Module 5 - DNA Repair
1/35
Earn XP
Description and Tags
included in exam 2
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
36 Terms
normal DNA replication
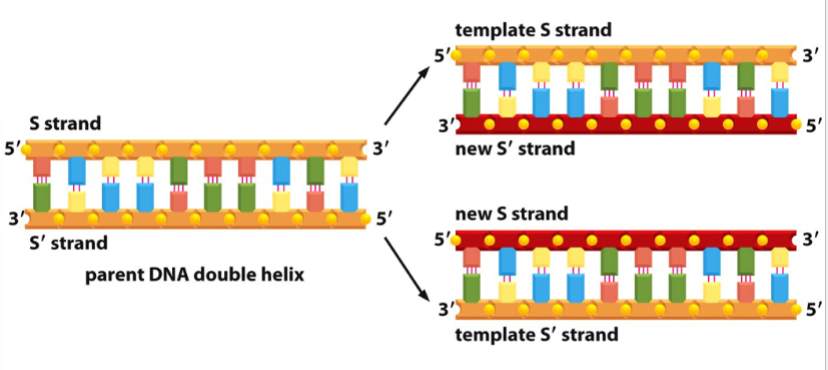
a strand of DNA serves as a template for the synthesis of complementary, antiparallel strands in the 5’ to 3’ direction
both strands of the original DNA serve as templates, yielding two total new DNA molecules
semiconservative replication (each new strand has half of the parent helix)
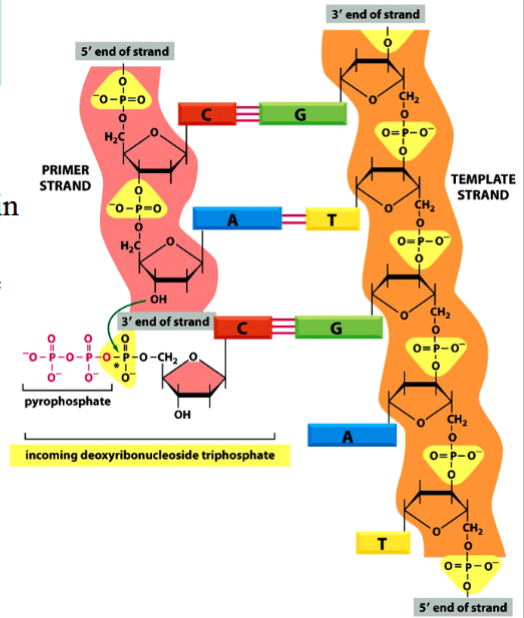
phosphodiester bond formation
requires energy in order for a new one to be formed
energy comes from breaking of a high-energy phosphoanhydride bond in dNTP
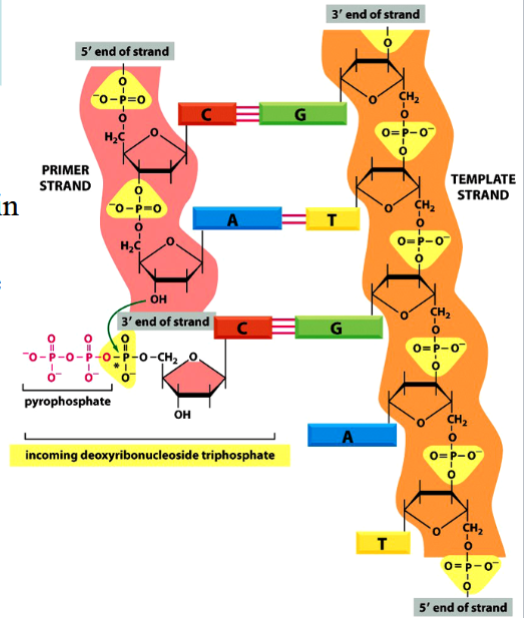
phosphodiester bond
the bond between different nucleotides in a nucleic acid (between bases in DNA)
requires energy to be formed
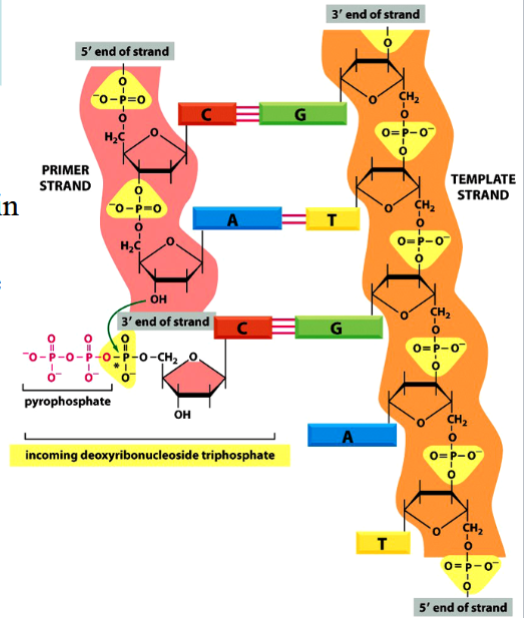
phosphoanhydride bond
the high-energy P-P bond between phosphates in an NTP or NDP
breakage of this bond provides energy required for a phosphodiester bond to form
(see the arrow in the picture)
proofreading during DNA synthesis
3’ to 5’ exonuclease activity done by DNA polymerase
if a wrong nucleotide is placed during synthesis (5’ to 3’), polymerase removes it (3’ to 5’) and replaces it with the correct nucleotide (5’ to 3’)
is made possible by 5’ to 3’ synthesis
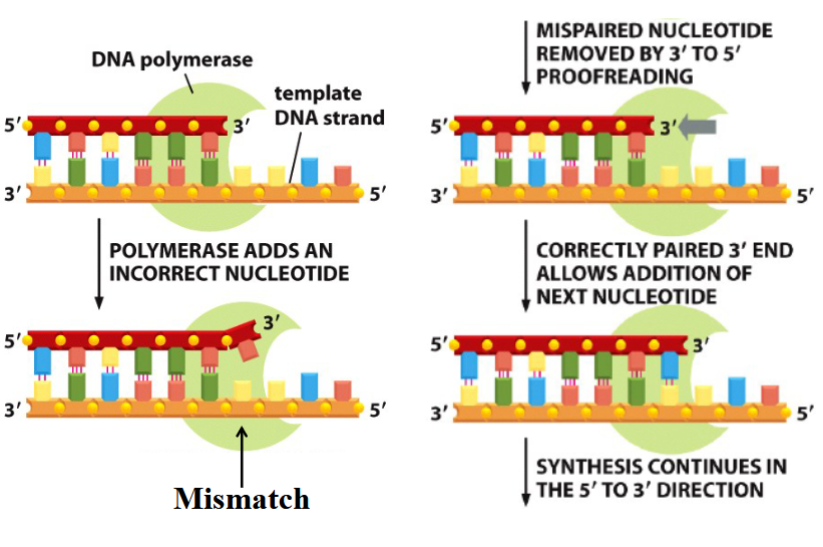
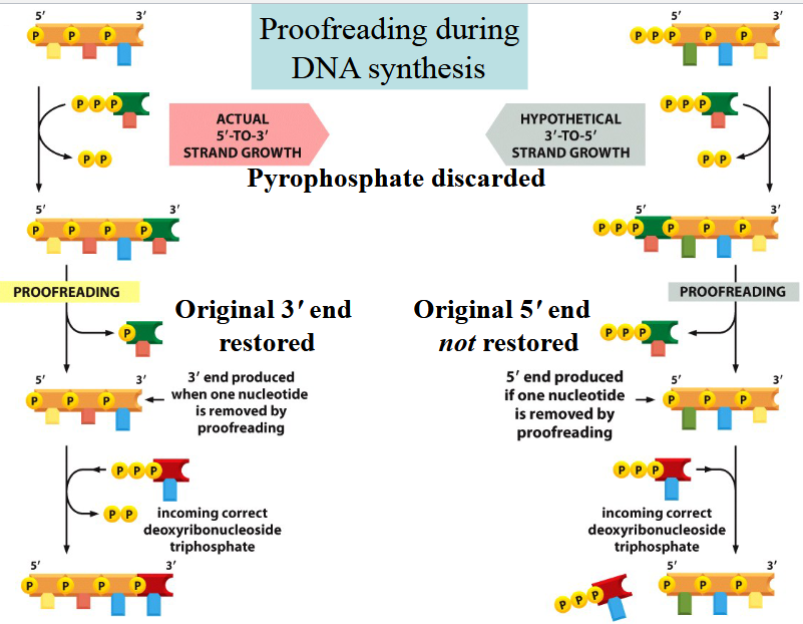
why must DNA synthesis occur in the 5’ to 3’ direction?
proofreading is only possible if synthesis is 5’ to 3’
if polymerization occured 3’ to 5’:
energy needed for a new phosphodiester bond to form would come from breaking of a phosphoanhydride bond at the 5’ end of the chain
but if the last nucleotide (dNTP) were removed during proofreading, there would be no P-P bond available to add the next molecule (not enough energy)
(if a mistake were made, the energy needed to fix the mistake would be removed with the mistake → could not be fixed)
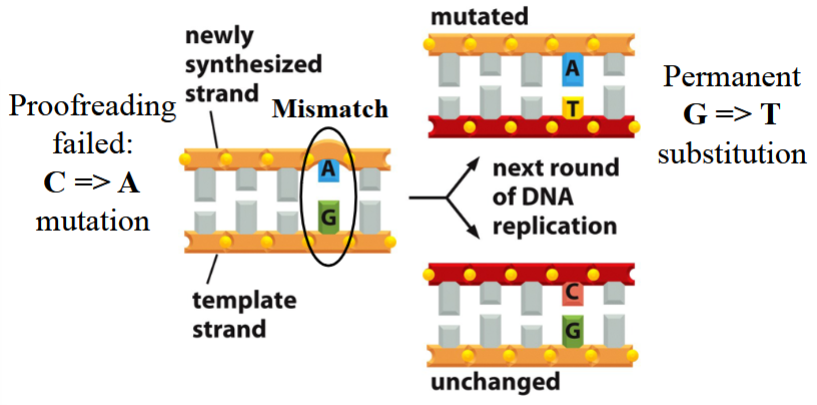
nucleotide mismatches
the consequence of errors in DNA replication
can lead to permanent changes if left unrepaired before the next round of DNA synthesis
(error without mismatch repair occurs every 1 per 107 nucleotides copied)
(error with mismatch repair occurs every 1 per 109 nucleotides copied)
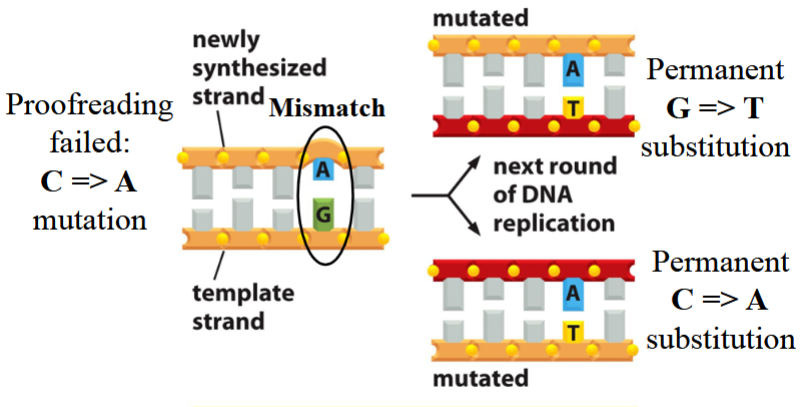
error with no mismatch repair
proofreading fails, a mutation occurs (e.g. C→A; A+G pair instead of C+G)
no repair occurs
with the next round of DNA replication:
one new strand matches with the nonmutated template, remains unchanged (C+G pair)
the other new strand matches with the mutated template, has a permanent G→T substitution (A+T pair instead of C+G)
error with incorrect mismatch pair
proofreading fails, a mutation occurs (e.g. C→A; A+G pair instead of C+G)
repair occurs, but the correct template is “fixed” to match the mutated template (G→T instead of A→C again)
excision and repair only occurs on the template (old) strand
with the next round of DNA replication:
both new strands match with the doubly mutated template (both A+T instead of C+G)
one strand has a permanent C→A substitution because of the false repair
the other strand has a permanent G→T substitution because of the original proofreading fail mutation
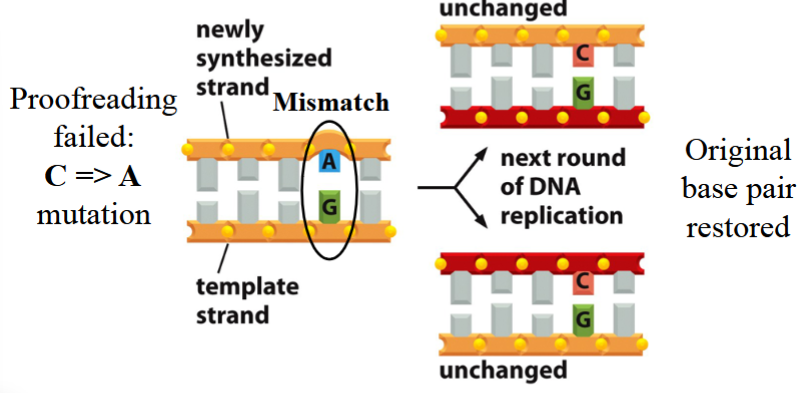
error with correct mismatch repair
proofreading fails, a mutation occurs (e.g. C→A; A+G pair instead of C+G)
repair occurs, the mutated template is fixed to match the original correct template (A back to C again, G is unchanged)
excision and repair only occurs on the newly synthesized strand
with the next round of DNA replication:
original base pair is restored (both new strands are C+G)
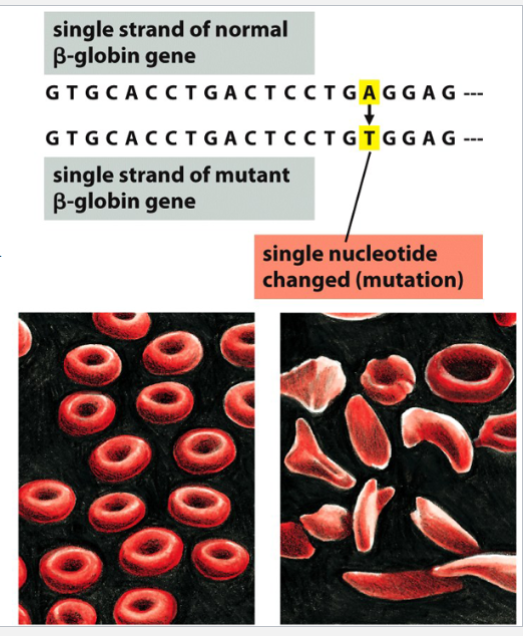
sickle-cell anemia
an example of a consequence of a point mutation in the beta-globin gene
a single nucleotide substitution occurs in DNA (A→T) → causes an amino acid change
the changed amino acid alters the tertiary structure of beta-globin
blood cells are deformed (sickled) and function/efficiency is inhibited
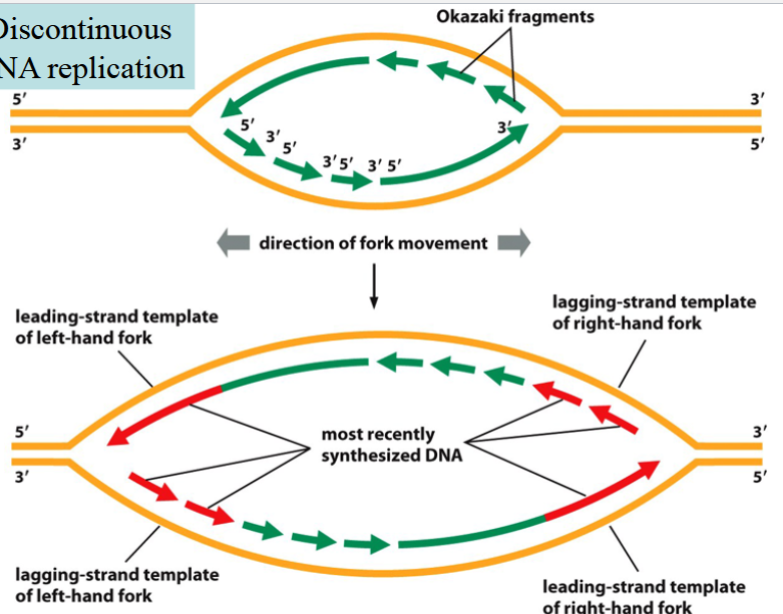
problem and solution of mismatch repair in eukaryotes
problem: how does the cell know which DNA strand is synthesized, and which one of the two nucleotides is wrong in a mismatched pair?
solution: the newly synthesized strand is interrupted due to discontinuous replication → results in Okazaki fragments with nicks/breaks
mismatch repair must be done before the nick is repaired by ligase
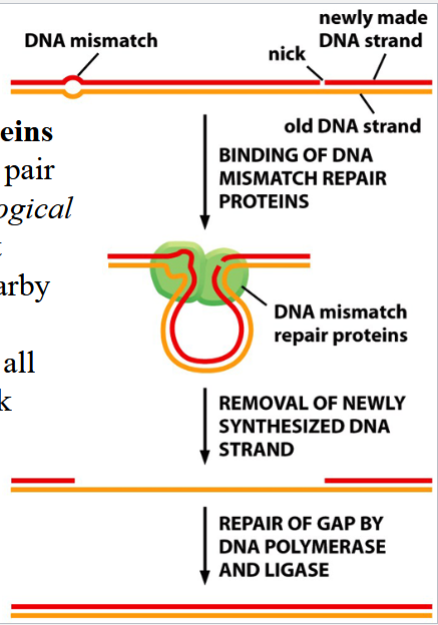
model for mismatch repair in eukaryotes
DNA mismatch repair proteins recognize the mismatched pair (due to topological disturbance caused by imperfect fit) and bind to it
the DNA is scanned for a nearby break
the nicked strand is digested from the break to the mismatch site
DNA polymerase and DNA ligase complete the repair (polymerase replaces the removed DNA, ligase seals the nick)
hereditary nonpolyposis colorectal cancer (colon cancer)
aka HNPCC
affects ~1 in 200 people
caused by defects in MutS or MutL genes; both code for DNA mismatch repair proteins
results in higher frequency of mutations in other genes, e.g. genes that regulate cell proliferation (unregulated cell proliferation → cancer)
spontaneous mutations
categories: deaminations or radiation-induced dimerization
consequences: distorted strands or mismatches with unusual bases
occur because DNA exists in the environment of the cell (an open system) → thus, DNA is surrounded by radiations and molecules that can react with and alter it
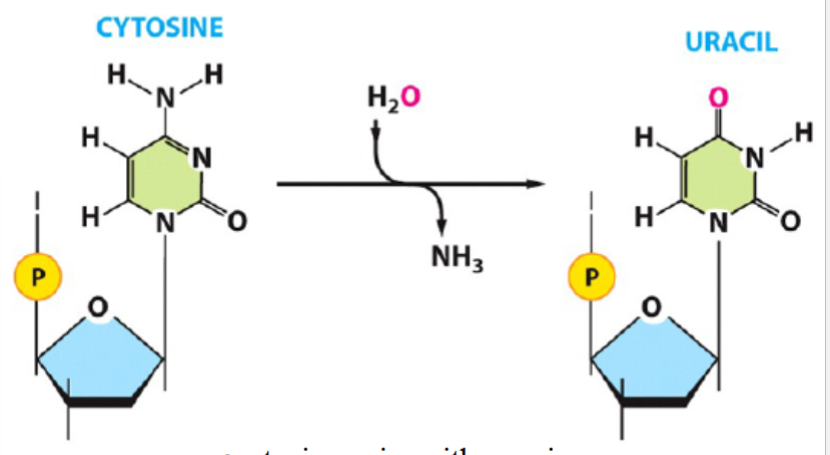
deamination
e.g. cytosine deamination
amino group (NH2) of a base (e.g. cytosine) is removed and replaced by a carbonyl (cytosine) (with addition of water, to yield NH3)
uracil then pairs with adenine (instead of normal cytosine-guanine pair)
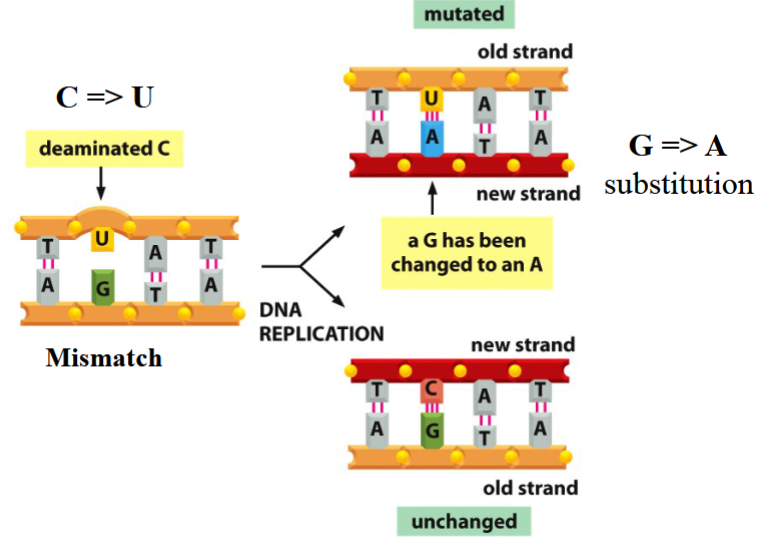
consequences of deamination
a new strand is not like the template strand
e.g. C→ substitution yields to a G→A substituted new strand
DNA should not have U in it
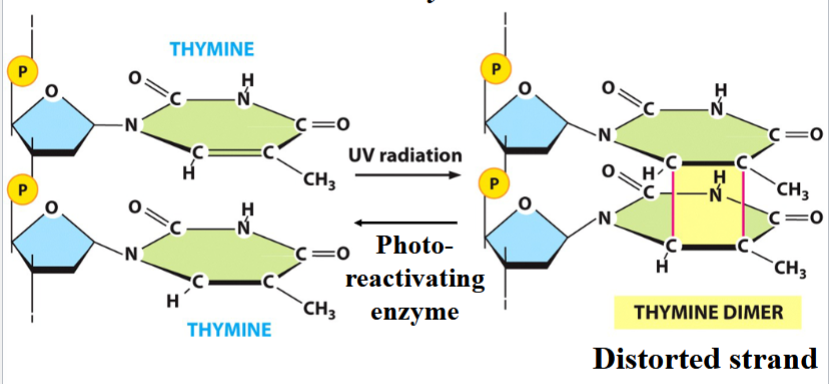
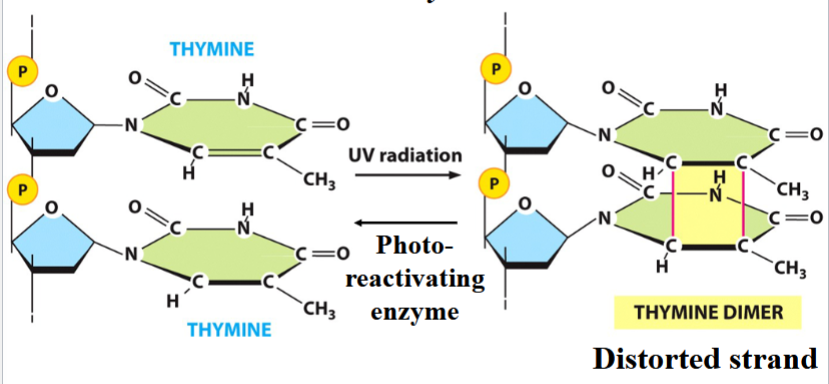
radiation-induced mutation
e.g. UV-induced thymine dimer
a cyclobutane ring forms between adjacent T bases (distorted strand)
the joined Ts cannot pair with As
can be reversed by photo-reactivating enzyme
the basis for UV decontamination
2 categories of repair mechanisms for spontaneous mutation
direct reversal of DNA damage
base excision repair
direct reversal/repair
a category of repair mechanisms for spontaneous mutations
an altered molecule is fixed by reversing the chemical transformation
requires specific enzymes for each individual lesion
e.g. thymine dimers can be reacted, using a specific photoreactivating enzyme
(in picture, work backward)
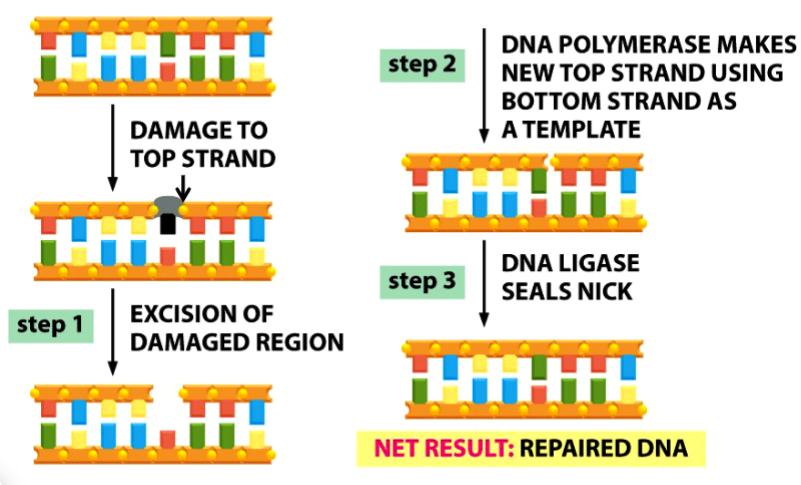
base excision repair
a general mechanism for repairing nucleotide mismatches caused by spontaneous mutations
damaged base(s) are replaced
unpaired base is recognized and replaced before DNA replication occurs
chemical transformation is NOT reversed
has 3 steps
involves 5 enzymes
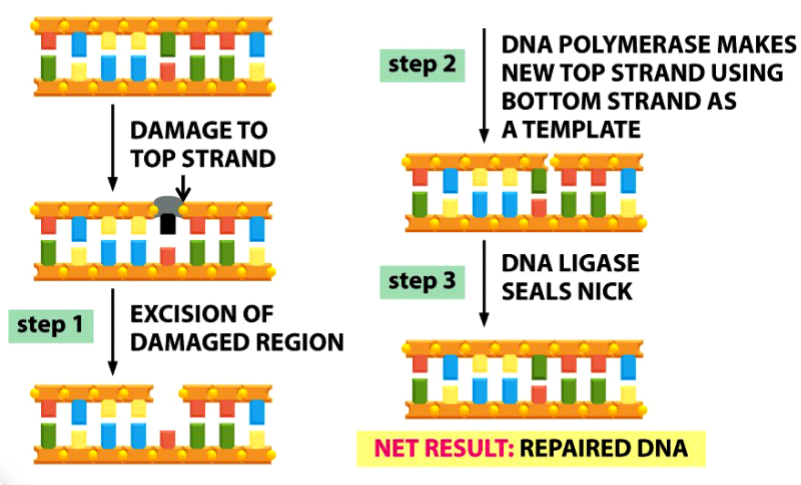
steps to base excision repair
1) DNA glycosylase removes the damaged base (black in picture) → leaving only sugar and phosphate backbone
endonuclease recognizes site and cleaves phosphodiester bond (arrow in picture)
deoxyribosephosphodiesterase removes the remaining sugar (gray in picture) and phosphate (yellow dot in picture)
2) DNA polymerase places a new nucleotide
3) DNA ligase seals the nick
DNA rearrangements
recombination events that alter arrangement of genes within chromosomes
can be site-specific
site-specific recombination
a type of DNA rearrangement that occurs between specific DNA sequences that share partial sequence homology (similarity)
specific proteins recognize the homologous sequences and mediate somatic recombination
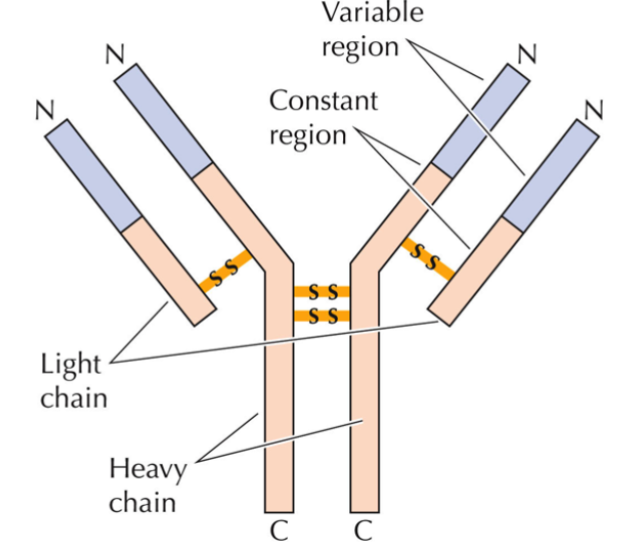
immunoglobin structure (antibody structure)
production occurs in B lymphocytes as they differentiate and mature
2 heavy chains, 2 light chains; joined by disulfide bonds → produce Y shape
all chains consist of variable N region and constant C region
rearrangement of genes by site-specific recombination and then specific splicing of primary RNA transcript is what gives them diversity
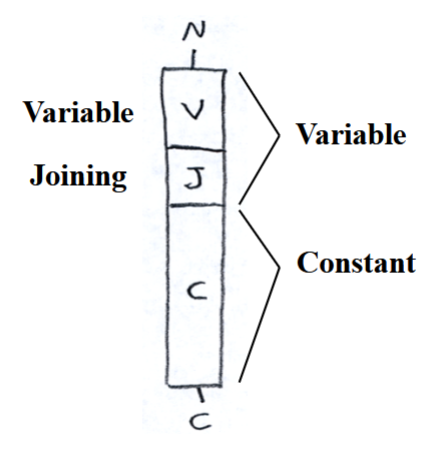
structure of immunoglobin light-chain genes
variable (V) region, joining (J) region, constant (C) region
(going from N terminus to C terminus)
250 V regions that code for the N-terminal end of the variable region
4 J regions that code for the C-terminal end of the variable region (not the C-terminal end of the entire chain)
single C region that codes for the constant region
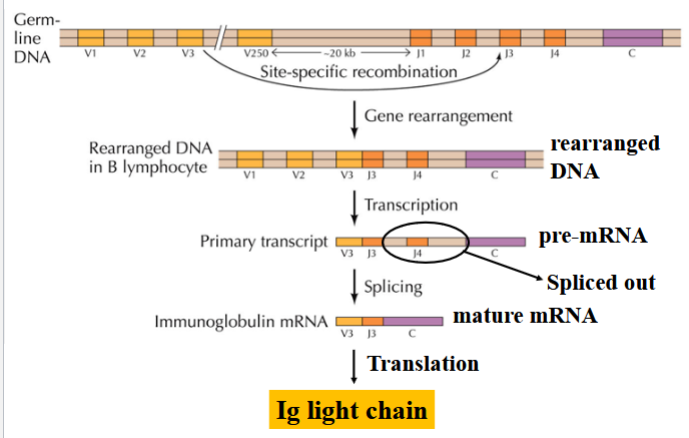
rearrangement of immunoglobin light-chain genes
an example of site-specific recombination
in maturing B lymphocytes, site-specific recombination joins one of the V regions with one of the J regions
transcription occurs
all other J regions are removed by splicing to generate the mature mRNA
(depending on which J region is kept and which ones are spliced, a unique light chain is yielded → diverse immunoglobin)
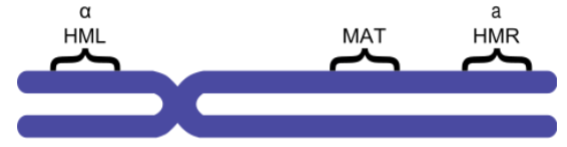
site-specific recombination on the yeast MAT locus
2 mating types in Saccharomyces cerevisiae: MAT alpha and a
both MATalpha and MATa are alleles of the MAT locus on chromosome 3 (controls mating type)
HML and HMR (hidden MAT left/right) contain silent copies of MATalpha and MATa
mating type can be switched by replacing the allele on the MAT locus with one of the copies in HML or HMR by site-specific recombination
Barbara McClintock
a pioneer in cytogenetics
created a model for transposition in 1950, on pigmentation in maize kernels

Barbara McClintock’s transposition model (1950)
expression of a gene that is necessary for Anthocyanin (a pigment) formation can be suppressed by a dissociator
if dissociator is transpositioned, gene is released → anthocyanin can be made
dissociator transposition is controlled by the activator (also can be replicated and transpositioned)
more copies of activator = more pigment expression and color → results in mosaic color patterns in kernels
(spots on the kernel with more color have more activator copies in that region)
(dissociator and activator are both called transposable control elements)
transposable element
a chromosome segment that can move
if a control element, its transposition can affect gene expression
e.g. dissociator and activator in Barbara McClintock’s model
insertion of simple bacterial transposons
nonreplicative transposition or replicative transposition
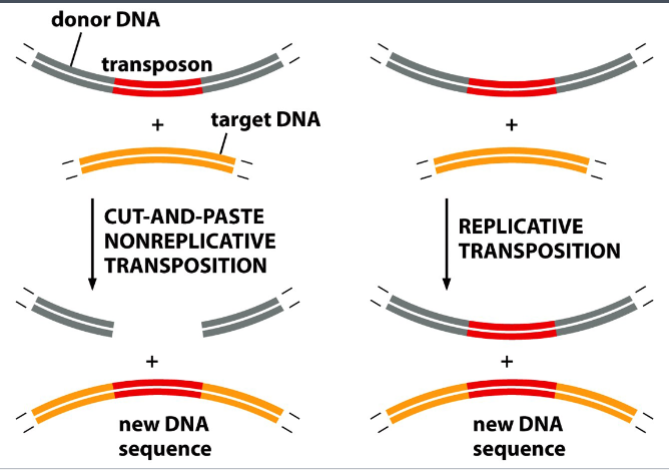
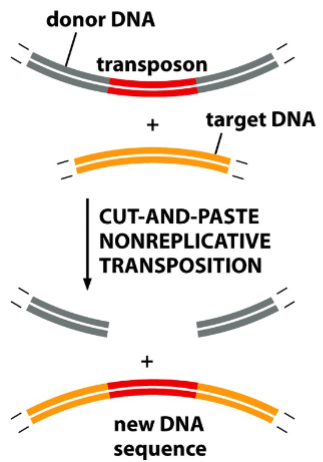
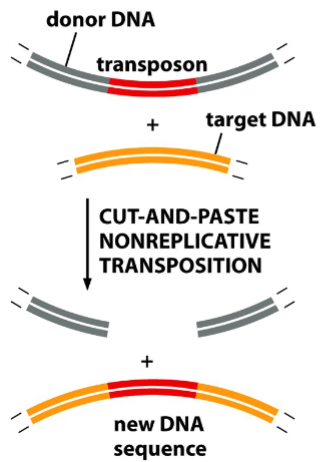
nonreplicative transposition
“cut and paste”
a method of insertion of simple bacterial transposons
transposase (an enzyme) cleaves an insertion sequence specifically at the ends of inverted repeats, and makes cuts on the target site
(note: the target sequence is not specific)
the insertion sequence is cut from the donor site and pasted into the target site
result: the insertion sequence moves from one site of the genome to another
replicative transposition
“copy and paste”
a method of insertion of simple bacterial transposons
a copy of the insertion sequence is made by local DNA replication
that copy of the insertion sequence is pasted into the target site, without being removed from the original DNA
result: a new copy of the insertion sequence appears at the target site
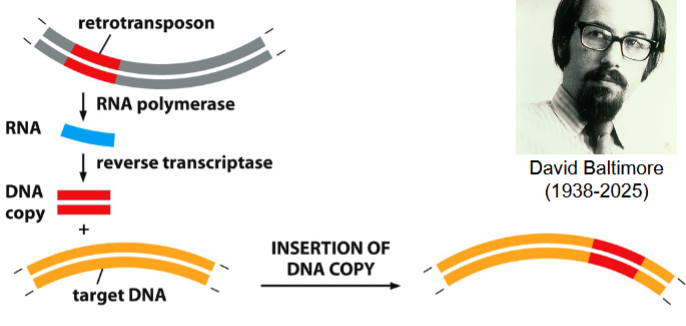
retrotransposition
a form of transposition that requires the synthesis of a copy of a retrotransposon
catalyzed by reverse trancsriptase (RT)
discovered by David Baltimore
can generate multiple copies of retrotransposons in the genome
many interspersed repeats in today’s eukaryotic genomes originated this way
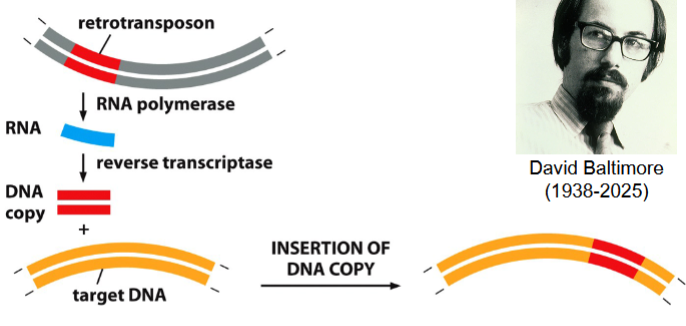
the process of retrotransposition
retrotransposon is transcribed to an RNA molecule (catalyzed by RNA polymerase)
complementary DNA strand is synthesized (catalyzed by reverse transcriptase (RT)) → result: DNA-RNA double stranded hybrid
RNA strand is destroyed, replaced with a new strand of DNA (catalyzed by RT) → result: double-stranded copy of the retrotransposon
double-stranded copy of retrotransposon can now be inserted somewhere else in the genome