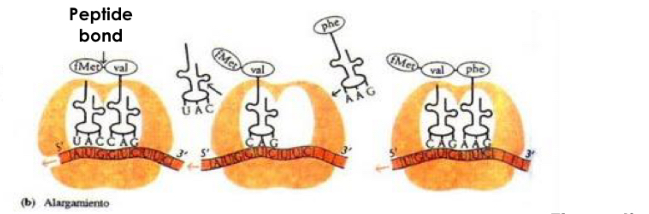
Step: Elongation
-elongation of peptide chain
Steps:
aminoacyl-tRNA binds to A site of ribosome
New peptide Bon formed between amino acid carried by this tRNA and growing amino acid chain
Translocation: ribosome moves a codon on mRNA
A-site: empty, P-site: polypeptidic chain, E-site: tRNA without amino acid
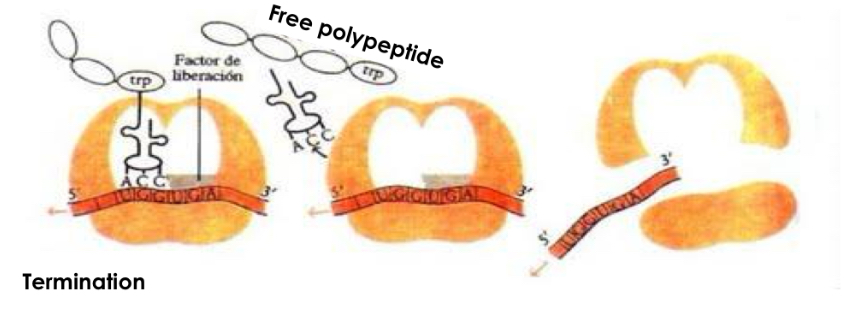
Step: Termination
-A site enters stop codon of mRNA which is not recognised by any tRNA
-no amino acid for new peptide bond
-polypeptide chain released from ribosome
-both ribosomal subunits separate
Protein syntheses in Eukaryotes
-Tales Place after mRNA maturation (splicing and addition of cap)
-Initation of protein synthesis more complex
-Elongation and termination similar
Protein degradation
Key step to recycle nitrogen and iron from proteins
-proteins in constant production and removal
-amount of protein maintained by synthesis and degradation
Main mechanisms to regulate protein degradation
1. PROTEASOMIC DEGRADATION (UBIQUITIN-PROTEASOME SYSTEM)
2. LYSOSOMAL DEGRADATION (AUTOPHAGY)
Proteasomic degradation
Takes place due to macromolecules complex called Proteasome Which hydralyzes proteins in cytosol
Complex (26S) formed by:
▪ core particle (20S)
▪ two regulatory particles (19S) in the ends.
Degrade: Damaged or oxidised proteins, short-lived proteins
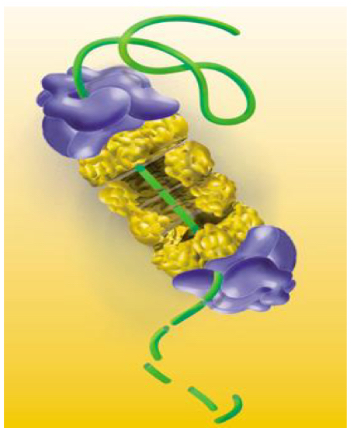
Mechanism
These proteins are labeled by incoparating small protein ubiquitins, proteins recognise ubiqitin and direct the labeled protein towards Proteasome
-ubiquitin capable of polymerising in form of chains
-binds covalently to protein to be degraded via ATP-consumption pathway involving 3 different enzymes (E1-3)
-Labeled proteins interact with regulatory particles
and are degraded by proteolysis in the core
particle.
Dependent of ATP
Lysosomal degradation (Autophagy)
Takes place in lysosomes
-Lysosomes are membrane-enclosed organelles containing hydrolytic enzymes
-responsible for normal destruction and replacement of long-lived proteins: membrane organiser cellular interior proteins that need to be recycled
-damaged proteins, big macrostructures and whole organelles
-also degrade other types of substances: sugars, nucleotides and lipids
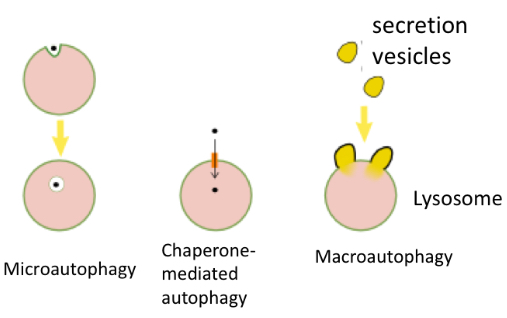
Types of lysosome degradation/autophagy
Microautophagy: the lysosome incorporates
soluble cytoplasmic proteins. The lysosome engulfs
small amounts of cytoplasm containing the proteins to
be degraded (never big structures)
Chaperone-mediated autophagy: lysosomes directly captures the proteins the be degraded by a receptor, called chaperone
the lysosome engulfs a vesicle contains large amount of protein or entire organelle