Cell Biology (copy)
1/226
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
227 Terms
What is the reason behind the differences in function and structure in cells?
Differences come from different accumulation of RNAs distributed in the cells. Some RNAs are common in all cell types (housekeeping genes).
Analysis of mRNA expression can pinpoint which cell type we are looking at.
Why is it important to look at protein levels instead of RNA levels when comparing cell types?
mRNA undergo multiple regulatory steps before being translated
mRNA can be translated multiple times, and are therefore do not always reflect of the protein abundance
proteins can be further modified, which the mRNA does not identify.
What is 2D page, and why is it used?
In 2D page the proteins are fist separated by isoelectric point, and then by molecular weight to isolate different isoforms of the same protein.
What are the different checkpoints in which cells can alter their gene expression (7 checkpoints)?
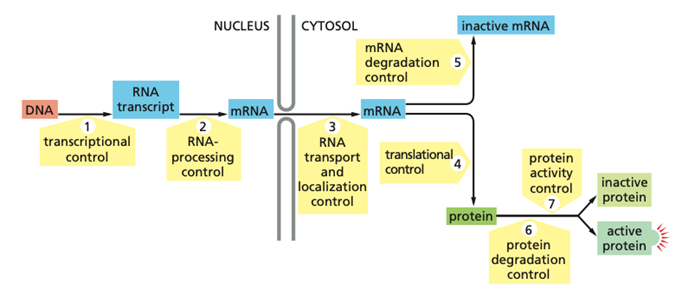
Transcriptional control – control of whether a gene is transcribed from DNA into an RNA transcript or not.
RNA processing control – regulation of how the primary RNA transcript is processed into mature mRNA, including splicing, capping, and poly-A tail addition.
RNA transport and localization control – control of when and where the mRNA is transported from the nucleus to the cytosol and where it is localized in the cell.
Translational control – regulation of whether and how efficiently the mRNA is translated into protein.
mRNA degradation control – control of mRNA stability and how quickly mRNA is degraded or inactivated.
Protein degradation control – regulation of protein lifetime through controlled degradation.
Protein activity control – regulation of whether the protein is active or inactive after it has been made, for example by modification, localization, or inhibitor binding.
Define:
co-activators and co-repressors
mediators
cis- and trans-regulatory sequences
activators
repressors
Co-activators and co-repressors are regulatory proteins that do not bind DNA directly but influence transcription by interacting with DNA-bound transcription factors. Co-repressors decresegene expression by condensing chromatine and inhibiding RNA polymerase binding. Co-activators increase transcription by recruiting RNAP and chromatne remodelling enzymes
Mediator is a large multiprotein complex that acts as a bridge/co-activtor between transcription regulators bound at enhancers and RNA polymerase II at the promoter. It integrates signals from many activators and repressors and helps assemble and regulate the transcription initiation complex. Th mediator is esential as RNA polymerase II does not interact directly with most transcription regulators.
Cis-regulatory sequences are DNA elements located on the same DNA molecule as the gene they regulate, such as promoters, enhancers, and silencers. They serve as binding sites for transcription regulators.
Trans-regulatory factors are usually proteins such as transcription factors, that can diffuse through the nucleus to bind cis-regulatory sequences and control gene expression.
Activators are transcription regulators that increase gene expression by helping recruit or stimulate RNA polymerase and the transcription machinery.
Repressors are transcription regulators that decrease gene expression by blocking RNA polymerase recruitment or by recruiting factors that inhibit transcription.
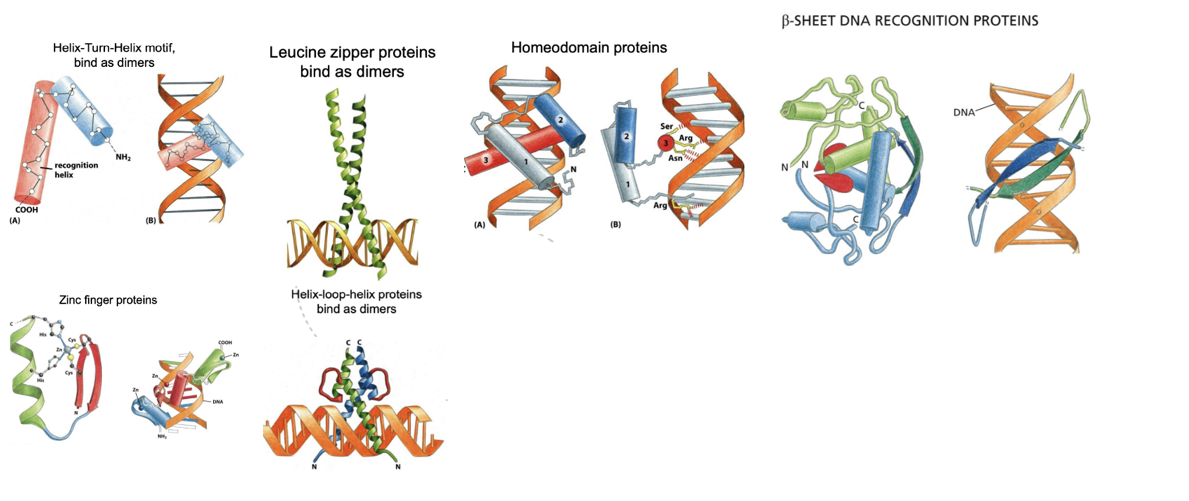
What are Helix–Turn–Helix, Leucine zipper, Homeodomain, Zinc finger, β-sheet, and Helix–Loop–Helix proteins examples of?
They are DNA-binding motifs in transcription factors.
These structural domains allow transcription regulators to recognize and bind specific DNA sequences, usually in the major groove, and often function as dimers to increase binding specificity and affinity.
· Helix-Turn-Helix motif proteins bind to DNA as dimers, and their structure consists of two 𝛼-helices connected by a short stretch of amino acids. They are kept in place by specific interactions that happen between the two of them. The recognition site is placed on the C-terminal helix.
· The Leucine zipper proteins bind as dimers as well to DNA.
· The Homeodomain proteins possess three 𝛼-helices, tightly packed by hydrophobic interactions. The important domains for the recognition of DNA are the helices 3 (major groove) and 1 (minor groove).
· 𝛽-sheet DNA recognition proteins have two 𝛽-sheet domains, which interact with the DNA thanks to the amino acid side chains that go into the major groove.
· The Zinc Finger proteins contain one or more atoms of zinc as structural components.
Helix-Loop-Helix proteins are related to the leucine zipper proteins and have two 𝛼-helix domains joined together by a loop, which allows one of the two helices two interact with the other forming the dimerization surface. The segment of the 𝛼-helices that emerges from there interacts with DNA.
Why do many TFs bind as dimers? what is cooperative binding and non-cooperative binding?
A monomer TF only recognizes a few nucleotides in a DNA sequence, and does not have a strong afinityy towards the sequence. Mnay form dimer as it guarantees extra affinity and more specificity. Dimers can be formed by two molecules of the same protein (homodimers), or by two different (heterodimers). Most of the transcription regulators can associate with a vast range of other proteins, which allows the cell to have more variability when it comes to recognize a specific cis-regulatory sequence.
Cooperative binding is when binding of one protein to DNA makes it easier for another protein to bind nearby, increasing overall binding strength and specificity than if they were monomers. This also produces a switch like system, where a gene is quickly turned on when they are over a certain threshold.
Non-cooperative binding occurs when proteins bind DNA independently of each other, i.e. binding of one molecule does not help another molecule bind.. Binding increases gradually with protein concentration and produces a weaker, more graded response.
Why do TFs bind DNA in nucleosiomes with a lower affinty than naked DNA? What is “breathing”, and how does it promote cooperative binding?
1) the surface of the cis-regulatory sequence (e.g. enhancer, promoter), may be hidden in the nuclosome
2) Transcription factor binding often requires DNA bending or distortion, which is opposed by the tight wrapping of DNA around histones.
Breating is a spontanious unwrapping of the DNA from the surface of a nuclosome due to thermal fluctiuations, which temperarily exposes DNA, allowing TFs to bind. This can support cooperative binding if one TF manage to bind when it sees a window of opportunity, and thereafter another TF can bind when the structure has loosened.
what are pioneer factors?
Pioneer factors are special transcription factors that can bind to DNA inside tightly packed, "closed" chromatin (nucleosomes), which other proteins can't easily access. By binding to these hidden DNA sites, they initiate chromatin remodeling, opening up the DNA to allow other transcription factors and regulatory proteins to bind
Explain the trp operon as an example of gene regulation in prokaryotes
The operon contains a promoter, an operator, and several structural genes that encode enzymes needed for tryptophan biosynthesis. When tryptophan levels in the cell are low, the trp repressor protein is inactive and cannot bind the operator. As a result, RNA polymerase can bind the promoter and transcribe the operon, leading to production of enzymes that synthesize tryptophan.
When tryptophan levels are high, tryptophan acts as a corepressor by binding to the trp repressor. This binding causes a conformational change that allows the repressor to bind the operator sequence. Binding of the repressor to the operator blocks RNA polymerase and shuts off transcription of the operon.
What is the purpose of DNA looping?
DNA looping allows transcription factors bound to cis-regulatory regions (such as enhancers) to interact with RNA polymerase at the promoter, thereby regulating gene expression. RNA polymerase binds to the promoter, not the enhancer, and transcription is activated or repressed through interactions with transcription factors.
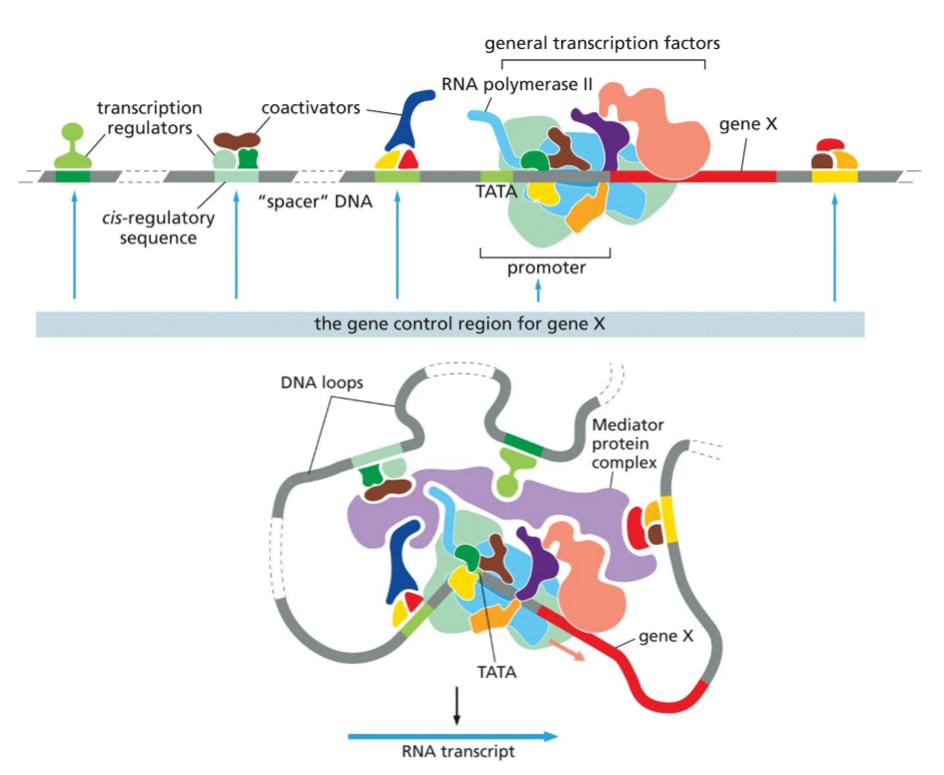
Use the image to explain gene regulation in eukaryotes:
what is gene control region?
what is function of mediator?
difference between general TFs and transcription regulators?
function of co-activators and co-repressors?
final assembly of all individual components decide…?
In eukaryotes, gene regulation depends on the gene control region, which includes the promoter and all cis-regulatory sequences that control expression of a specific gene. Transcription regulators bind these cis-regulatory sequences and influence transcription. Through DNA looping, regulators interact with the promoter-bound machinery via the Mediator complex, which acts as a bridge between transcription regulators and RNA polymerase II together with the general transcription factors. General transcription factors are required for transcription of all genes and assemble at the promoter, while transcription regulators are gene-specific and determine when and how strongly a gene is expressed. Co-activators and co-repressors, which do not bind DNA directly, are recruited by transcription regulators to enhance or inhibit transcription, often by modifying chromatin or stabilizing the transcription machinery. The final combination and assembly of all these components determines whether a gene is switched on or off and the level of its expression.
what is transcriptional synergy? what can activators do when RNAP stops transcribing?
Transcriptional synergy is when multiple activators work together to stimulate transcription much more strongly than the sum of their individual effects.
When RNA polymerase II pauses or stops after initiation, activators can recruit additional transcription factors that remodel the chromatine and support the elongation. the activators “restart” the RNAP
Activators help reshape local chromatin once bound to the cis-regulatory sequences. How and what is the consequence?
Activators reshape local chromatin by recruiting co-activator complexes such as histone acetyltransferases and chromatin-remodelling enzymes. These enzymes modify histones and reposition or remove nucleosomes, making the DNA more accessible. The consequence is a more open chromatin structure that allows general transcription factors and RNA polymerase II to bind the promoter more easily, thereby increasing transcription.
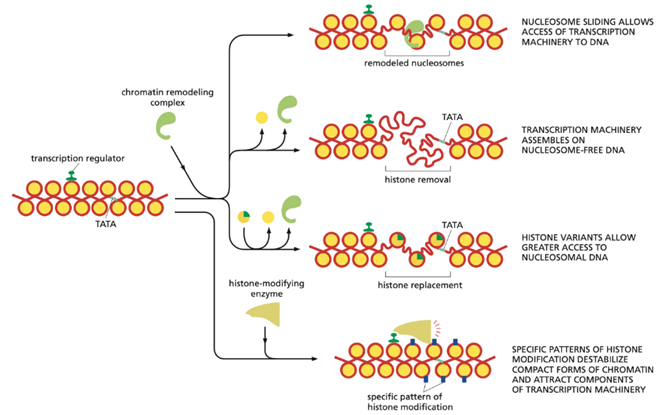
Explain this image:
Transcription activator proteins regulate gene expression by recruiting several proteins that modify chromatin structure at promoters.
These include histone-modifying enzymes, ATP-dependent chromatin-remodelling complexes, and histone chaperones. Together, they remodel nucleosomes by sliding them along DNA, removing them, or replacing standard histones with histone variants, which increases DNA accessibility.
Histone acetylation further promotes nucleosome removal by weakening histone–DNA interactions and facilitating the action of histone chaperones.
As a result, the chromatin becomes more open, allowing general transcription factors, RNA polymerase II, and the Mediator complex to bind efficiently and initiate transcription.
how do repressors work?
Repressors work by reducing or blocking transcription. They can bind to cis-regulatory sequences and prevent activators or the transcription machinery from binding. Many repressors recruit co-repressors such as histone deacetylases or chromatin-condensing complexes, which tighten chromatin and make DNA less accessible. As a result, RNA polymerase II and general transcription factors cannot assemble efficiently at the promoter, and gene expression is reduced or turned off.
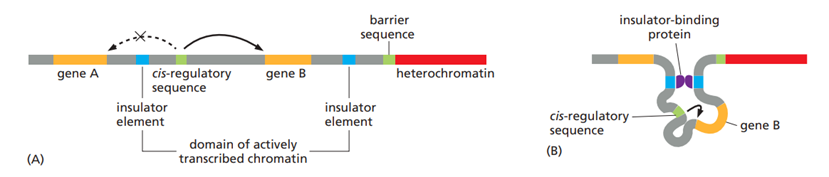
what is the function of insulators?
To avoid crosstalk between different control regions, several types of DNA elements compartmentalize the genome into discrete regulatory domains. Insulators prevent cis-regulatory sequences (activators and repressors) from activating/repressing inappropriate genes by forming loops, keeping a gene and its control region in rough proximity, and helping to prevent the control region from “spilling over” to adjacent genes.
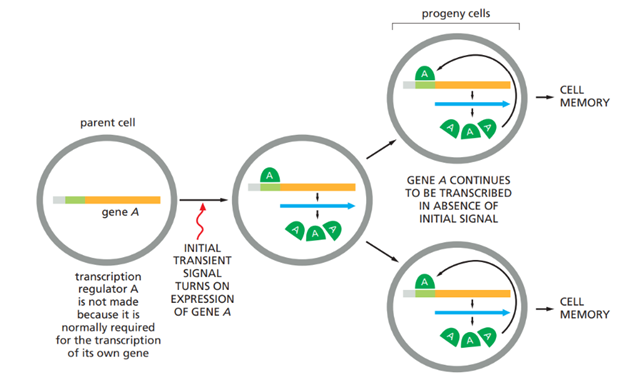
What is cell memory? What is the molecular basis of cell memory in mammals?
Cell memory in mammals is the ability of cells to maintain its gene expression through many rounds of cell divisions, even after the original signal promoting the gene expression is gone. This memory therefore allows differentiated cells to preserve their identity over time, through mechanisms such as a self-sustaining gene regulatory circuit combined with epigenetic mechanisms
Cell memory is often initiated by an external signal that activates a master cell-type–specific transcription regulator. This regulator activates transcription of its own gene as well as other genes required to maintain the differentiated state, creating a positive feedback loop. After cell division, the transcription regulator is inherited by both daughter cells, where it continues to reinforce its own expression and that of downstream target genes. There are several other circuits that cells use to control gene expression. A negative feedback loop is often used to keep the concentration of a protein within a defined range. A low level/lack of repressors cause a higher gene expression, while high levels/active repressors cause a lower gene expression.
If a cell has a misfolded protein and this is transported into the daughter protein, the daughter proteins right folded proteins, can be wrongly folded (prions)
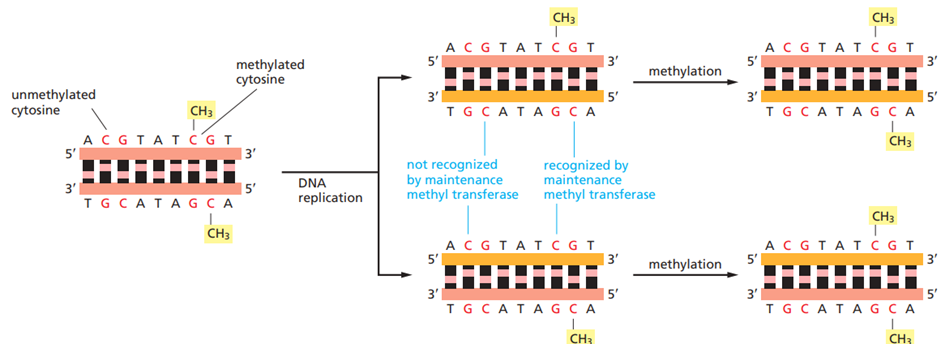
Cell memory is further reinforced by epigenetic mechanisms, particularly DNA methylation and chromatin modifications. In vertebrate cells, DNA methylation occurs mainly at CpG islands, and is associated with transcriptional repression. During replication, DNA methylation at CpG sites is copied onto newly synthesized DNA strands by maintenance methyltransferases, ensuring heritable gene repression. The whole point of methylation is to prevent the transcription of certain regions in the genome, as methylation blocks transcription factors from binding directly to the DNA.
Histone modifications act together with DNA methylation to stabilize gene expression states. When a gene needs to be silenced, repressor proteins can recruit enzymes that add specific chemical marks to histones. These histone marks make the chromatin more compact and can attract other proteins, including enzymes that add DNA methylation. DNA methylation then further strengthens gene silencing and helps recruit even more chromatin-modifying proteins. In this way, gene repression reinforces itself and can be copied each time the cell divides.

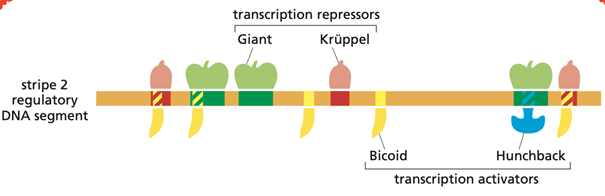
What are the molecular mechanisms that regulate the expression of the Eve gene (Even-skipped) in Drosophila?
The expression of the Even-skipped (eve) gene in Drosophila is regulated through a combinatorial transcriptional control that generates precise spatial patterns during early embryonic development.
When eve expression begins, the Drosophila embryo is a single large cell containing many nuclei within a shared cytoplasm. This cytoplasm contains transcription regulators that are distributed unevenly along the anterior–posterior axis, creating positional information that distinguishes different regions of the embryo. The regulatory DNA sequences of the eve gene have evolved to sense and interpret these concentration gradients, resulting in eve being expressed in seven sharply defined stripes.
Regulation is mediated by multiple cis-regulatory enhancer elements, each of which integrates positional information provided by transcription factors present in specific gradients or domains. Key regulators include Bicoid and Hunchback, which function as transcriptional activators, and Giant and Krüppel, which act as repressors. Each enhancer contains binding sites for both activators and repressors, and transcription is dependent on the concentration of each.
The combined action of these four transcription factors determines the precise spatial pattern of eve expression. The gene control region is extensive and contains multiple interacting cis-regulatory sequences that define the individual expression stripes. Maximal transcription requires the presence of both activators, whereas binding of either repressor is sufficient to shut off eve transcription.
Describe how pre-mRNA is processed and how that can affect the produced protein.
Pre-mRNA processing is a series of co- and post-transcriptional modifications that convert a newly synthesized pre-mRNA into a mature mRNA that can be translated. These processing steps strongly influence which protein is produced, how much of it is made, and how stable the mRNA is.
The first major step is intron removal by splicing. Introns are removed and exons are joined together by the spliceosome, a large RNA–protein complex that recognizes conserved sequences at the 5′ splice site, the branch point, and the 3′ splice site. Many genes undergo alternative splicing, in which different combinations of exons are joined. This allows a single gene to produce multiple mRNA variants and therefore different protein isoforms with altered domains, functions, or localization. Alternative splicing is regulated by splice activators and splice repressors, which control which exons are included or skipped.
A second key processing step is RNA cleavage and polyadenylation at the 3′ end of the transcript. The pre-mRNA is cleaved downstream of a poly-A signal, after which a poly-A tail is added. This process requires several protein factors, including CPSF, which recognizes the polyadenylation signal, and CstF, which cleave the strand. Alternative polyadenylation can generate mRNAs with different 3′ ends, affecting protein length or regulatory sequences in the 3′ UTR, thereby influencing mRNA stability and translation efficiency.
The 5′ cap, a 7-methylguanosine cap, is added shortly after transcription begins. Together with the polyA tail, it protects the mRNA from degradation and is essential for efficient translation. Cap-binding proteins at the 5′ end and polyA-binding proteins at the 3′ end interact to promote ribosome recruitment and stabilize the mRNA
Pre-mRNA processing can also involve mRNA editing, which changes the nucleotide sequence after transcription. Two types of such RNA editing occur: the A-to-I editing (ADARs) and C-to-U editing. If the edit occurs in a coding region, it can either change the amino acid sequence of the protein or produce a truncated protein by creating a premature stop codon. If outside the coding region it can affect mRNA from the nucleus to the cytosol, the efficiency with which the RNA is translated, and miRNA binding.
How are induced pluripotent stem cells produced, and how can a few master regulators reprogram cells?
Induced pluripotent stem cells (iPSCs) are produced by taking fully differentiated somatic cells, e.g. fibroblasts, and forcing them to express a small set of transcription factors that are normally only active in embryonic stem cells. Classically, this is done by introducing the master regulators Oct4, Sox2, and Klf4 e.g. using plasmids. When these factors are expressed for a sufficient period, the cells lose their differentiated identity and acquire properties similar to embryonic stem cells, including self-renewal and pluripotency.
A small number of master regulators can reprogram cells because they act at the top of gene regulatory networks. These transcription factors directly activate large sets of genes required for pluripotency while repressing genes associated with the differentiated state. They also regulate each other’s expression through positive feedback loops, which stabilize the new cell identity. In addition, master regulators recruit chromatin-modifying enzymes and chromatin-remodeling complexes that alter DNA accessibility, leading to widespread epigenetic reprogramming. Differentiated cells are not the result of irreversible DNA changes, but rather transcriptional and epigenetic changes in the sequence. This means that it can be reversed!
Attenuation and riboswitches are examples of post-transcriptional controls. how do they work?
Attenuation works by coupling transcription to translation in bacteria. The mRNA forms alternative secondary structures depending on ribosome movement. When the metabolite (e.g. an amino acid) is abundant, rapid translation causes formation of a terminator hairpin in the mRNA, stopping transcription early. When the metabolite is scarce, ribosome stalling allows formation of an anti-terminator structure, so transcription continues.
Riboswitches are regulatory RNA elements within mRNAs that directly bind small molecules. Ligand binding changes the RNA structure, which either terminates transcription or blocks translation. In this way, riboswitches control gene expression without the need for regulatory proteins.
What are examples of translational control
xamples of translational control include regulation of whether and how efficiently an mRNA is translated into protein:
Regulatory proteins that bind mRNA (often in the 5′ UTR) and block or promote ribosome binding. e.g. initiation factors
microRNAs (miRNAs) that bind target mRNAs and inhibit translation or promote mRNA degradation.
Phosphorylation of translation initiation factors, which globally increases or decreases translation.
mRNA localization, where translation occurs only at specific locations in the cell.
Explain how X-chromosome inactivation operates?
Alteration in the chromatin structure of an entire chromosome can modulate the levels of expression of most genes on that chromosome.
Males and females differ in their sex chromosomes. Females have two X chromosomes, whereas males have one X and one Y chromosome. Mammals have evolved a dosage compensation mechanism to ensure that the same amount of most of the X-chromosome gene products is made in both male and female cells, despite the fact that females contain twice as many X-chromosome genes. This is done through X-inactivation, where one of the chromosomes is tightly condensed heterochromatin, making it mostly inactive
In humans, the process begins from a site called XIC (X-inactivation center) with the synthesis of a long non-coding RNA, called Xist, whose gene lies on the X chromosome. This transcript is synthesized by only one of the two X chromosomes in females. Once a Xist RNA molecule is synthesized, it does not leave the X chromosome and diffuses and coat it.
Xist does not silence genes directly but recruit’s chromatin-modifying complexes, including DNA methylases, histone-modifying enzymes, and structural components specific to the inactive X chromatin. An example is Polycomb, which add repressive histone modifications which leads to chromatin compaction, exclusion of transcription machinery, and recruitment of DNA methyltransferases that methylate promoters of X-linked genes. The tight heterochromatin is silenced.
Some of the genes, however, are not silenced since they are present inside loops, called escapee, delimited by insulator elements.
How and where are mRNAs degraded?
mRNA degradation is an essential process that regulates gene expression by controlling how long an mRNA persists and how much protein it can produce.
In bacterial cells, mRNAs are generally very unstable, with half-lives of only a few minutes before being degraded by nucleases. As bacterial mRNA is rapidly synthesized and degraded, they can quickly adjust their gene expression in response to environmental changes.
In eukaryotes, mRNA is more stable, but they are also degraded. mRNAs strt degrading as soon as they come into the cytosol, where their poly-A tail is shortened. Once the poly-A tail reaches a critical length, two main degradation pathways occur (sometimes at the same time):
· The 5′ cap is removed, exposing the mRNA to rapid degradation from the 5′ end.
· Degradation continues from the 3′ end through the shortened poly-A tail and into the coding region.
The 3′ UTR sequences are especially important in controlling mRNA lifetimes, and they often carry binding sites for specific proteins that increase or decrease the rates of poly-A shortening, decapping, or 3′-to-5′ degradation. Translation efficiency also affects mRNA stability, as mRNAs that are efficiently translated tend to be degraded more slowly.
In addition to exonuclease-mediated decay, some mRNAs are degraded by endonucleases, which cleave the mRNA internally. This leads to rapid degradation of the fragments
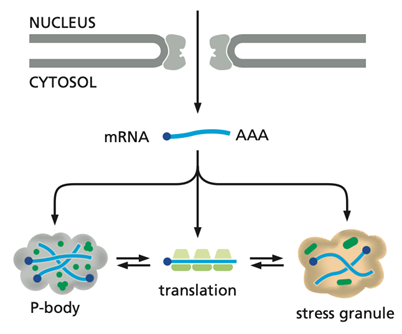
When mRNAs are no longer actively translated, they can accumulate in cytoplasmic structures called P-bodies, which are enriched in mRNA-degrading enzymes. In P-bodies, mRNAs may continue to be degraded or be stored in a repressed state and later returned to the cytosol for translation. mRNAs stored in this way often code for proteins that the cell needs quickly, and this strategy bypasses the time-consuming steps of de novo mRNA production. During cellular stress, untranslated mRNAs can also accumulate in stress granules, where they are temporarily stored. Once stress conditions are relieved, these mRNAs are released back into the cytosol and resume translation.
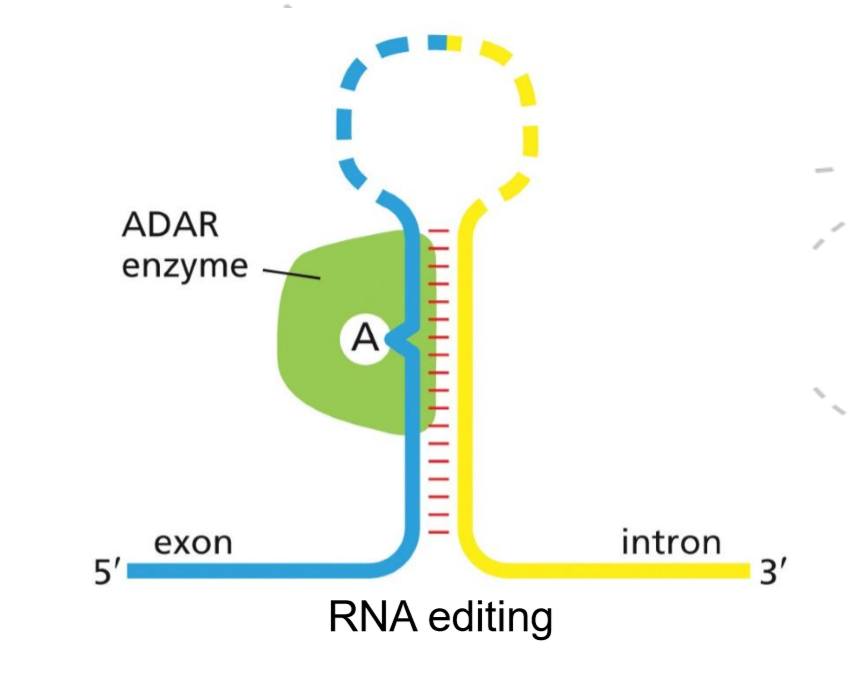
What is RNA editing and how is it carried out?
RNA editing is a post-transcriptional process in which the nucleotide sequence of an RNA molecule is altered after it is made, so the RNA sequence differs from the DNA template.
It is carried out by editing enzymes that chemically modify specific bases in the RNA. The most common types are adenosine-to-inosine (A→I) editing by ADAR enzymes and cytidine-to-uridine (C→U) editing by deaminases. These changes can alter codons, affect RNA stability or splicing, or change how the RNA is translated, increasing the diversity of proteins produced from a single gene.
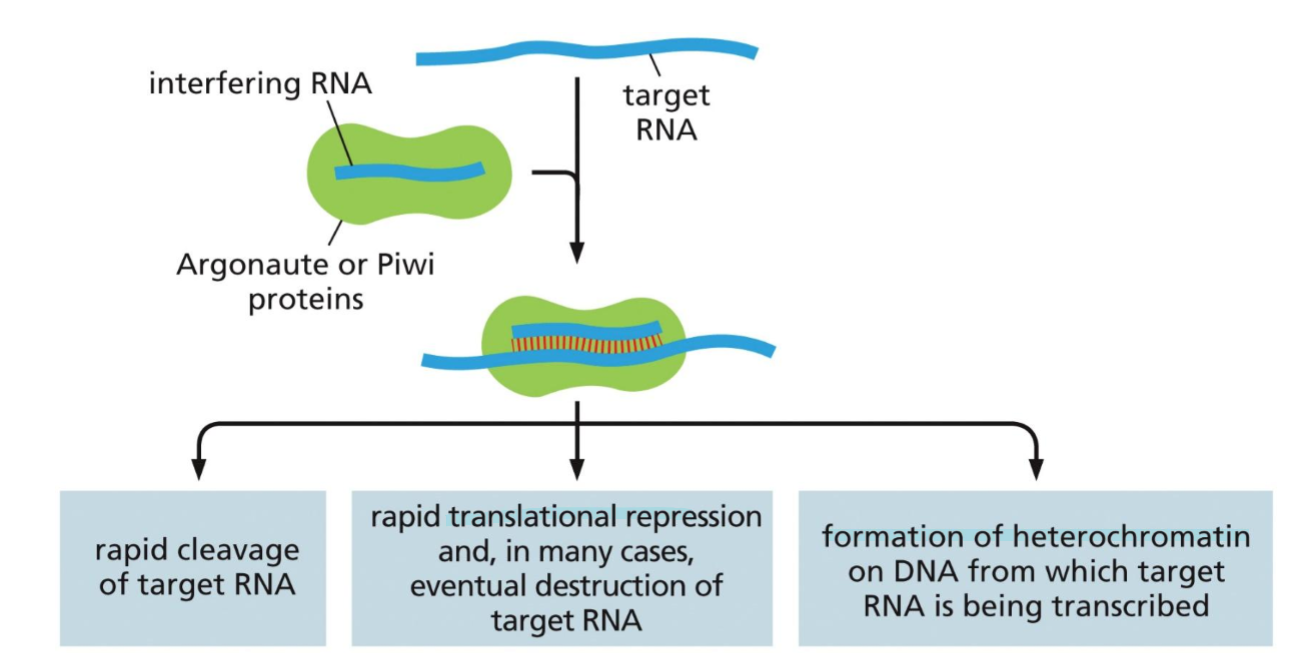
what are iRNAs? WHat are the three main classes? similarities between them?
RNAs that serve as a defence against viruses. they are short ssRNAs which serve as a guide, which bind to other RNAs in the cell. If the target is mRNA, the iRNA can target its destruction or inhibition. If the target is about to be transcribed, the iRNA can bind and form heterochromatine.
There are three classes of iRNAs; siRNA, ppiRNAs, and miRNAs. even though they difer in fnction they have some similarities, they all bind through RNA-RNA interaction and reduce gene expression. once produced they associate with either the enzyme Piwi or Argonaute
what are miRNAs? How are they processed?
MicroRNAs (miRNAs) are short non-coding RNAs, typically 21–23 nucleotides long. The primary function of miRNAs is to control gene expression by binding to complementary sequences, usually in the 3′ UTR of target (poly-A tail)
miRNAs are transcribed as long precursor molecules which form characteristic stem-loop structures, which are recognized and processed/cut by the enzyme Dicer. The resulting mature miRNA is then assembled with a set of proteins to form the RISC complex. Within RISC, the miRNA guides the complex to target mRNAs by base-pairing with complementary sequences.
This process is greatly facilitated by the Argonaute protein, a component of RISC. If the base-pairing is exact, the mRNA is cleaved by the Argonaute protein, removing the mRNA’s poly-A tail and exposing it to exonucleases. If the base-pairing between the miRNA and the mRNA is not exact, the enzyme does not slice the mRNA. Rather, translation of the mRNA is repressed.
miRNAs present several advantages. First, a single miRNA can control hundreds of different mRNAs. Second, regulation by miRNAs can be combinatorial. Different miRNAs can bind cooperatively to their target mRNAs if their recognition sites are spaced appropriately. Third, an miRNA occupies relatively little space in the genome when compared with a protein.
Short:
· Post-transcriptional regulation of gene expression
· Repression of translation of target mRNAs
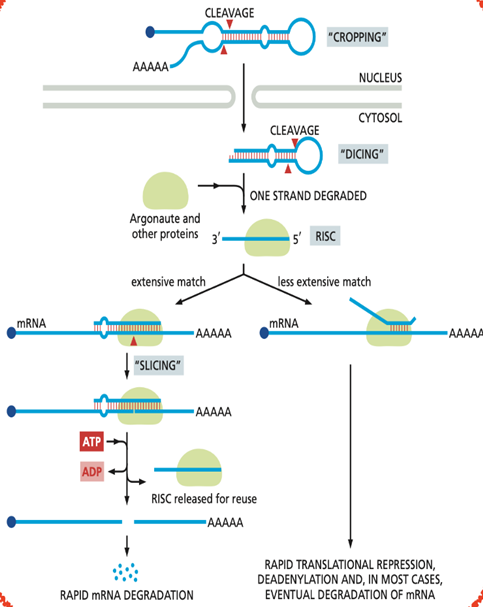
· Promotion of mRNA deadenylation and degradation
what are siRNAs?
siRNAs (small interfering RNAs) are short double-stranded RNA molecules (~21–23 nucleotides) that regulate gene expression by silencing specific mRNAs. They are also produced by the Dicer complex
They work by being incorporated into the RISC complex, where one RNA strand guides the complex to a complementary mRNA. The target mRNA is then cleaved and degraded, preventing translation and effectively turning off gene expression.
what is the RITS complex?
The RITS complex mediated transcriptional gene silencing
It contains small interfering RNAs (siRNAs) that guide the complex to complementary DNA or nascent RNA transcripts. Once targeted, RITS recruits chromatin-modifying enzymes that add repressive histone modifications, leading to heterochromatin formation and stable silencing of gene transcription. this happends while the RNA is being transcribed, shutting the synthesis of the target RNA off, not just destroying the mRNA.
The RITS complex also uses argonaute
What are piRNAs and their function?
Piwis are small non-coding RNAs which bind to Piwi
Their main function is to silence transposable elements, especially in germ cells. They do this by guiding Piwi proteins to transposon-derived RNAs or genomic regions, leading to RNA degradation and/or formation of repressive chromatin. This protects genome stability and prevents harmful mutations during gamete formation.
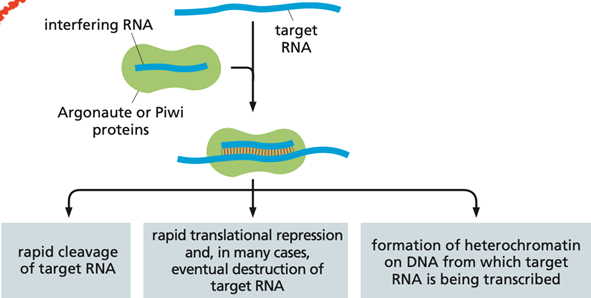
What re the three main functions of lncRNAs?
Function as scaffold RNA molecules, holding together groups of proteins to coordinate their functions.
Serve as guide sequences, binding to specific RNA or DNA target molecules through base-pairing.
ability to organize biomolecular condensates, the non-membrane-bound assemblies of proteins and nucleic acids
Explain RNA intererence in bacteria through the CRISPR-system:
RNA interference in bacteria through the CRISPR system provides adaptive immunity against viruses.
Short RNA sequences derived from invading viral DNA are stored in the CRISPR locus and later transcribed into guide RNAs (crRNA + tracrRNA). These guide RNAs direct Cas proteins to matching viral DNA or RNA, which is then cut and destroyed, preventing infection.
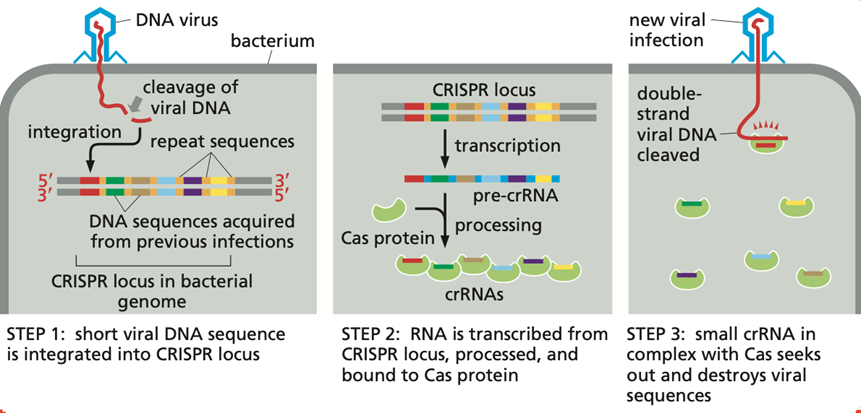
What are transcription circuts and genetic switches?
Genetic circuits are networks of interacting genes and regulatory proteins that process information and control gene expression, similar to electronic circuits. They are built from regulatory elements such as promoters, transcription factors, and feedback loops.
Molecular switches are simple genetic circuits that toggle genes between “on” and “off” states. They often rely on positive feedback or mutually inhibitory regulators to create stable states, allowing cells to make clear, robust decisions such as committing to a cell fate or responding to environmental changes.
PART 2
What is the colonial flagellate hypothesis? What is so special with choanoflagellates?
The colonial flagellate hypothesis is a classic evolutionary idea about how multicellular animals originated. It proposes that animals evolved from unicellular flagellated protists that lived together to form an aggregate. Over time, the cells in these colonies became more tightly integrated, became adhered, and began to specialize for different functions, folding in to make tissues. This increase in cooperation and specialization eventually produced multicellular organisms with distinct tissues.
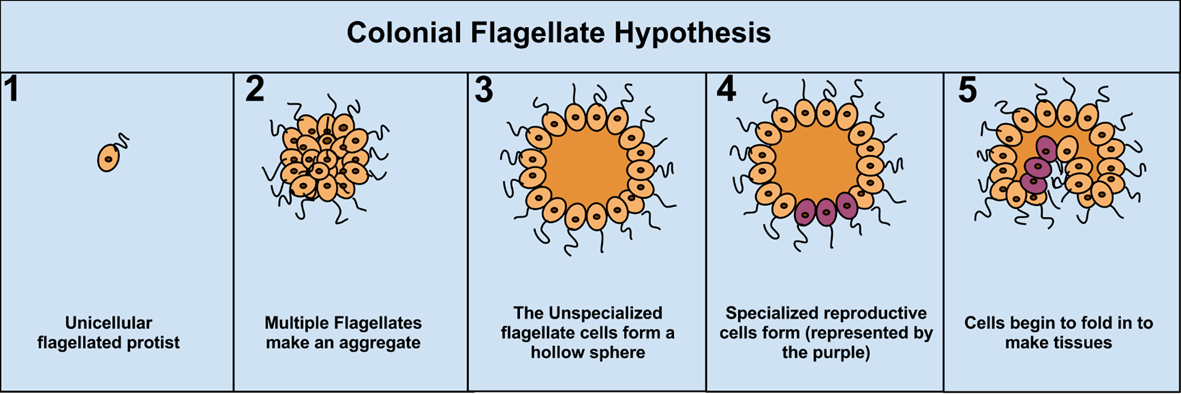
Choanoflagellates are special because they are the closest known unicellular relatives of animals and provide strong support for the colonial flagellate hypothesis. Members of Choanoflagellata have cells with a single flagellum surrounded by a collar of microvilli, which is structurally and functionally almost identical to cells in sponges. Many choanoflagellates can form colonies, illustrating a plausible intermediate stage between unicellular and multicellular life. In addition, they possess genes for cell adhesion and signaling that were once thought to be unique to animals, indicating that the genetic toolkit for multicellularity evolved before the origin of true animals.
What is the principle of bacterial quorum sensing?
Unicellular organisms have a mechanism for responding to physical and chemical changes in their environment, including signals produced by other cells. In bacteria, this takes the form of quorum sensing, a process in which cells secrete small chemical signaling molecules that accumulate in the environment as population density increases. When the signal concentration reaches a critical threshold, it is detected by the bacteria and triggers coordinated changes in gene expression across the population. Through quorum sensing, bacteria can effectively measure their population density and synchronize behaviors such as motility, antibiotic production, spore formation, and sexual conjugation in a population density–dependent manner.
What is the steps in communication in multicellular organisms?
A cell emits a signal —> reciver cell binds signal molecule to receptor —> receptor activation —> activates signalling pathway —> prouce second messengers —> effector proteins manipulate host cell functions (e.g. TFs)
what are the four main types of signalling between cells?
Type of signaling | Mechanism |
|---|---|
Contact-dependent | Signal molecule remains bound to the signaling cell and activates receptors on a neighboring cell through direct contact via its receptor. |
Paracrine | Secreted signaling molecules called local mediators act locally on nearby target cells. uses mechanisms that involve rapid uptake of the signal or immobilzation. cells can also produce signals that they themselves respond to celled autocrine signalling. |
Endocrine | Hormones are produced by endocrine cell or gland, and are released into the bloodstream and act on distant target cells. the signalign is slow. |
Synaptic (neuronal) | Neurons release neurotransmitters across synapses to specific target cells. work over long distance and give a rapid response. |
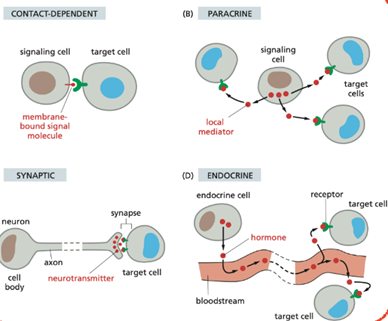
What are general examples of signaling molecules? explain the terms hydrophilic and hydrophlic and how it affects the sigalling molecules
General examples of signaling molecules include hormones, growth factors, neurotransmitters, cytokines, and lipid-derived signals.
Hydrophilic signaling molecules are water-soluble and cannot cross the plasma membrane. They bind to receptors on the cell surface, which then trigger intracellular signaling pathways. Examples include peptides, proteins, and many neurotransmitters.
Hydrophobic (lipophilic) signaling molecules are lipid-soluble and can diffuse across the plasma membrane. They bind to intracellular receptors, often in the cytosol or nucleus, which directly regulate gene expression. Examples include steroid hormones and thyroid hormones. Needs carrier protein in the bloodstream
Each cell displays a distinct set of receptors, meaning they can respond to a distinct set of signal molecules. How does this affect the cell?
why do different cell types respond differently to the same signal molecule?
Because each cell expresses a distinct set of receptors, it can sense and respond only to specific signals. This determines the cell’s behavior, differentiation, and function, allowing the same signaling molecule to produce different responses in different cell types.
Different cell types respond differently to the same signal molecule because they express different receptors and intracellular signaling proteins, which activate different signaling pathways and cellular responses.
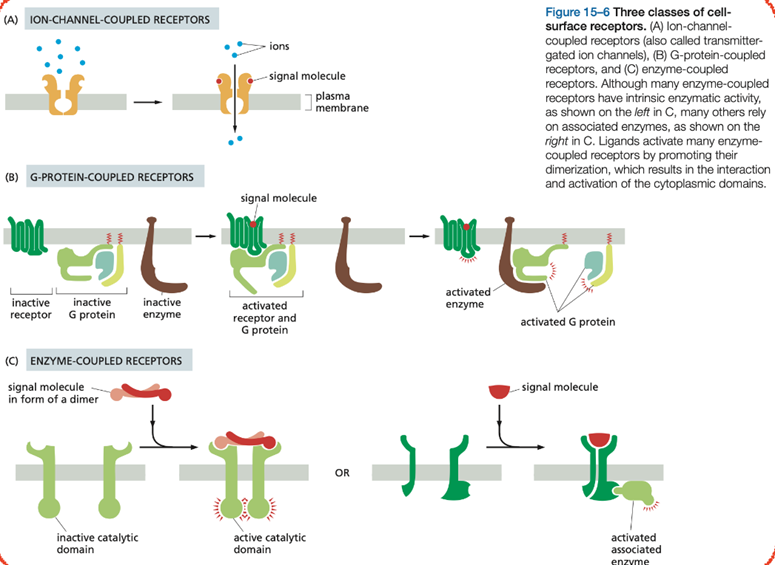
what are the three main cell surface receptors? explain them short
Ion-channel-coupled receptors are involved in rapid synaptic signaling. This type of signaling is mediated by a small number of neurotransmitters that open or close an ion channel formed by the protein to which they bind, briefly changing the ion permeability of the plasma membrane and changing the excitability of the postsynaptic target cell. Most ion-channel-coupled receptors belong to a large family of homologous, multipass transmembrane proteins.
G-protein-coupled receptors (GCRs) act by indirectly regulating the activity of a separate plasma-membrane-bound target protein, which is generally either an enzyme or an ion channel. A heterotrimeric GTP-binding protein (G protein) mediates the interaction between the activated receptor (bound with ligand) and this target protein.
Enzyme-coupled receptors either function as enzymes or associate directly with enzymes that they activate. They are usually single-pass transmembrane proteins that have their ligand-binding site outside the cell and their catalytic or enzyme-binding site inside. The great majority, however, are either protein kinases or associate with protein kinases, which phosphorylate specific sets of proteins in the target cell when activated.
In cell signalling, why do we say that signaling molcules behave like molecular switches?
Most signalling molcules are proteins which either generate second messengers, or activate the next protein in the cascade. many of these proteins behave like a switch, as they can turn from inactive to active when reciving a signal. Often this happeds via phosphorylation, where a protein kinase turns them on, and a protein phosphatase turns their activity off.
What are the functions of GAP and GEF proteins when it comes to regulating GTPases?
what is the differnece between a GAP. GEF, and GTP-binding protein?
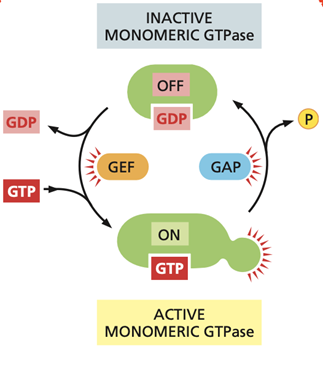
Small GTPases act as molecular switches by cycling between an active GTP-bound state and an inactive GDP-bound state. GAPs and GEFs regulate this cycle in opposite ways.
· GEFs (guanine nucleotide exchange factors) activate GTPases by promoting the release of GDP from the GTPase. Because GTP is present at a higher concentration than GDP in the cytosol, GTP binds spontaneously once GDP is released. This switches the GTPase into its active, signaling state.
· GAPs (GTPase-activating proteins) inactivate GTPases by stimulating GTP hydrolysis activity, leading to rapid conversion of bound GTP to GDP. This switches the GTPase off and terminates signaling.
A GTP-binding protein (G protein) is a molecular switch that is active when bound to GTP and inactive when bound to GDP.
A GEF (guanine nucleotide exchange factor) activates a GTP-binding protein by promoting the exchange of GDP for GTP.
A GAP (GTPase-activating protein) inactivates a GTP-binding protein by stimulating its GTP hydrolysis to GDP.
other than GEFs and GAP, phosphorylation can also change the activity of a protein. explain:
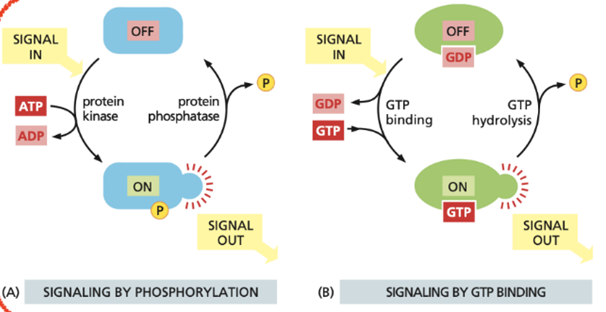
Phosphorylation changes protein activity by the reversible addition of a phosphate group to specific amino acids. This is carried out by protein kinases (turns ON) and reversed by protein phosphatases (turns OFF).
Describe how adaptors and scaffold proteins cooperate in receptor signaling complexes.
It is inevitable that signaling molecules sometimes interact with components of other pathways, which can lead to unwanted cross-talk and interference. Adaptor and scaffold proteins work together to ensure that receptor signaling is fast, efficient, and highly specific, and avoids cross-talk. These proteins organize receptor signaling by assembling specific signaling complexes at activated receptors.
Adaptor proteins are small, non-enzymatic proteins that contain interaction domains such as SH2, SH3, or PTB domains. After receptor activation, adaptor proteins bind to phosphorylated sites on the receptor or on associated proteins and recruit downstream signaling molecules to the receptor complex. In this way, adaptors act as molecular connectors that link receptors to intracellular enzymes such as kinases or GTPase regulators.
Scaffold proteins bind multiple signaling proteins simultaneously and hold them together in stable complexes. By physically organizing several components of a signaling pathway in close proximity, scaffold proteins increase the speed and efficiency of signal transmission and enhance pathway specificity. They also help prevent cross-talk by keeping the correct signaling proteins together while separating them from components of other pathways.
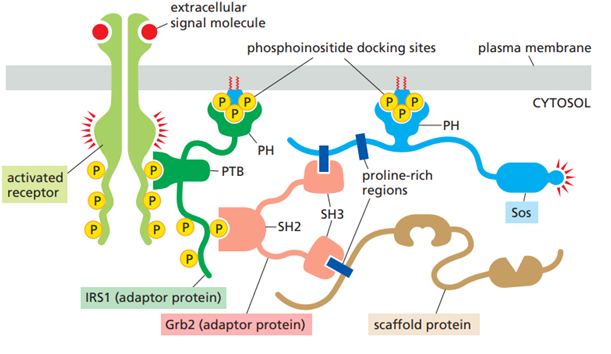
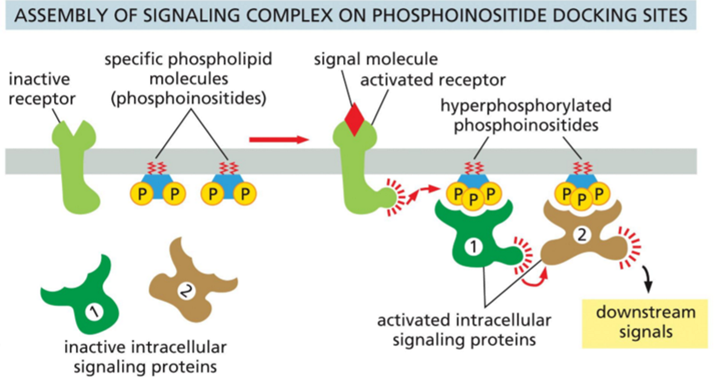
Why are phosphoinositides important in many signaling systems?
Phosphoinositides are a small group of membrane lipids made from phosphatidylinositol, which is found mainly on the inner side of cell membranes. They have a lipid part that anchors them in the membrane and a sugar head group (inositol) that can be modified by adding or removing phosphate groups at different positions.
When receptors are activated, enzymes such as PI3K produce specific phosphoinositides next to the receptor. These modified lipids act as docking sites in the membrane. The activation of the receptor often leads to creation of multiple phosphorylated sites in the plasma membrane next to the receptor through activation of PI3K. These phosphorylated sites work as docking sites which recruit other proteins involved in the signaling pathway and activate them.
In this way, phosphoinositides provide spatial control of signaling by ensuring that signaling proteins are activated only at the correct membrane location and time.
How can a cell avoid crosstalk with other pathways?
Cells avoid cross-talk between signaling pathways by using
1) highly specific interactions and spatial organization of signaling components. Signaling proteins recognize each other through precise binding domains, ensuring that only the correct partners interact.
2)adaptor and scaffold proteins assemble pathway-specific signaling complexes, keeping components of the same pathway together and physically separated from others.
3) Localization of signaling to specific membranes or subcellular regions further increases specificity.
Explain the SH2, SH3, PTB, and PH domains:
SH2, SH3, PTB, and PH are interaction domains found in adaptor and scaffold proteins.
SH2 and PTB domains are phosphate binding domains. They allow adaptor proteins to bind activated, phosphorylated receptors or signaling proteins.
SH3 domains are protein–protein interaction domains that bind proline-rich sequences, enabling adaptors and scaffolds to recruit additional signaling proteins.
PH domains are lipid-binding domains that bind specific phosphoinositides, targeting adaptor or scaffold proteins to particular membrane sites.
How can biomolecular condensates be important during receptor cell signaling?
Biomolecular condensates can be important during receptor cell signaling because they help organize signaling molecules into concentrated, membrane-associated assemblies without the need for a surrounding membrane. When a receptor is activated, many signaling proteins are recruited to the same region of the cell. By forming biomolecular condensates, these proteins can cluster together, creating a localized signaling point.
This increases the efficiency and speed of signaling, because enzymes and their substrates are brought very close to each other. Condensates also enhance signaling specificity by selectively including the correct signaling components while excluding unrelated proteins, which helps reduce unwanted cross-talk between pathways.
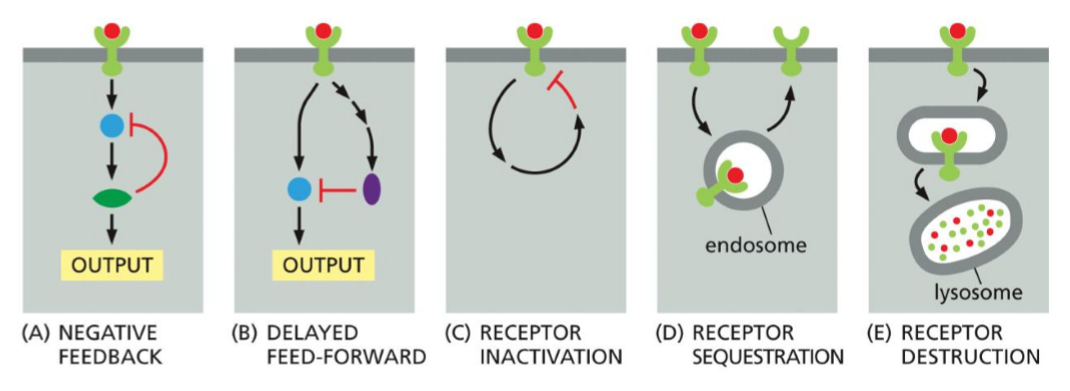
Why is negative feedback regulation important during receptor adaptation and desensitization?
Negative feedback is a regulatory mechanism where activation of a signaling pathway triggers inhibitory processes that reduce or shut down the same signal, i.e. the pathway output from the activated receptor, feeds back to inhibit the receptor itself, or a protein in the pathway.
Receptor adaptation is the process by which a cell reduces its response even though there is a constant or prolonged stimulus. Desensitization is a related process in which continued stimulation makes a receptor less responsive or inactive.
Negative feedback is important for adaptation and desensitization because it prevents overstimulation and maintains signaling within a useful range. The cell can therefore sense if there is a change in the signal strength, over a wide range of concentrations. Negative feedback loops with short time decays are important for such adaptive responses as because they allow cells to respond quickly to a signal and then rapidly turn the response off, even if the signal is still present.
What factor determines the speed of the signaling response?
The proteins that are available in the cell already. when the response rewuires changes in proteins already present in the cell, the response will be fast. if the response involves changes in gene expression and the synthesis of new protiens, it usually takes longer.
How does a protein keep its relative amount if it has a long and short halflife, and if the synthesis rate is low/high?
· If the molecule is produced at a low rate, it must also have a slow degradation to maintain a relative concentration
· If it has a short half-life, the molecule must be produced at a high rate to maintain a concentration
· If the synthesis rate is decreased 10 times, the molecules with short half-lives will drop fast (because not enough is synthesized vs. degraded)
· If the synthesis rate increased by 10 times, the molecule with short half-lives will increase rapidly
What is the difference between a sigmoidal and a all-or-noen response in cell signaling?
A sigmoidal response is a gradual, S-shaped increase in signaling output as signal concentration rises. It reflects cooperative interactions and allows cells to fine-tune their response over a range of signal strengths.
An all-or-none response is switch-like: once the signal crosses a threshold, the pathway is fully activated, and below that threshold there is little or no response. This produces decisive, irreversible or stable cellular outcomes.
Which feedback loops does a cell utalize, and why?
Positive feedback amplifies signaling output. Weak positive feedback steepens the response, while strong positive feedback creates an all-or-none, self-sustaining switch that can remain active even after the signal decreases.
Negative feedback reduces signaling output. With long delays it can cause oscillations, and with short delays it produces transient responses that decay during sustained stimulation, leading to adaptation or desensitization and allowing cells to respond to changes in signal strength.
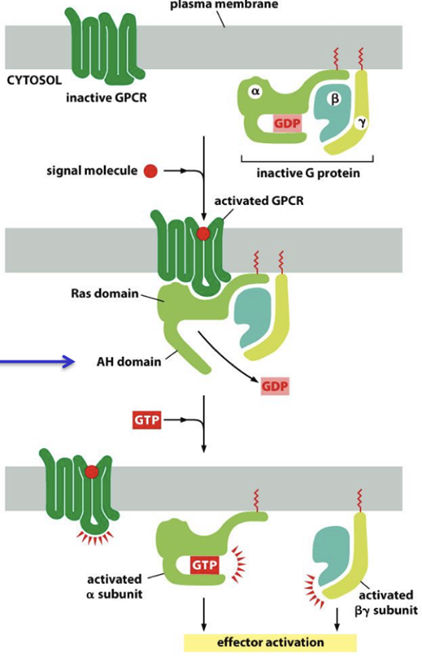
How do G-protein coupled receptors (GPCRs) activate G-proteins?
G-protein-coupled receptors (GPCRs) are the largest family of cell-surface receptors. Despite responding to a wide variety of signals, including hormones, neurotransmitters, light, and odorants, all GPCRs share a common structure of a single polypeptide with seven transmembrane α-helices. They transmit signals into the cell by activating G proteins, and the same signaling molecule can activate multiple GPCRs.
When an extracellular signal molecule (e.g. light, olfactory, acetylcholine) binds to a GPCR, the receptor undergoes a conformational change that enables it to activate a heterotrimeric GTP-binding protein (G protein). G proteins are composed of three protein subunits: α, β, and γ. The α subunit is the catalytic one, while β, and γ are regulatory. Moreover, both α and γ are post-transcriptionally modified attach them to the cell membrane.
In the unstimulated state, the α subunit has GDP bound. When a GPCR is activated upon ligand binding, it acts like a guanine nucleotide factor (GEF) and makes the α subunit release its bound GDP, allowing GTP to bind in its place. This causes a conformational change in the Gα subunit, releasing the G protein from the receptor and triggering dissociation of the GTP-bound Gα subunit from the Gβγ pair. Both the α subunit and the βγ subunits can activate various proteins downstream.
Signal termination occurs when the intrinsic GTPase activity of the Gα subunit hydrolyses GTP to GDP, a process accelerated by target proteins or regulators of G-protein signaling (RGS).How do G-protein coupled receptors (GPCRs) activate G-proteins?
How are the concentration of cAMP regulated within the cell? what is PKA and AKAB? What is CREB and CRE?
Why is cAMP such an important second messenger?
cAMP levels are regulated by adenylyl cyclase, which is activated or inhibited by GPCR signaling. cAMP is also broken down into AMP by PDE.
Many extracellular signals work by increasing the intracellular cAMP consentration. cAMP mainly excerts its function by activating PKA.
When cAMP levels rise, cAMP binds to PKA’s regulatory subunits, releasing the active catalytic PKA. These then phosphorylate target proteins, including metabolic enzymes, ion channels, and transcription factors, thereby altering cell activity and gene expression.
AKAP is a scaffold protein that binds PKA and anchors it to specific locations inside the cell. By localizing PKA near its substrates and signaling components, AKAPs ensure fast, specific, and localized cAMP signaling, reducing unwanted cross-talk with other pathways.
CREB (cAMP response element–binding protein) is a transcription factor that is activated by phosphorylation (often by PKA). Once activated, CREB stimulates transcription of specific genes.
CRE (cAMP response element) is a short DNA sequence in the promoter of target genes. Activated CREB binds to CRE to increase gene transcription in response to cAMP signaling.
cAMP is an important second messenger because it efficiently amplifies and spreads signals inside the cell. A single activated receptor can lead to the production of many cAMP molecules, greatly amplifying the original signal. cAMP can rapidly diffuse through the cytosol and activate multiple targets,
What is phospolipases? what do they do? describe the pathway?
Phospholipases are enzymes that cleave phospholipids in cellular membranes.
They cut specific bonds in phospholipids, which can remodel membranes and, importantly, generate signaling molecules.
THE WHOLE AIM OF THE PATHWAY IS TO INCREASE THE INTRACELULAR LEVELS OF Ca2+ TO FURTHER ACTIVATE DOWNSTREAM PROTEINS
Ligand binding – An extracellular signal binds a GPCR (via Gαq) or an RTK.
Receptor activation – The activated receptor stimulates phospholipase C (PLC).
PIP₂ cleavage – PLC cleaves PIP₂ into IP₃ (soluble) and DAG (membrane-bound).
IP₃ diffusion – IP₃ diffuses through the cytosol to the ER.
IP₃ receptor activation – IP₃ binds IP₃ receptors on the ER, causing Ca²⁺ release.
Ca²⁺ amplification (positive regulation) – Released Ca²⁺ can activate ryanodine receptors, leading to further Ca²⁺ release (Ca²⁺-induced Ca²⁺ release).
DAG signaling – DAG remains in the membrane and, together with Ca²⁺, activates PKC.
Ca²⁺ sensing – Ca²⁺ binds calmodulin, forming a Ca²⁺–calmodulin complex.
Downstream activation – Ca²⁺–calmodulin activates CaM kinases (CaMKs) and other Ca²⁺-dependent enzymes, altering cell activity and gene expression.
IP₃ regulation (negative regulation) – ITPKA phosphorylates IP₃, and 5-phosphatases (5PTase) dephosphorylate IP₃, reducing IP₃ levels and limiting Ca²⁺ release.
Ca²⁺ removal (negative regulation) – Ca²⁺-ATPase pumps (SERCA in the ER and PMCA in the plasma membrane) actively pump Ca²⁺ back into the ER or out of the cell.
Signal termination – Reduced IP₃ levels and Ca²⁺ clearance shut down Ca²⁺ signaling and PKC/CaMK activity.
What is calmodulin and what is the function of CaM kinase?
Calmodulin is a small, Ca²⁺-binding regulatory protein that acts as a calcium sensor. When intracellular Ca²⁺ levels rise, Ca²⁺ binds to calmodulin, causing a conformational change that allows it to interact with and regulate target proteins.
CaM kinases are protein kinases that are activated by the Ca²⁺–calmodulin complex. Once activated, they phosphorylate specific target proteins, leading to changes in cell activity, including metabolism, secretion, and gene expression.
Describe two important functions phosphoinositides have during cell signaling.
They recruit signaling proteins to membranes.
Specific phosphoinositides act as docking sites for proteins with lipid-binding domains (such as PH domains), ensuring that signaling proteins are localized to the correct membrane at the right time.They serve as precursors for second messengers.
Certain phosphoinositides can be cleaved to generate second messengers, such as IP₃ and DAG, which trigger downstream signaling events like Ca²⁺ release and kinase activation.
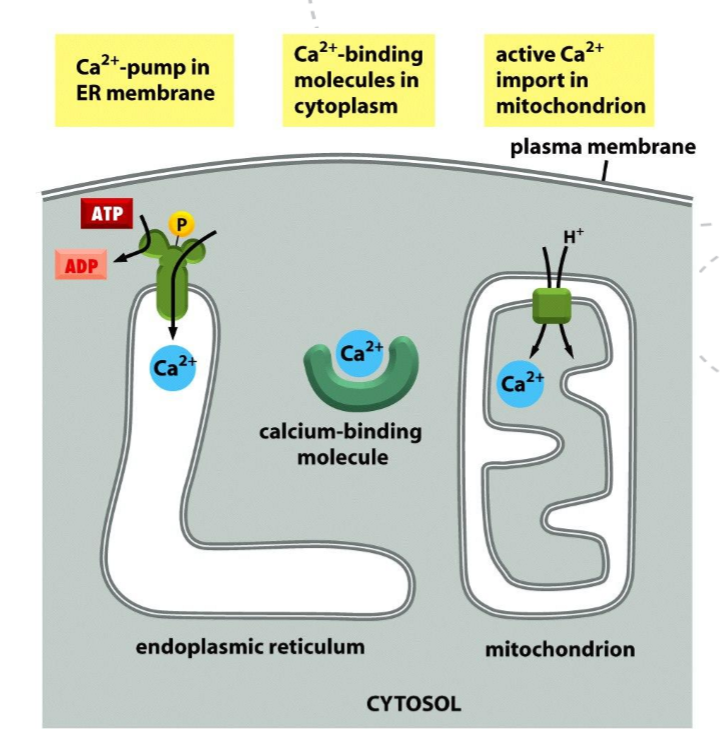
Explain some of the processes that operate to regulate Ca2+ levels in the cell?
Release from intracellular stores via IP₃ receptors and ryanodine receptors in the ER
Ca²⁺ reuptake into the ER by Ca²⁺-ATPase pumps
Ca²⁺ extrusion from the cell by plasma membrane Ca²⁺-ATPase
Ca²⁺ buffering by calmodulin’
Explain how an odorant is detected by a GPCR and how this leads to membrane depolarization. what is the result of memebrane depolarization
An odorant is detected when it binds to a specific olfactory GPCR on the membrane of an olfactory sensory neuron.
Timeline for odorant detection and membrane depolarization (including CaCCs):
Odorant binding – An odorant binds to an olfactory GPCR on the cilia of an olfactory sensory neuron.
GPCR activation – The receptor changes conformation and becomes active.
G protein activation – The GPCR activates the G protein Golf (GDP → GTP on Gα).
Adenylyl cyclase activation – Gα–GTP activates adenylyl cyclase.
cAMP production – Adenylyl cyclase converts ATP into cAMP.
CNG channel opening – cAMP opens cyclic nucleotide–gated (CNG) cation channels.
Na⁺ and Ca²⁺ influx – Na⁺ and Ca²⁺ enter the cell through CNG channels.
CaCC activation – The rise in Ca²⁺ activates Ca²⁺-activated Cl⁻ channels (CaCCs).
Cl⁻ efflux – Cl⁻ flows out of the cell, amplifying depolarization.
Membrane depolarization – Ion movements depolarize the membrane and trigger an action potential. This electrical signal propagates along the neuron and transmits the information.
Explain how the photoreceptor cell is desensitized?
Photon absorption – Light activates rhodopsin (opsin + retinal) in the photoreceptor membrane.
Signal initiation – Activated rhodopsin activates the G protein transducin, which activates phosphodiesterase (PDE).
cGMP decrease – PDE hydrolyzes cGMP, causing cGMP-gated Na⁺ channels to close and the cell to hyperpolarize.
Ca²⁺ levels fall – Closure of cGMP-gated channels reduces Ca²⁺ influx, lowering intracellular Ca²⁺.
Ca²⁺-dependent feedback (desensitization begins) –
Low Ca²⁺ activates guanylyl cyclase (via GCAPs), increasing cGMP synthesis.
Low Ca²⁺ increases sensitivity of cGMP-gated channels to cGMP.
Rhodopsin inactivation – Activated rhodopsin is phosphorylated by rhodopsin kinase and bound by arrestin, preventing further transducin activation.
Response reduction – cGMP levels partially recover, some channels reopen, and the response to continued light is reduced.
Adapted state – The photoreceptor becomes less sensitive to sustained light but remains responsive to changes in light intensity.
desensitation of GPCR occur through:
Receptor inactivation: GPCRs are altered so they can no longer interact with G proteins.
Receptor sequestration: GPCRs are temporarily internalized, preventing access to extracellular ligands.
Receptor destruction: Internalized GPCRs are targeted to lysosomes and degraded
in each of the three ponis above desensitation depends on phosphorylation by PKA, PKC, or GPCR kinases (GRKs), including RK.
Activated GPCRs stimulate GRKs to phosphorylate multiple sites on the receptor Arrestin binds phosphorylated GPCRs and promotes desensitization by:
Blocking further interaction between the receptor and G proteins
Acting as an adaptor that links the receptor to enzymes initiating endocytosis
How is light detected by a GPCR and explain how this results in decrease of neurotransmitter release?
Light is detected in photoreceptor cells by a GPCR called rhodopsin, which contains the light-sensitive molecule retinal. When a photon is absorbed, retinal changes shape, activating rhodopsin. Activated rhodopsin then activates the G protein transducin, which in turn activates phosphodiesterase (PDE). PDE breaks down cGMP, causing cGMP-gated Na⁺ and Ca²⁺ channels in the plasma membrane to close.
Closure of these channels leads to membrane hyperpolarization, which reduces Ca²⁺ entry into the synaptic terminal. Because neurotransmitter release is Ca²⁺-dependent, the drop in intracellular Ca²⁺ causes a decrease in neurotransmitter release from the photoreceptor cell.
What are enzyme coupled deceptors, and what are RTKs?
Enzyme coupled receptors are trnasmembrane proteins with a ligand binding somain and a kinase domain. the kinase domain either has enzymatic activity or associate directly with an enzyme.
RTKs (receptor tyrosine kinases) are cell-surface receptors that transmit signals by phosphorylating tyrosine residues on themselves and other proteins.
When a ligand (such as a growth factor) binds, RTKs dimerize, activating their kinase domain. They then autophosphorylate on tyrosines, creating docking sites for intracellular signaling proteins, which initiates downstream signaling pathways that regulate cell growth, survival, and differentiation.
Signaling through the PDGF receptor results in dimerization of the receptor and autophosphorylation. Describe the signaling pathways activated through the PDGF receptor and how signaling can be desensitized.
Binding of PDGF to the PDGF receptor (an RTK) induces receptor dimerization, which activates the intrinsic tyrosine kinase activity of the receptor. The two receptor subunits then autophosphorylate specific tyrosine residues on their cytosolic tails. These phosphotyrosines act as docking sites for intracellular signaling proteins containing SH2 or PTB domains, initiating multiple signaling pathways.
Major signaling pathways activated by the PDGF receptor:
Ras–MAPK pathway: Grb2 aactivating Ras, which triggers the MAP kinase cascade and leads to changes in gene expression and cell proliferation.
PI3K–Akt pathway: PI3K binds to the activated receptor and produces PIP₃, which recruits and activates Akt, promoting cell survival and growth.
PLCγ pathway: PLCγ binds to the receptor and is activated, cleaving PIP₂ into IP₃ and DAG, leading to Ca²⁺ release and PKC activation
Each signaling proteins associates with a particular phosphorylated site on the RTK
Signaling is desensitized through several mechanisms:
Receptor internalization by endocytosis, reducing receptor availability at the cell surface
Dephosphorylation of receptor tyrosines by protein tyrosine phosphatases
Ubiquitination and lysosomal degradation of the receptor
Negative feedback from downstream kinases that inhibit upstream signaling components
What is the function of the SH2 domain in RTKs
The SH2 domain in RTK signaling functions to bind specifically to phosphorylated tyrosine residues on activated receptor tyrosine kinases.
By recognizing phosphotyrosines in a specific amino-acid context, SH2 domains allow intracellular signaling proteins to dock onto activated RTKs, ensuring recruitment of the correct signaling molecules and initiation of downstream signaling pathways.
How can Ras signaling be activated through receptor tyrosine kinases? describe the steps.
Why are signaling pathways associated with Ras often dysregulated in cancers?
Ligand binding – A growth factor binds to a receptor tyrosine kinase (RTK), causing receptor dimerization.
Autophosphorylation – The activated RTK phosphorylates tyrosine residues on its cytosolic tail.
Adaptor recruitment – The adaptor protein Grb2 binds to phosphotyrosines on the RTK via its SH2 domain.
GEF recruitment – Grb2 recruits SOS (a GEF) through its SH3 domains.
Ras activation – SOS promotes GDP–GTP exchange on Ras, activating Ras.
MAPKKK activation – Active Ras–GTP activates Raf (MAP kinase kinase kinase).
MAPKK activation – Raf phosphorylates and activates MEK (MAP kinase kinase).
MAPK activation – MEK phosphorylates and activates ERK (MAP kinase).
Cellular response – ERK phosphorylates cytosolic and nuclear targets, including transcription factors, leading to changes in gene expression, cell growth, and proliferationi If Ras is continuously sending growth-promoting signals, this leads to uncontrolled cell division, resistance to apoptosis, and tumor development even in the absence of external growth signals.
What determines the time and place that a certain gene is transcribed in the cell?
The specific combination of transcription regulators present in the nucleus
The type of cis-regulatory sequences associated with it
The arrangement of various cis-regulatory sequences associated with it
The relative position of cis-regulatory sequences associated with it
What is Rho? How is Rho siganaling activated?
Rho is imporatn in regulating both the actin and microtubule cytoskeletons, controlling cell shape, polarity, motility, and adhesion. They also regulate cell-cycle progression, gene transcription, and membrane transport
Rho signaling is activated through regulation of Rho family small GTPases (Rho, Rac, Cdc42), which act as molecular switches.
Steps of Rho signaling activation:
Extracellular signal (e.g. growth factors, integrin binding, GPCR or RTK activation) activates upstream receptors.
Activation of Rho-GEFs – Receptors stimulate specific guanine nucleotide exchange factors (GEFs).
Rho activation – Rho-GEFs promote exchange of GDP for GTP on Rho, switching it to the active form.
Effector binding – Active Rho–GTP binds downstream effector proteins (e.g. ROCK, formins).
Cellular response – Effectors regulate the actin cytoskeleton, cell shape, adhesion, migration, and gene expression.
How is PI 3-kinase signaling terminated?
PI3-kinase signaling is terminated mainly by reversing PIP₃ production and inactivating downstream components:
PTEN phosphatase dephosphorylates PIP₃ back to PIP₂, removing the membrane docking sites for Akt and other PH-domain proteins.
Protein phosphatases dephosphorylate and inactivate Akt and other downstream targets.
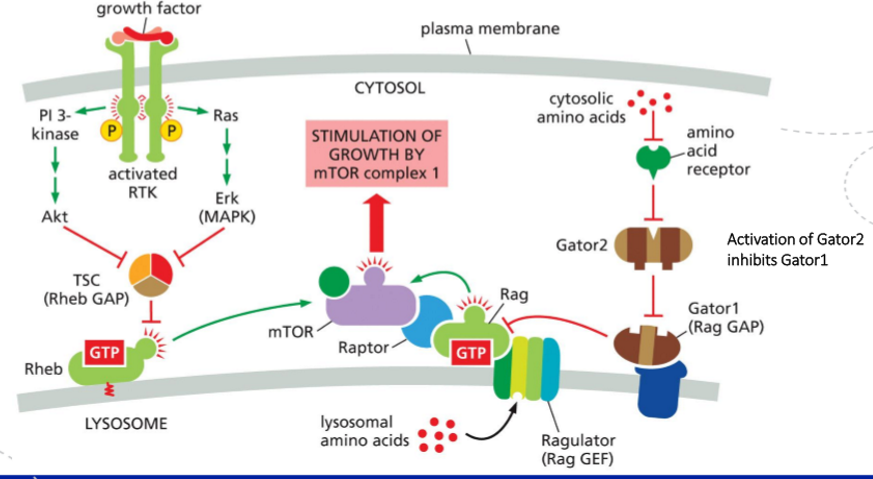
How is the mTORC1 complex activated? What’s the function of TSC complex and Rheb?
How mTORC1 is activated
Amino acids activate Rag GTPases, which recruit mTORC1 (mTOR + Raptor) to the lysosomal membrane.
Growth factors (via RTK → PI3K → Akt) inhibit the TSC complex, which allows Rheb to stay GTP-bound.
Rheb–GTP at the lysosome directly activates mTORC1 once mTORC1 has been brought there by the Rag proteins.
So: Rag = brings mTORC1 to lysosome (permission), Rheb–GTP = switches mTORC1 on (activation).
Function of TSC complex and Rheb
Rheb is a small GTPase that directly activates mTORC1 when GTP-bound.
TSC complex (TSC1/2) is a GAP for Rheb, thereby turning mTORC1 off.
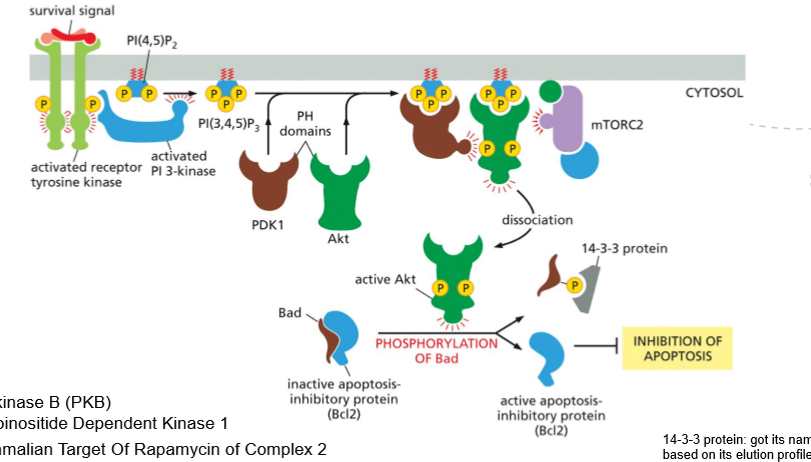
What is the purpuse of the PI3K/Akt/mTOR pathway, and what are the steps for inhibiting apoptosis?
The PI3K/Akt/mTOR pathway promotes cell survival, growth and metabolism in repsonse to nutrients and growth factors
STEPS
Growth factor binding – A ligand binds a receptor tyrosine kinase (RTK).
RTK activation – The receptor dimerizes and becomes autophosphorylated.
PI3K recruitment/activation – PI3K binds to phosphotyrosines on the RTK and becomes active.
PIP₃ production – PI3K converts PIP₂ into PIP₃ in the plasma membrane.
Akt recruitment – PIP₃ recruits Akt (and PDK1) to the membrane via PH domains.
Akt activation – Akt is phosphorylated and activated by PDK1 (and mTORC2).
AKt phosphorylates Bad, making it release Bcl2
Bcl2 becomes active and can inhibit apoptosis
What is the purpuse of the JAK-STAT pathway, and what are the steps for in the signaling pathway?
Purpose of the JAK–STAT pathway
The JAK–STAT pathway provides a direct and rapid route from cell-surface receptors to gene expression. It is mainly used by cytokines and growth factors to control immune responses, cell proliferation, differentiation, and survival.
Steps in the JAK–STAT signaling pathway:
Ligand binding – A cytokine or growth factor binds to its cell-surface receptor.
Receptor dimerization – Ligand binding brings two receptor chains together.
JAK activation – Receptor-associated JAK kinases phosphorylate each other and become active.
Receptor phosphorylation – Activated JAKs phosphorylate tyrosines on the receptor cytosolic tails.
STAT recruitment – STAT proteins bind to phosphotyrosines on the receptor via SH2 domains.
STAT phosphorylation – JAKs phosphorylate the bound STATs.
STAT dimerization – Phosphorylated STATs dissociate and form dimers.
Nuclear translocation – STAT dimers move into the nucleus.
Gene regulation – STATs bind DNA and regulate transcription of target genes.
This pathway is efficient because STATs act as both signaling proteins and transcription factors, eliminating the need for complex signaling cascades.
How is the JAK-STAT pathway inhibited?
The JAK–STAT pathway is inhibited by negative feedback mechanisms, mainly:
SOCS proteins, which block JAK activity and promote receptor degradation
Protein tyrosine phosphatases, which dephosphorylate JAKs, receptors, or STATs
What is the purpuse of the TGF-b pathway, and what are the steps for in the signaling pathway?
Purpose of the TGF-β pathway
The TGF-β pathway regulates cell development
Steps in the TGF-β signaling pathway:
Ligand binding – TGF-β binds to the type II TGF-β receptor.
Receptor complex formation – The type II receptor recruits and phosphorylates the type I receptor.
Smad activation – The activated type I receptor phosphorylates R-Smads (Smad2/3).
Smad complex formation – Phosphorylated R-Smads bind the co-Smad (Smad4).
Nuclear translocation – The Smad complex moves into the nucleus.
Gene regulation – Smads cooperate with other transcription factors to regulate target gene expression.
How is the TGFb pathway regulated?
Redgulated through negative feedback.
The genes that smads code for, also code for inhibitory smads which bid tot the receptor tail and inhibits siganlling by competing with R-smads and recruiting Smurf leading to receptor ubiquination.
Describe the principle of lateral inhibition. Use Notch-Delta as an example.
Lateral inhibition is a mechanism that amplifies small differences between neighboring cells so that they adopt different fates. When one cell begins to differentiate, it actively inhibits neighboring cells from adopting the same fate, creating a patterned tissue with alternating cell types.
In Notch-Delta this relies on contact-dependent signaling activated by Delta, a single-pass transmembrane signal protein found on the future neural cell's surface. By binding to its receptor on a neighboring cell, Delta instructs the neighbor not to become neural. If a small difference in signal strength exists between two cells, the lateral inhibition will reduce the signaling from the receiving cell. This results in a positive feedback that can amplify the initial differences between the cells and push the cells to adopt new cell fates.
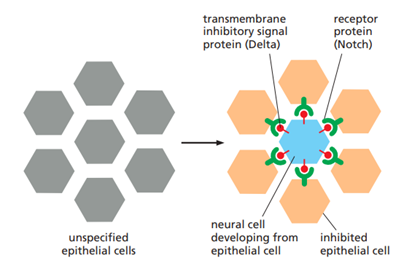
What is the purpose of the Notch pathway, and what are the steps for in the signaling pathway?
Notch plays an important role in contorlling cll fate and plays an important role in the development of tissues, including cell-fate, patern formation, and cell renewal. Notch itself is a single pass transmembrane protein, in need of proteolytic processing for function.
Steps:
Notch ligand (delta) binds the notch receptor on aneighbouring cell
binding exposes cleavage site
notch is cleaved three times consequetly on different spots by proteases
the activated cytoplasmic tail of the Notch protein goes into the nuclus and binds Rbphus
Rbphus activates gene transcription
What is the purpose of the Wnt/b-cateinin pathway, and what are the steps for in the signaling pathway?
The Wnt pathway mediate siganlling from local mediators and morphagen. it is itself a morphagen.
Steps:
No signal:
b-catenin is degraded by rpotein complex
wnt-repsonsive genes are inhibited by complex and corepressor
signal:
wnt binds fizzled and LRP
dishvelled is recruited and breaks down complex which breaks down b-catenin
b-catenin moves to nuclus and breaks down inhibiotry protein
b-catenin binds proteins to activate protein transcription
What is the purpose of the Hedgehog pathway, and what are the steps for in the signaling pathway?
What is the purpose of the NFkB pathway, and what are the steps for in the signaling pathway?
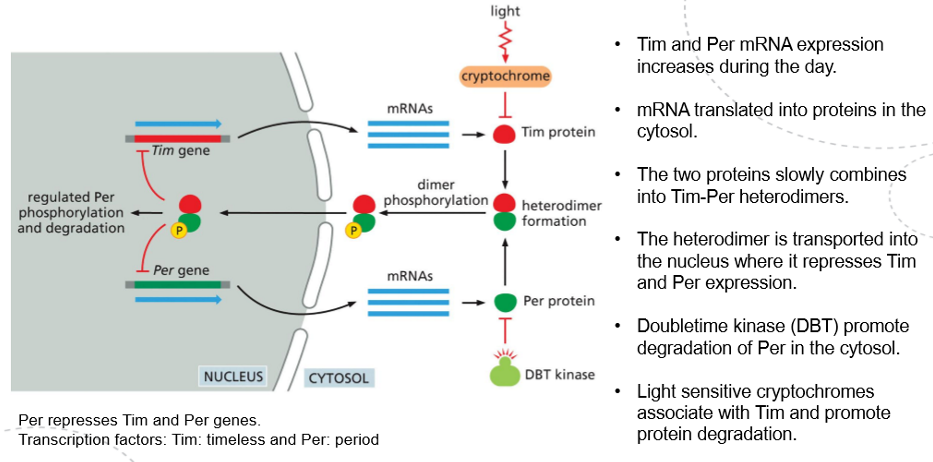
A fundamental principle of circadian clocks revolves around delayed negative feedback loops. Certain gene products accumulate to eventually switch off the transcription of their own genes, but this occurs with a time delay, causing the cell to oscillate between a state where gene products are present with transcription turned off and another state where they are absent, allowing transcription to resume.
In some cell types, the circadian clock is constructed using proteins that regulate their activities through post-translational mechanisms. In Drosophila, for example, during the day Tim and Per expression increases via mRNA translation, leading to the slow formation of Tim/Per heterodimers in the cytosol, which migrate into the nucleus where they down-regulate their own expression, creating a negative feedback loop that decreases their levels during the night. The stability of the two proteins is, however, regulated light sensitive cryptochromaes as well as DBT . Both of the proteins lead to the degradation of their respective targets, delaying the formation of the heterodimer, therefore altering the length of the circadian oscillations.
What are the similarities and differences between TGF-β and cytokine receptor (e.g. JAK-STAT) signaling?
Similarities
Both TGF-β receptors and cytokine receptors transmit extracellular signals directly to the nucleus to regulate gene expression.
Both pathways use latent cytoplasmic transcription regulators that become activated, move into the nucleus, and control transcription (Smads for TGF-β, STATs for cytokines).
Both are commonly involved in growth control, differentiation, immune regulation, and development.
Differences
Receptor type:
TGF-β receptors are serine/threonine kinase receptors with intrinsic kinase activity.
Cytokine receptors lack intrinsic kinase activity and rely on associated JAK kinases.
Signal transducers:
TGF-β signaling uses Smad proteins.
Cytokine signaling uses STAT proteins.
Activation mechanism:
In TGF-β signaling, the receptor directly phosphorylates Smads.
In cytokine signaling, JAKs phosphorylate STATs after receptor activation.
What is the plants immune system? How does it work?
Plants are constantly batteling a diverse set of potental invarders, above and below ground, e.g. bacteria, fungi, and insects.
The plants innate immunity allow the plants to sense the presence of microbes and start a precise and localized repsonse to prevent disease.
The plant recognize specialized molecular pattterns called MAMps (e.g. flagillin) by specialized receptors called RLK and RLKs. Upon detection a sinalling cascade is initiated, and activated calsium channelaes, MAP kinase and the production of ROS to change hormone levels which often suffice in bacterial defence by e.g. activates signaling pathways that strengthen the cell wall, produce antimicrobial compounds, and induce defense genes. this is called PTI
If not, a second defence layer is initiated. the same MAMps are recognized by specialized intracellular receptors called NLRs which produce the same outcome just with a greated intensity. this leads to regulated cell death. this is called ETI
How does PTI activation by MAMPs work (in detail). What is the outcome?
the innate immunity owrks through membrane bound receptors which recongiced MAMPs.
Steps in PTI:
RLK receptor bind flagellin
binding tirggers RLK to associate with co-reseptor BAK1, activating the receptor complex
RLK and BAK1 phosphorylate eachother and recruits ans phosphorylates kinase BIK1
BIK1 dossociate from the complex to propagate a sognaling cascade ownstream.
A crutial and visible outcome of this immunity is the closure of the stroma, and act as a physical defence.
What is the ZAR1 resistosome?
The plant immune receptrs, RLK, can come together to form complexes made from multiple receptor units by oligomerization.
One example is ZAR1. ZAR1 is a pentameric complex that forms a tunnel shape in the plasma membrane allowing Ca influx. Ca is required for triggering cell death and antibacterial immunity.
How is cell death regulated in plants?
Cell death in plants is tightly regulated and mainly occurs as programmed cell death (PCD), which is essential for development and defense. Unlike animals, plants do not use caspases or apoptosis but rely on plant-specific regulatory mechanisms.
Some examples of plant programed cell death are:
suspensor PCD: suspensor which helps transfer nutrient in embryo development dies when role is fufilled
HR-PCR: immune defence. death of one cell hinders spread
TE-PCD: during xylem development. tracheary cells die to make hollow tubes for water transport throughout the plant
PART 3
What are and why are accessory proteins essential?
Accessory proteins are proteins that bind to cytoskeletal filaments (actin filaments, microtubules, or intermediate filaments) and regulate their behavior and function.
They do not form the filaments themselves, but instead control organization, stability, interactions, and movement
They regulate where and when filaments assemble and disassemble (nucleation, growth, capping, severing).
They organize filaments into higher-order structures such as bundles and networks.
They link cytoskeletal filaments to membranes, organelles, cell junctions, and the ECM.
Motor accessory proteins generate force and movement using ATP.
What is the cytoskeleton? What is the main roles of Actin filametnts, microtubules, and intermediate filaments?
The cytoskeleton is a dynamic network of protein filaments that extends throughout the cytoplasm of a cell. It provides the cell with shape, mechanical strength, internal organization, and the ability to move
Actin filaments are thin, flexible filaments that mainly function in cell shape and cell movement. They form the cell cortex beneath the plasma membrane, generate protrusions such as lamellipodia and filopodia, drive muscle contraction with myosin, and form the contractile ring during cytokinesis.
Microtubules are long, stiff, hollow filaments that function in intracellular transport, cell polarity, and chromosome segregation. They serve as tracks for motor proteins (kinesins and dyneins), form the mitotic spindle during cell division, and build cilia and flagella for motility and sensing.
Intermediate filaments are strong, rope-like filaments that provide mechanical strength and resistance to stress. They help cells withstand stretching forces, maintain tissue integrity, and reinforce cell–cell and cell–matrix junctions in epitehelial cells.
How are actin filaments made up?
actin subunits form aggregares called seeds, which further assmeble into a long tightly a-helix known as f-actin.
the subunits in the filament are asuymettrical, with a slow growing - side, and a fast growinf + side. the ATP binding cleft is towards the - end in each filament.
once a seed is formed it can rapidly elongate by adding more subunits. it goes through three phases:
Lag (nucleation) phase – small, unstable actin oligomers form until a stable nucleus (seed) is created.
Elongation (growth) phase – rapid filament growth occurs as actin subunits add to the filament ends.
Steady-state (equilibrium) phase – the rate of subunit addition equals the rate of subunit loss, resulting in constant filament length. (critical consentration)
Actin association is must faster in the + end.
Actin filaments can exist in two forms. what are they and how does this affect actin filament assemby/disassembly?
ATP hydrolysis si slow in subunits, but drastically increase when the subunits form filaments.
Actin filaments exist in two forms: the ATP-bound (T form) and the ADP-bound (D form).
In the T form, actin subunits are bound to ATP, which makes the filament more stable and favors polymerization. The critical concentration is lower (minimum concentration of free actin monomers required for filament growth —> filament can grow even when free actin concentration is relatively low.), so subunits are more likely to add to the filament ends, promoting filament growth.
In the D form, actin subunits contain ADP after ATP hydrolysis. This form is less stable, has a higher critical concentration, and therefore favors depolymerization, leading to filament shrinkage.
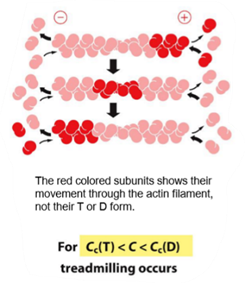
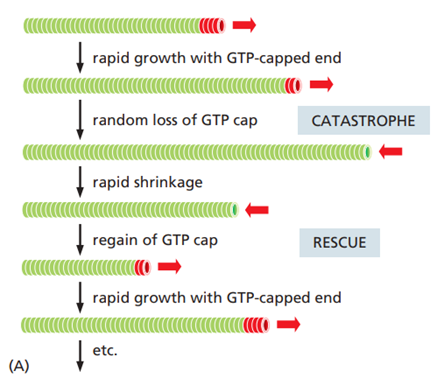
What is threadmilling?
Treadmilling is a dynamic steady state in which an actin filament grows at one end and shrinks at the other at the same time, so the overall filament length stays constant.
It occurs when:
The plus end is in the ATP-bound (T) form and adds subunits.
The minus end is in the ADP-bound (D) form and loses subunits.
Treadmilling requires a continuous supply of ATP, because ATP hydrolysis drives the conversion from T form to D form as subunits move through the filament.
Explain T- and D-tips in actin filaments. What does this mena in practice?
T-tips and D-tips describe the nucleotide state of actin subunits at the ends of an actin filament, and they determine whether that end tends to grow or shrink.
A T-tip is an actin filament end capped with ATP-bound actin (T form). This occurs when new actin subunits are added faster than ATP is hydrolyzed. T-tips are stable and favor polymerization, so filament growth occurs at that end.
A D-tip is an actin filament end composed of ADP-bound actin (D form). This occurs when ATP hydrolysis happens before new subunits are added. D-tips are less stable and favor depolymerization, so filament shrinkage occurs at that end.
In practice:
High actin concentration → T-tips at both ends → filament growth
Low actin concentration → D-tips at both ends → filament shrinkage
Intermediate actin concentration → T-tip at plus end and D-tip at minus end → treadmilling
What is the role of Arp2/3 complexes and formins? What is the role of profilin?
Arp2/3 complex and formins are actin-nucleating proteins, meaning they help start new actin filaments, but they generate very different actin structures.
The Arp2/3 complex nucleates branched actin filaments. It binds to the side of an existing actin filament and initiates growth of a new filament. It remains attached to the minus end of the new filament, while rapid elongation occurs at the plus end.
Formins nucleate unbranched, linear actin filaments. They form dimers that stabilize actin nuclei and stay associated with the growing plus end, allowing continuous elongation. Formins also associate with profilin.
Profilin regulate actin monomers and filament behavior. Profilin binds to actin monomers and promotes actin polymerization at the plus end, stimulating filament growth
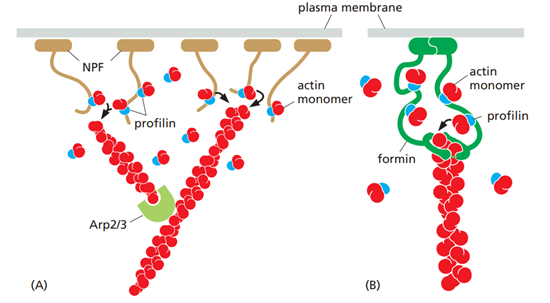
What is the structure of myosin?
Myosin is a motor proteins and, is composed of two heavy chains with globular heads at one end, and coiled tail at the other. In muscle cells myosin assemble into thick filaments. Each globular head can bind ATP
How does myosin move along the actin filament?
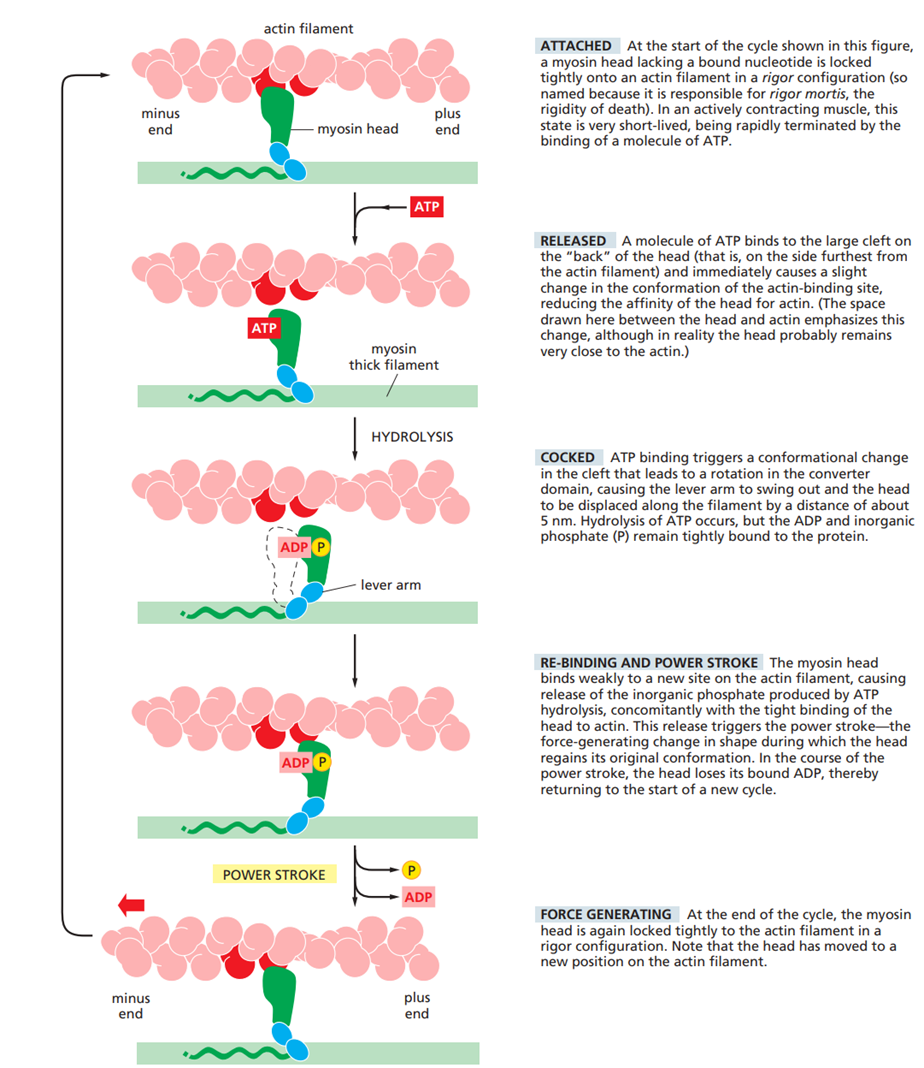
Myosin moves along an actin filament by converting ATP hydrolysis into mechanical force through a cyclic sequence of binding, conformational change, and release.
First, a myosin head binds to an actin filament. When ATP binds to the myosin head, myosin detaches from actin. The ATP is then hydrolyzed to ADP + P, which “cocks” the myosin head into a high-energy conformation.
Next, the myosin head rebinds to a new position on the actin filament closer to the plus end. Release of phosphate triggers the power stroke, during which the myosin lever arm swings and pulls the actin filament relative to the myosin. Finally, ADP is released, leaving myosin tightly bound to actin until a new ATP binds and the cycle repeats.
What is the composition of skeletal muscles?
Skeletal muscle is made up of mushle fibers. Inside each muscle fiber are numerous myofibrils, which run along the length of the cell. Myofibrils are composed of repeating contractile units called sarcomeres, the fundamental units of muscle contraction.
Each sarcomere contains:
Thin filaments made primarily of actin, along with the regulatory proteins tropomyosin and troponin
Thick filaments composed of myosin II
Sarcomeres are organized between Z-discs, where actin filaments are anchored, and capped by CapZ at the + end and tropomodulin at the - end, which stabilizes their length. Myosin filaments are centered in the sarcomere, and are cross-linked at the M-line. Myosin overlap with actin filaments, allowing contraction through filament sliding.
Two giant accessory proteins play key structural roles.
Nebulin, a large actin binding protein, stabilizes actin filaments along their length, acting as a molecular ruler, and stretches from the Z-disc to the actin filament minus end. Titin, a gigantic protein, extends from the Z-disc to the M-line, and a keeps thick filaments centered, pushing them towards the center, as if it was a spring.
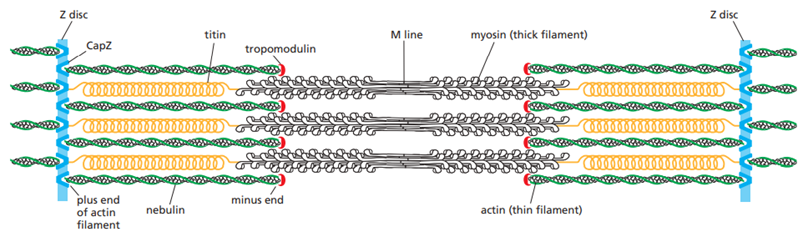
What happens during a muscle contraction?
An action potential reaches the muscle fiber and travels along the sarcolemma and down the T-tubules.
The sarcoplasmic reticulum releases Ca²⁺ into the cytosol.
Ca²⁺ binds to troponin C (part of the troponin complex on actin filaments).
Binding of Ca²⁺ causes troponin to change conformation.
This conformational change moves tropomyosin away from myosin-binding sites on actin.
Myosin heads bind to exposed actin sites (cross-bridge formation).
Release of Pi and ADP triggers the power stroke, pulling actin toward the M-line.
ATP binds to myosin, causing detachment from actin.
ATP hydrolysis re-cocks the myosin head for another cycle.
Repeated cycles cause sarcomere shortening and muscle contraction.
Ca²⁺ is pumped back into the sarcoplasmic reticulum, troponin releases Ca²⁺, tropomyosin blocks actin again, and the muscle relaxes.
What is a microtubule catastrophy? How does it occur?
A microtubule catastrophe is a sudden transition in which a microtubule switches from growth to rapid shrinkage.
It occurs when the GTP cap at the plus end is lost. While tubulin subunits are added with GTP bound, GTP is hydrolyzed after incorporation into the microtubule. As long as new GTP-tubulin is added faster than hydrolysis, a stabilizing GTP cap is maintained and the microtubule grows.
If tubulin addition slows or stops, GTP hydrolysis catches up, the GTP cap disappears, and the microtubule end is left in the GDP-bound (D) form, which is unstable. This causes the protofilaments to peel outward and the microtubule to undergo rapid depolymerization — the catastrophe.
Microtubuli have dynamic instability