Genetics - Lecture 25: COVID 19 Vaccines
1/27
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
28 Terms
Coronavirus
named after their appearance which looks like a sun
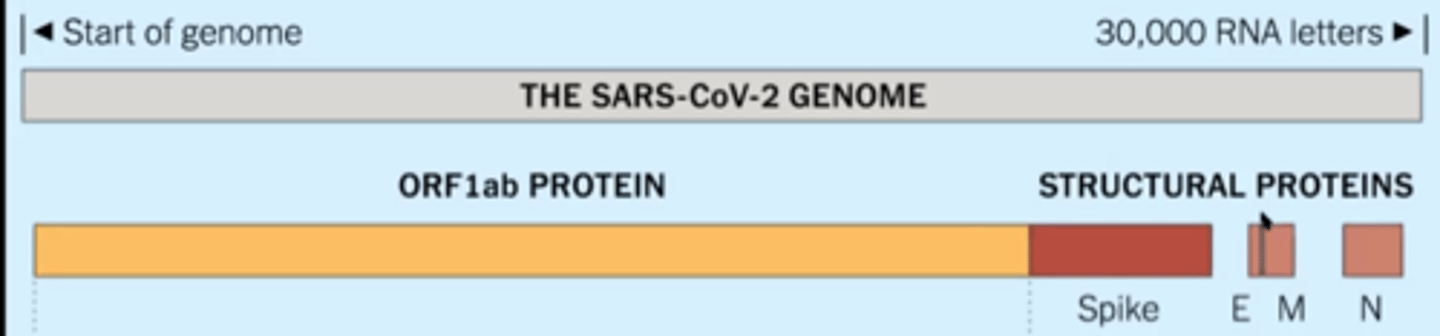
Genome of a coronavirus
single stranded positive sense RNA
Zoonotic coronavirus
jump from animals to humans; all beta-coronaviruses
ORF1ab
large part of SARS=CoV-2 genome that codes for a mega-protein that is then cleaved into 16 non-structural proteins
ACE2
Human COVID-19 receptor protein
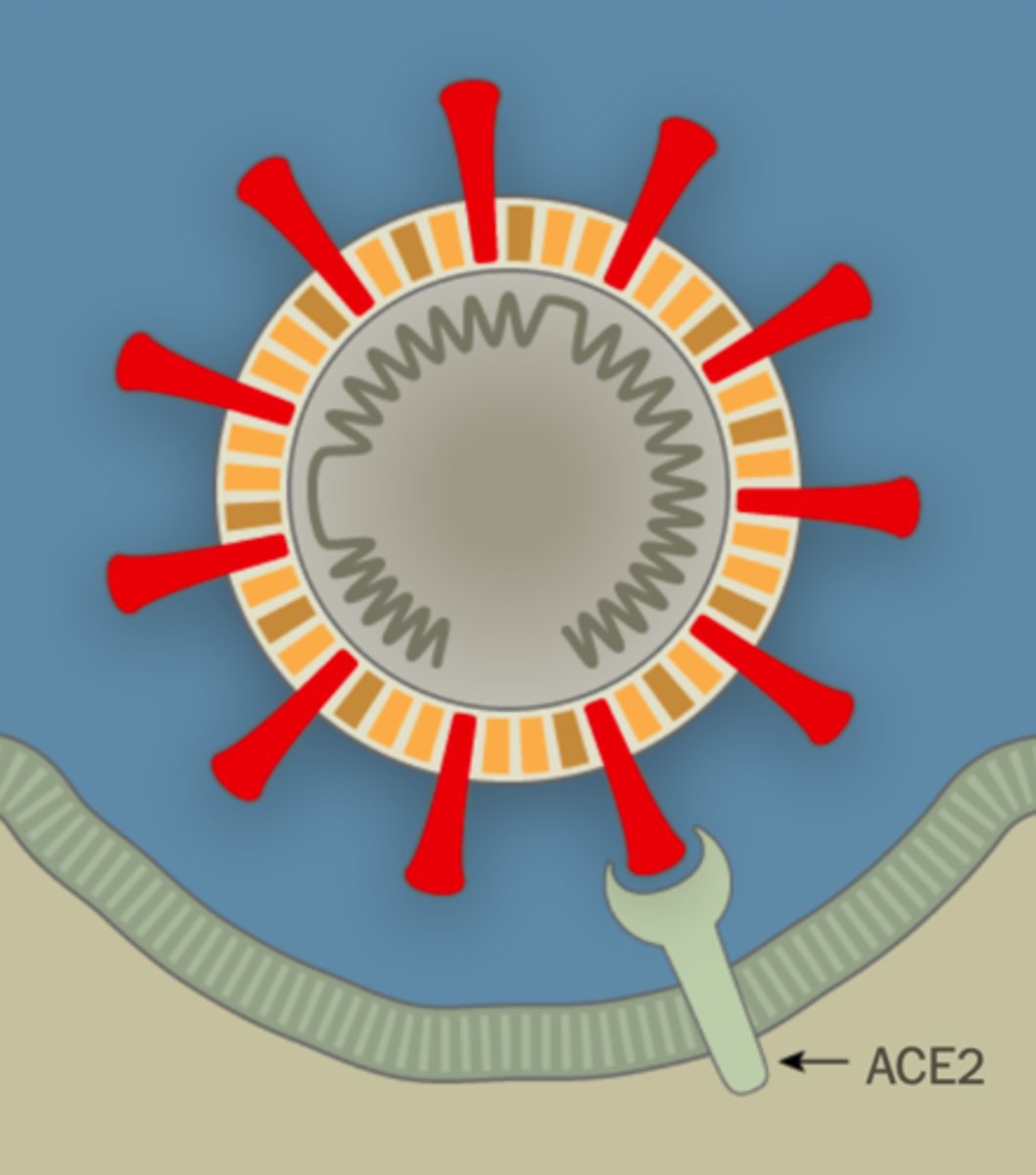
TMPRSS2 protease
Efficient viral entry requires cleavage of spike by this protease

Lateral flow immunoassays
-Labeled antibody reacts with the antigen as the sample moves through the pads and membranes of the test strip
-As the mixture migrates, antigen-antibody complexes are captures at a test line by immobilized antibody

Non-neutralizing antibodies
clump-up virus particles - large clumps more easily cleared by phagocytes
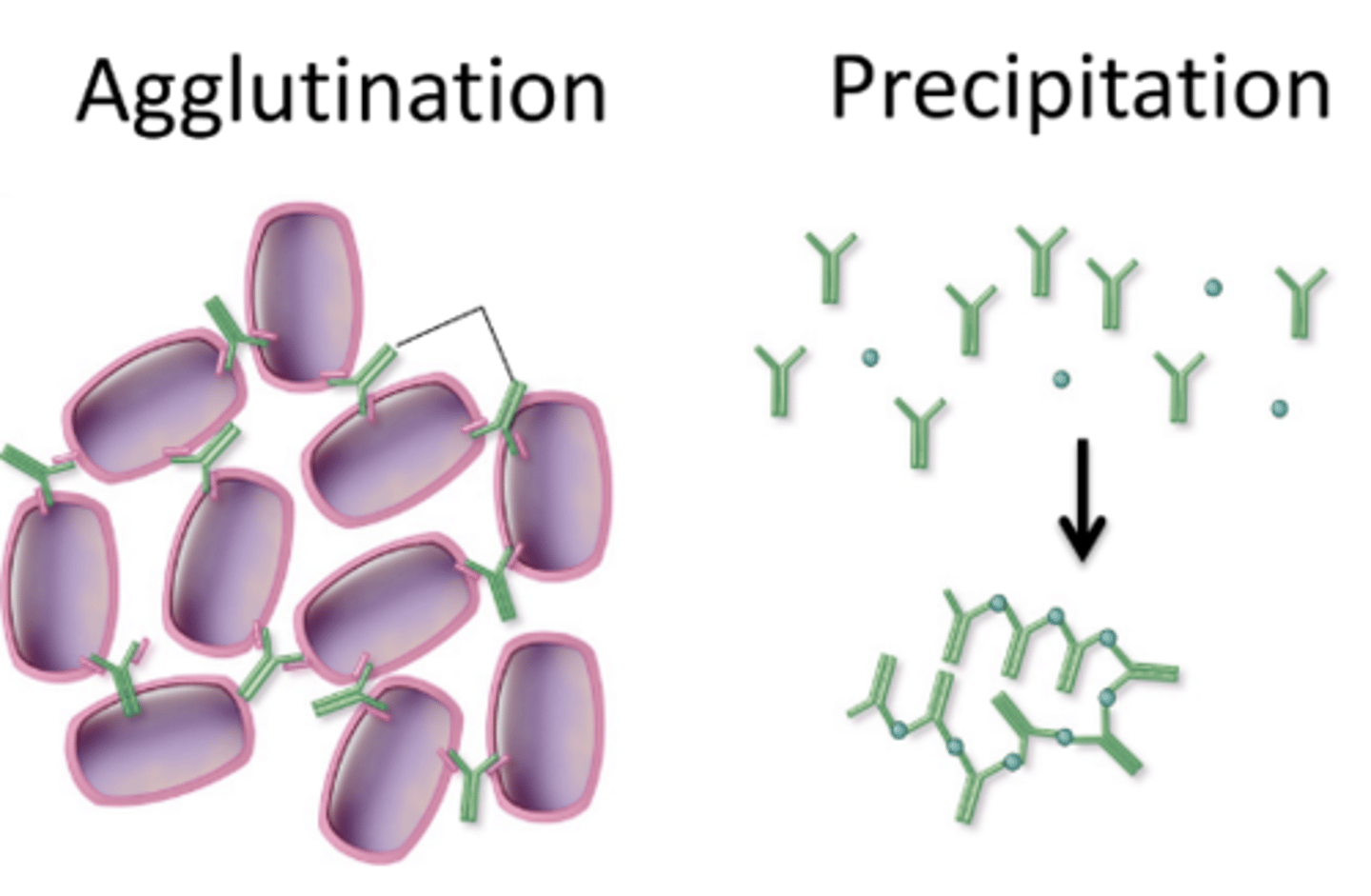
Neutralizing antibodies
block pathogen function directly
Traditional vaccines
weak or dead pathogens
Immunogenicity
Ability of pathogens to induce an immune response
Major challenge with traditional vaccines
need to grow lots of virus and kill it or inactivate it without loosing immunogenicity and without harming people
3 Modern strategies for making vaccines
1. Protein-based
2. Viral vector
3. mRNA
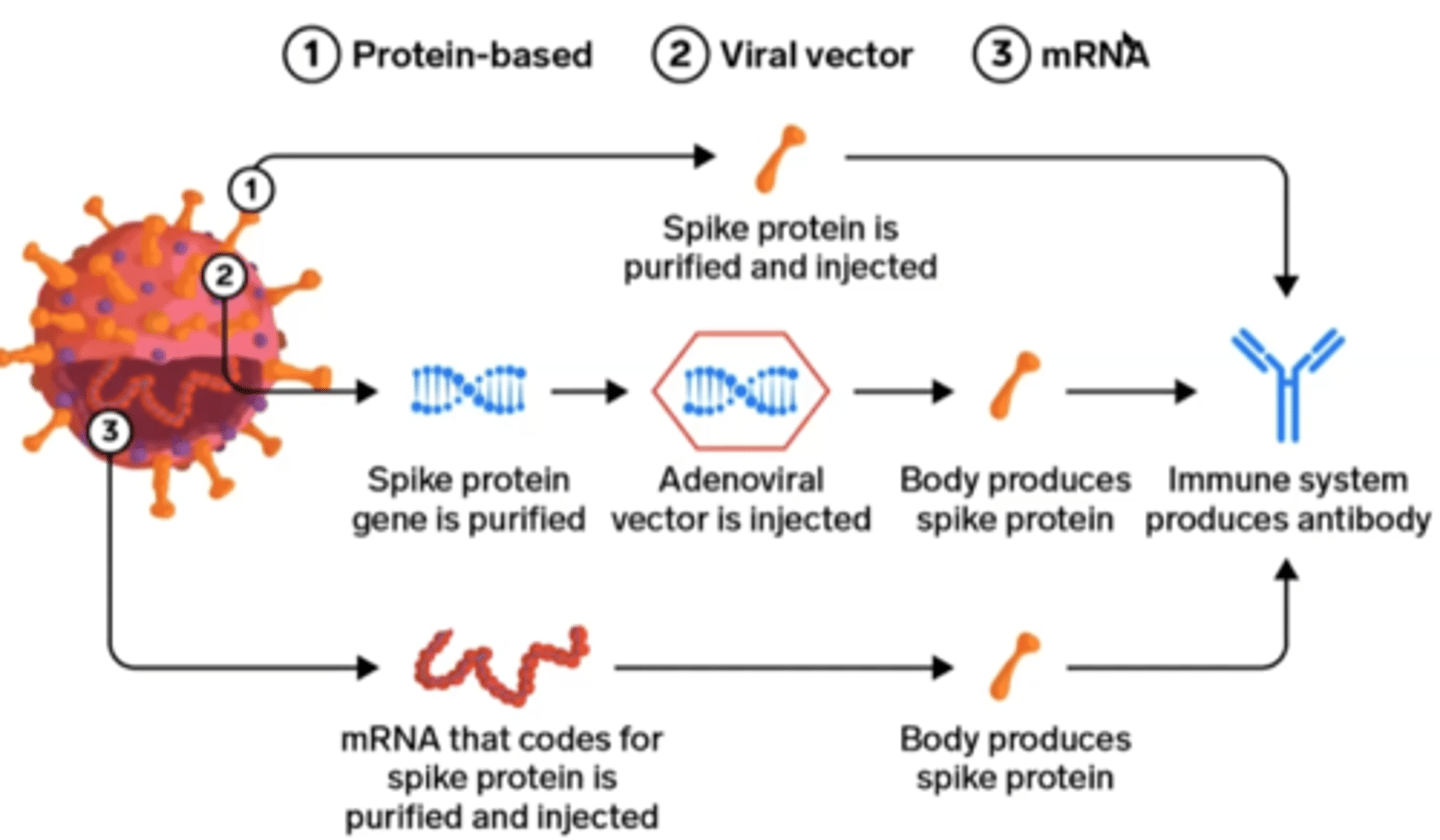
Main vaccine antigen
spike protein
Protein vaccines
recombinant proteins from virus injected to elicit immune response

Problem with protein vaccines
proteins do not invade cells, so may not get a strong T-cell response
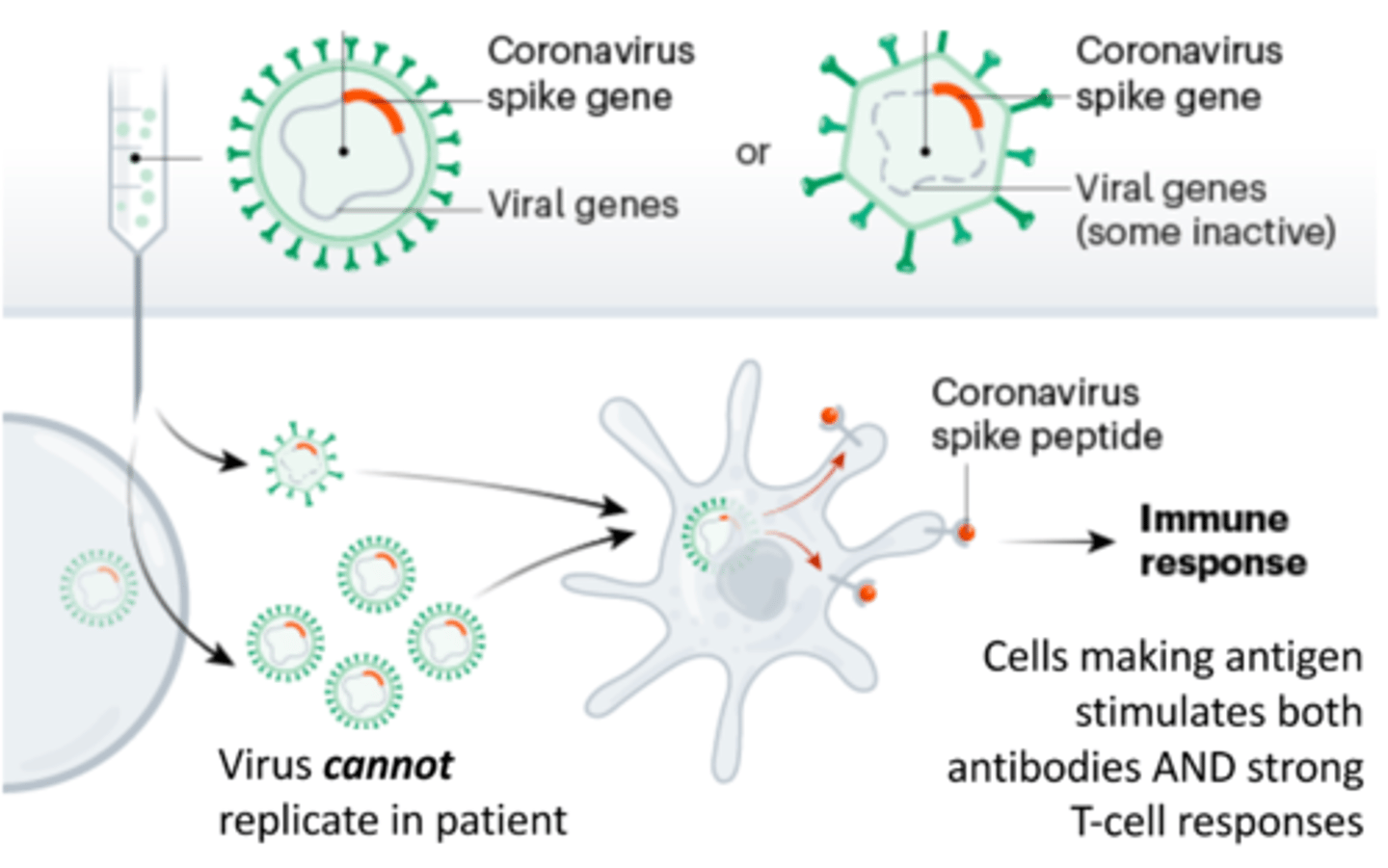
Viral vector vaccines
Put SARS-CoV-2 spike protein gene into crippled adenovirus genome
mRNA vaccines
mRNA from virus placed into lipid nanoparticle which delievers the mRNA to body cells
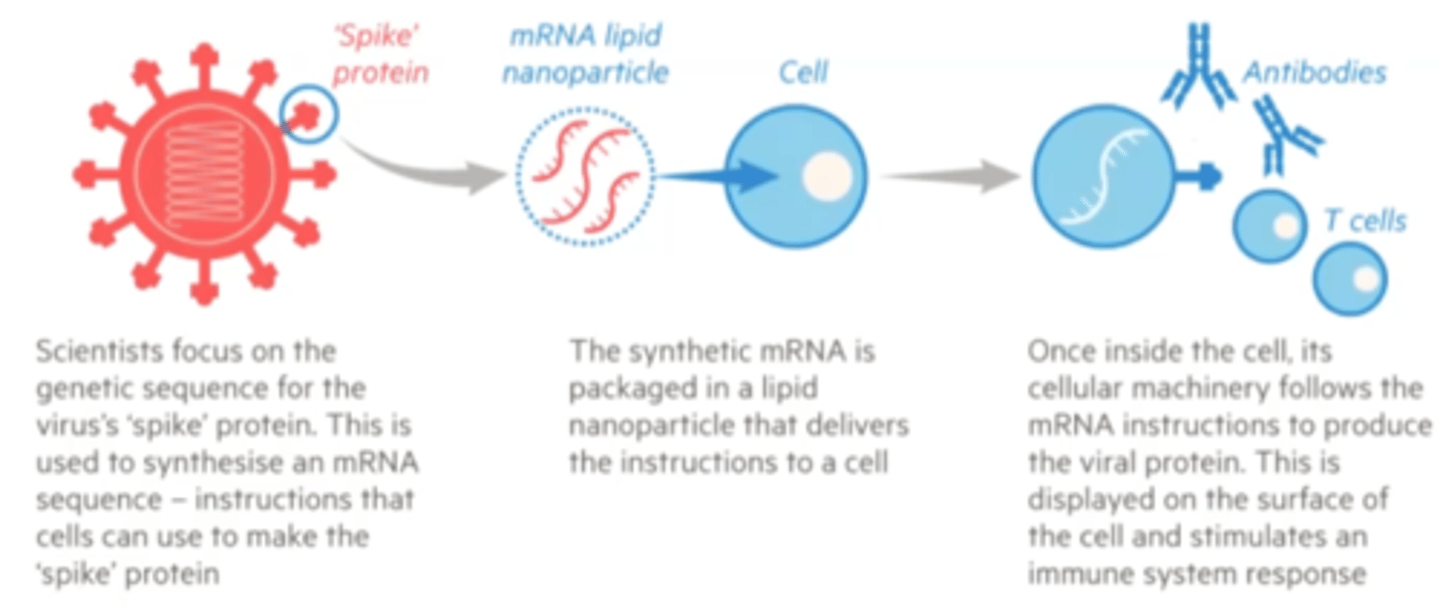
Benefits of mRNA vaccine
Faster and potentially safer than viruses to make: RNA and shell are completely synthetic
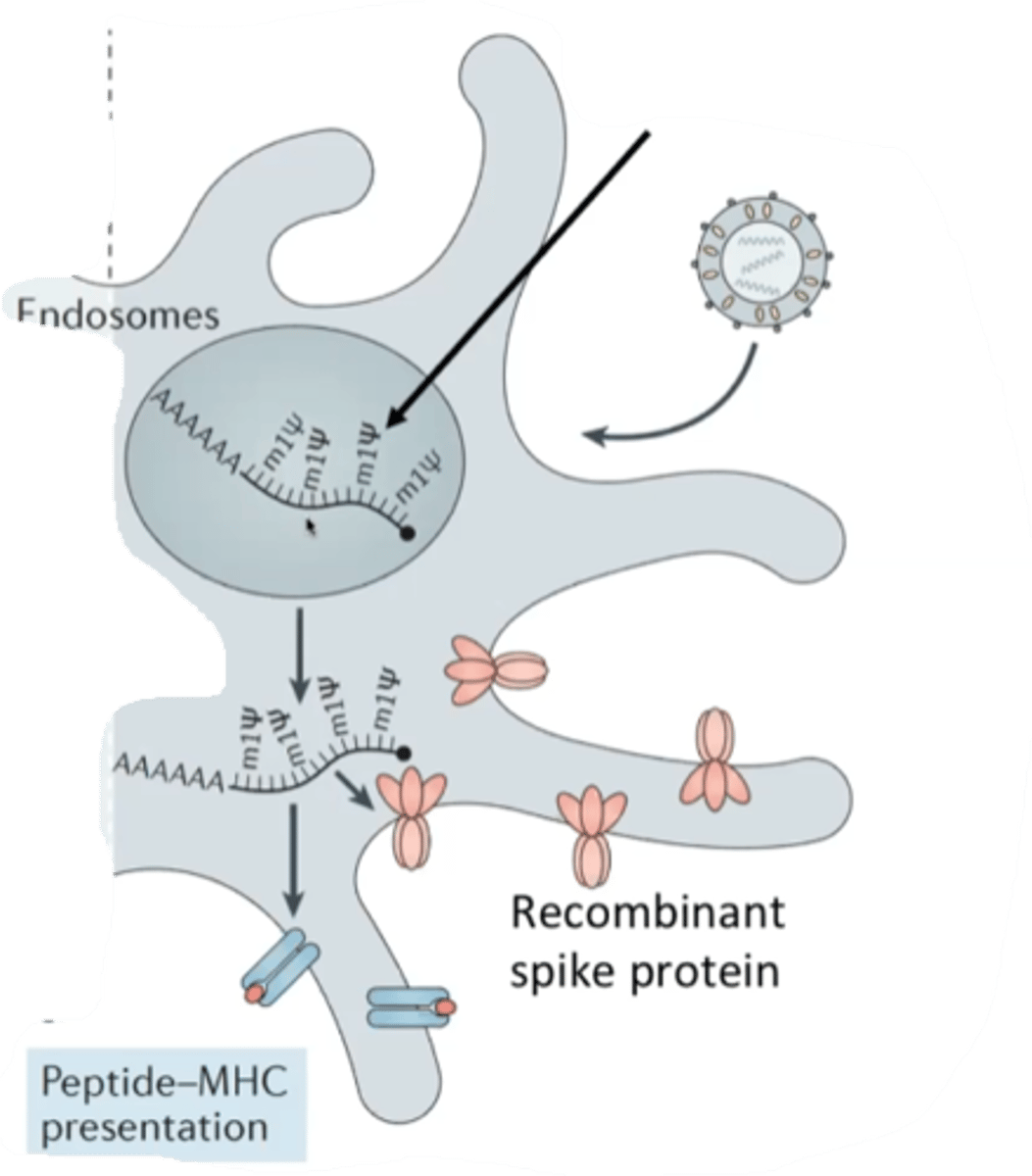
Two challenges of mRNA vaccines
- creating a lipid nanoparticle than can survive to fuse with a cell
- mRNA degredation in cells
Method to prevent premature degredation of mRNA from vaccines
Modified urasil bases that translate correctly, but are resistant to degredation
Spike protein with S-2P mutation
locked into pre-fusion conformation
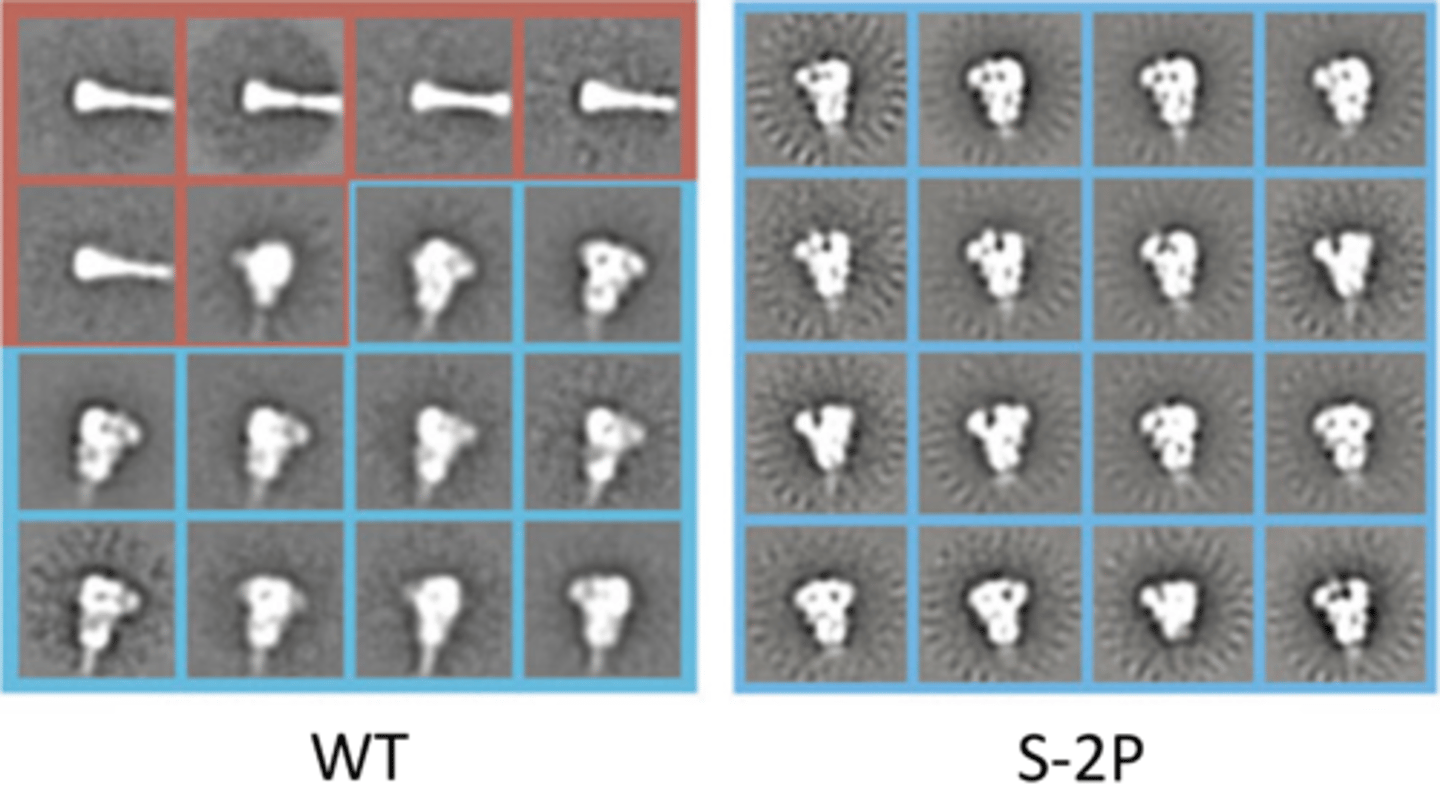
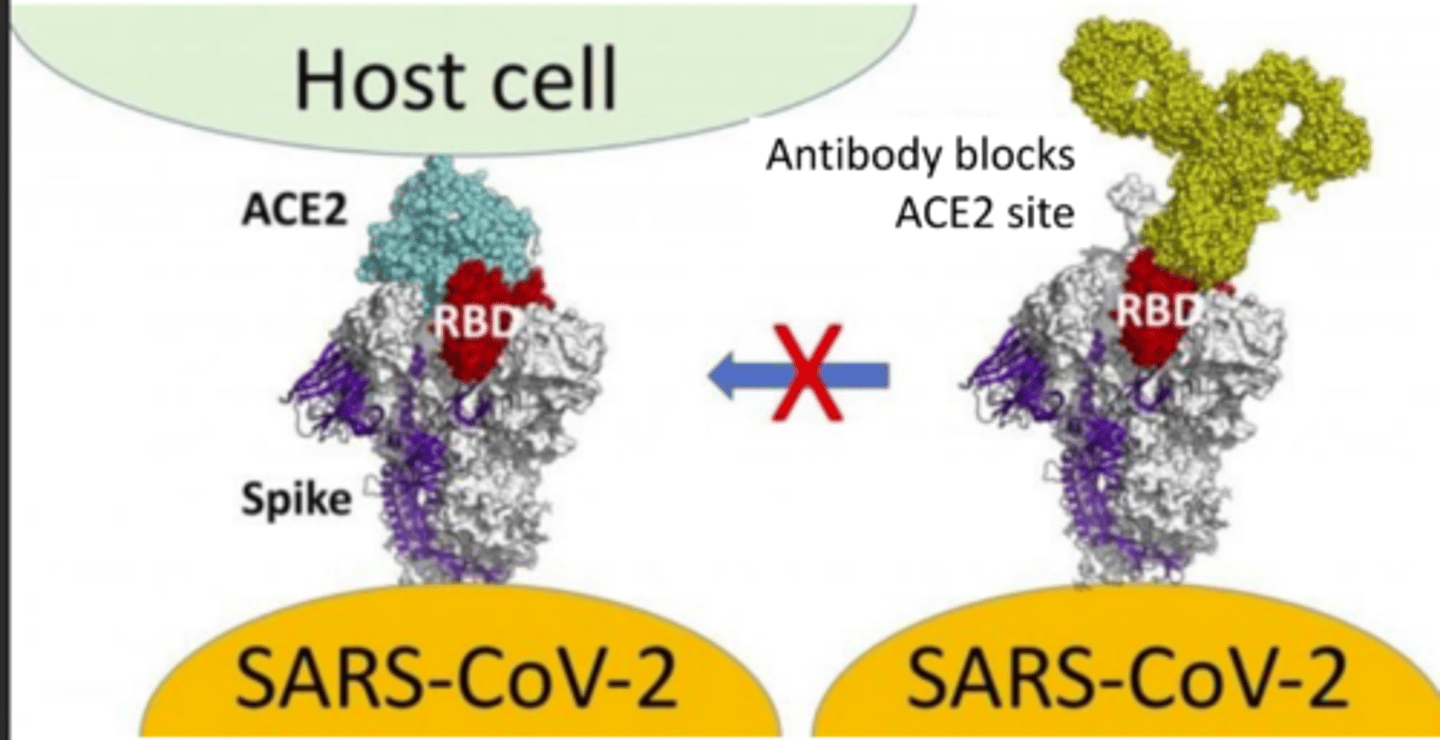
Vaccine with S-2P:
elicit anibodies to close spike protein (pre-fusion conformation) to prevent binding to ACE2, thus neutralizing virus
Vaccine-escape spike protein variants:
- reduce binding to neutralizing antibody
- maintain binding to ACE2 receptor
Difference between Omicron and the previous variants
Omicron spike did not require TMPRSS2 cleavage for efficient entry
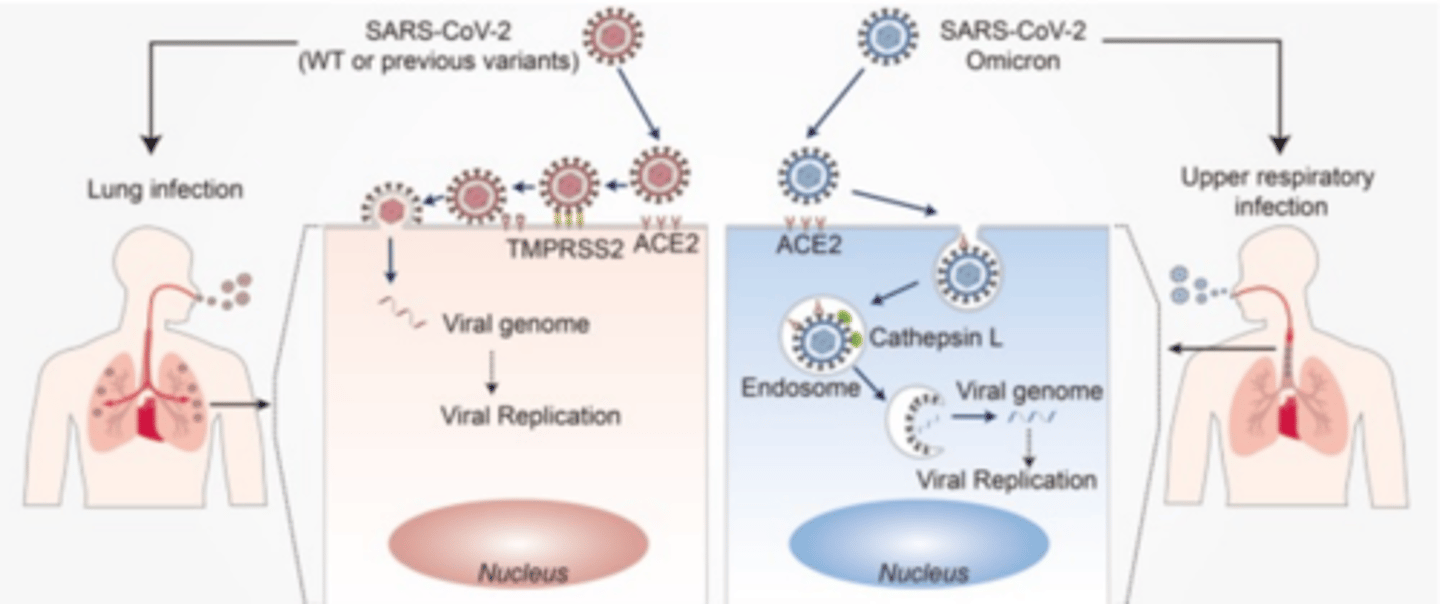
Why did variants before omicron infect deep into the lungs?
TMPRSS2+ cells are found deep in lungs
Where did omicron primarily infect?
upper resperatory tract, because those cells do not have TMPRSS2
Why was omicron more virulent, but less deadly?
more virus in the upper respiratory tract, thus easier to cough out and spread, BUT, not found deep in lungs and therefore less pneumonia