Cell Biology Exam 1 Review (Chps 1-4)
1/86
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
87 Terms
Define cytoskeleton and describe its functions
Network of different protein fibers that provide many functions
Functions:
Adopt different shapes
Organize organelles in specific positions
Interact with the environment
Carry out directed movement (cytoplasm and vesicle within cell)
Replicate
Support volume of cytoplasm
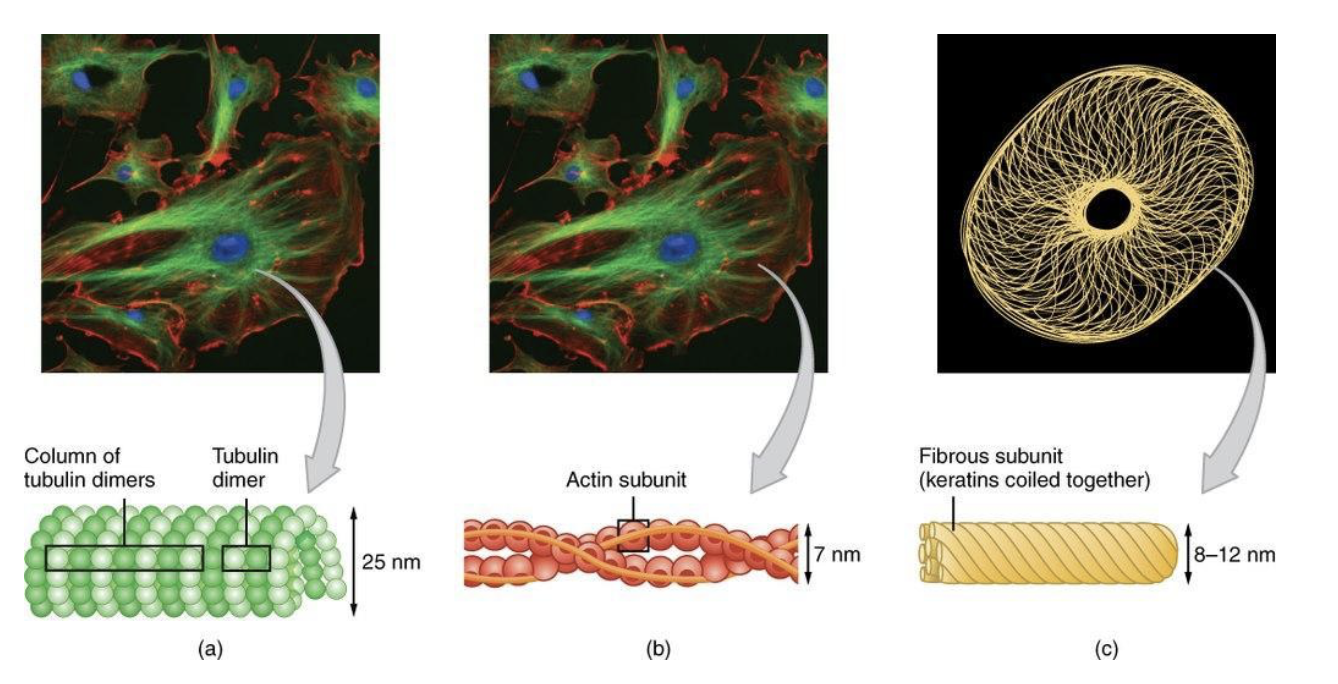
What are the three major types of protein filaments?
Microtubule:
Hollow cylinders of protein tubulin
Largest
Intermediate filament:
Rope-like fibers of intermediate filament proteins
Actin/microfilament:
Helical polymers of protein actin
Smallest
Describe the characteristics of Intermediate filaments
Forms mesh like network in cells; provide tensile strength
Type of cytoskeleton that makes up nuclear envelope
Monomers twist together to form “rope-like” polymer
Ex.) keratin and lamins
Do intermediate filaments have a role in cell movement?
No, main function is structural where tensile strength enables cells to withstand mechanical stress
What role do intermediate filaments play in epithelial cells?
Skin cells have high concentration of keratin
Intermediate filaments from one cell interact w/ others from adjacent cells via desmosomes
Desmosomes:
cell-cell junction that joins neighboring cells together
How do intermediate filaments protect cells from mechanical stress?
Intermediate filaments prevent the rupture of cells, due to desmosomes, allowing the cell to remain intact and together
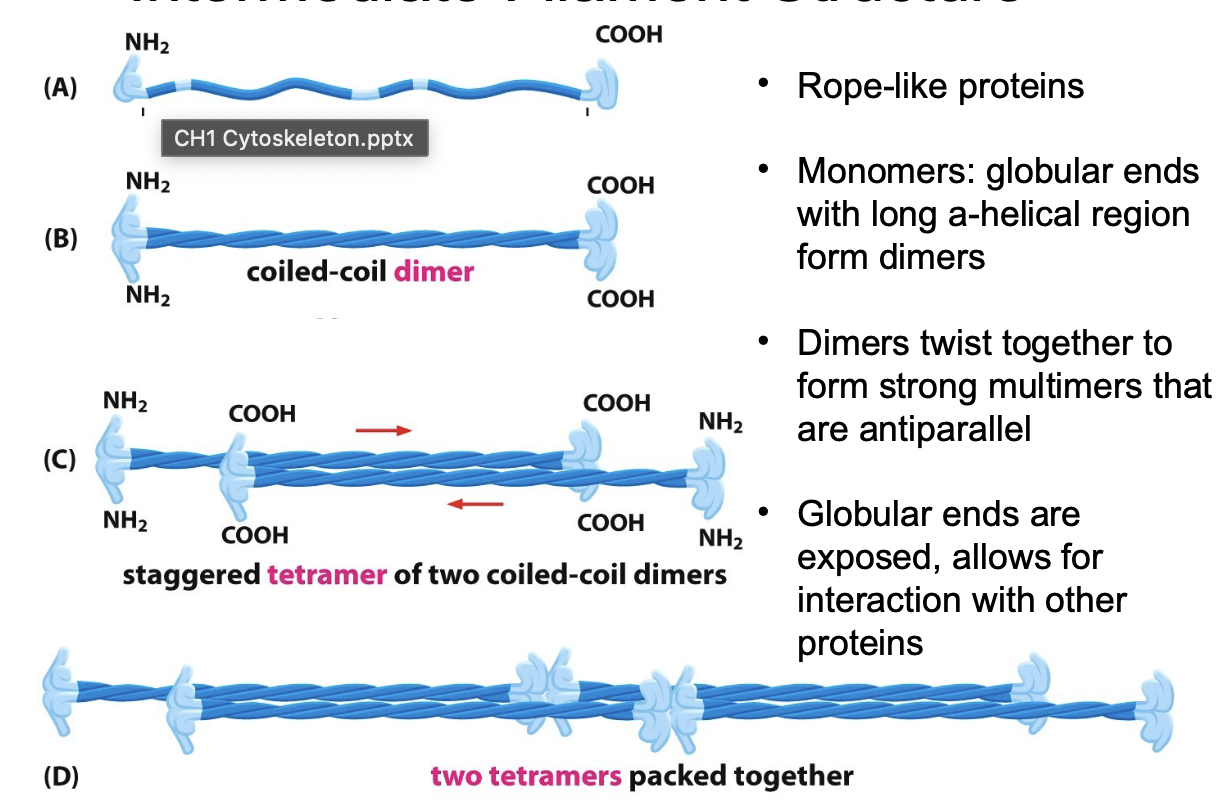
Describe the intermediate filament structure
Rope-like proteins
Monomers: globular ends w/ long a-helical region form dimers
Dimers twist together to form strong multimers that are antiparallel
Rope-like filament made of tetramer that are packed into a helical array of 8 tetramer strands
Globular ends exposed to allow interaction w/ proteins
What can mutations in keratin genes lead to and why?
Epidermolysis bullosa simplex (EBS):
rare genetic disorder caused by defects in gene coding for keratin proteins
Causes extremely sensitive skin, blisters, and skin to breakdown easily
Why?
mutation leads to loss of function in keratins causing the inability of cell to maintain structure under pressure
How would intermediate filaments be categorized?
Cytoplasmic
Keratins (in epithelia):
Diverse every kind of epithelia in body
Neurofilaments:
in nerve cells
Nuclear
Nuclear lamins:
in all animal cells
strengthen the nuclear envelope
Describe Lamins
Type of intermediate filament that makes up structural elements of nuclear lamina
Essential in maintaining its structure
Plays an important role in cellular functions, mitosis, where nuclear envelope breaks down during prophase and reforms in telophase
What are results of defects in nuclear lamin proteins?
Progeria:
premature aging causing irregular-shaped nuclear envelopes
potentially associated with defects in mitosis, causing unstable cell division
What are characteristics of microtubules?
Associated with motor proteins
Thickest type of cytoskeleton
Made of tubulin monomers
Pulls chromosomes apart during anaphase
Involved in vesicle trafficking/ movement
Describe microtubules and their cellular functions
Hollow, tube-like filaments
polymer of tubulin
extend from microtubule organizing centers (MTOC), like centrosomes, spindle poles, and basal bodies
Cellular functions:
Spindle formation
Cilia/flagella movement
Intracellular transport
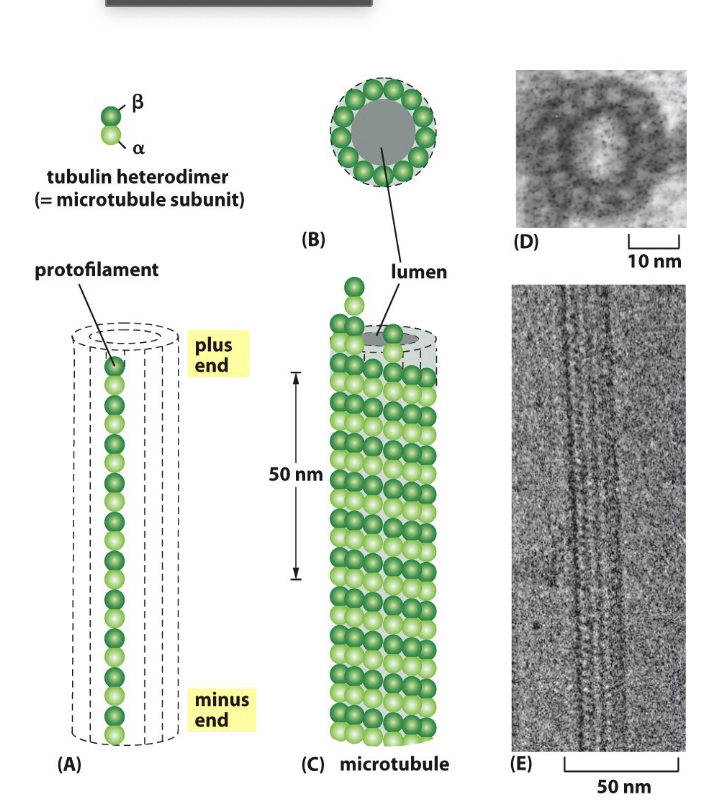
Describe the structure of microtubules
aB dimers arranged/stacked together in the pro filament oriented in the same direction
13 parallel pro filament has a structural polarity; one need B-tubulin (plus end) and a-tubulin ( minus end)
Pro filaments make up “hollow tube”
Dimers add to plus end faster than minus end
Primary structural elements: flagella, cilia, centrioles
Define dynamic instability
Microtubules rapidly extend (polymerization) and shrink (depolymerization)
Grows outward from an organizing center by the addition of aB tubulin dimmers to end
Crucial for rapid remodeling
GTP hydrolysis controls dynamic instability and dimers can hydrolyze GTP
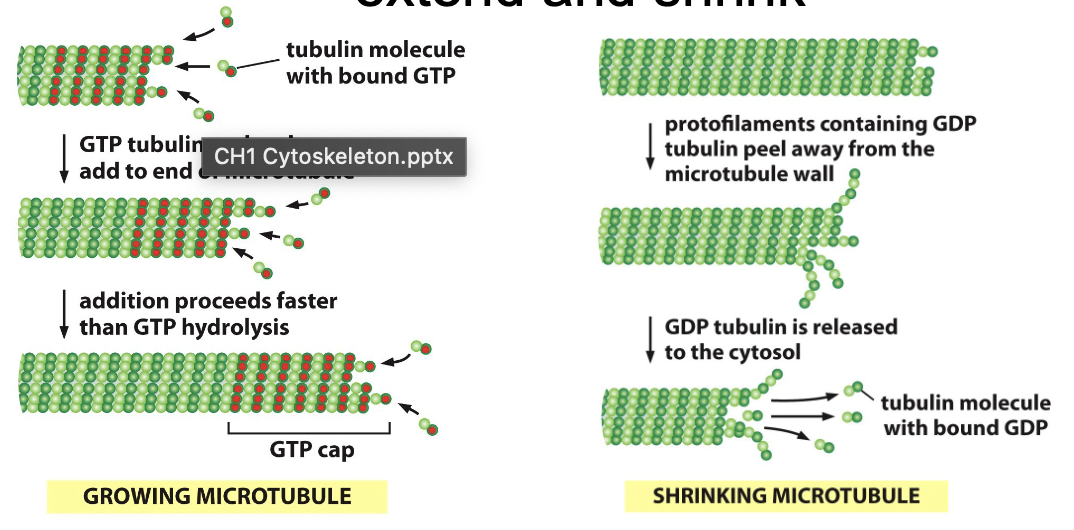
What is one example of dynamic instability?
movement of chromosomes to opposite poles of a dividing cell during mitosis called spindle fibers
Describe the guide transport of microtubules
Many cells are polarized (microtubules extend in one direction)
Example: neurons
Outward transport:
vesicles with membrane or secretory proteins
Inward transport:
Damaged proteins and organelles
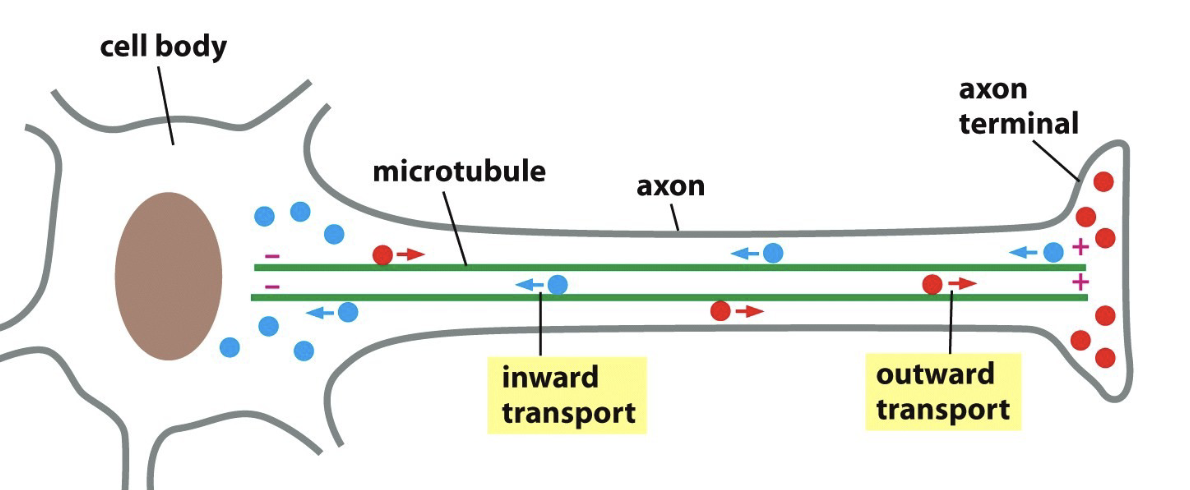
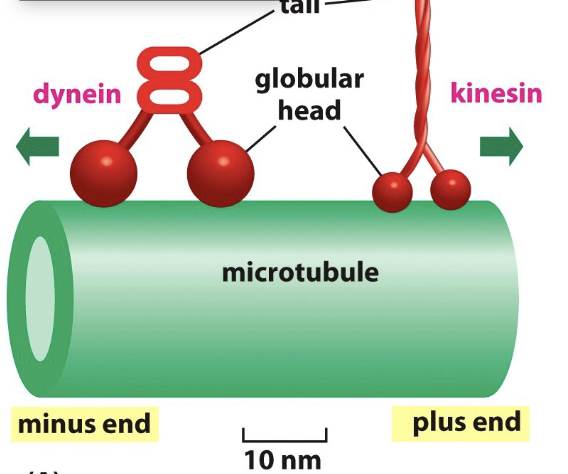
Define motor proteins and the two types in microtubules
Motor proteins:
move along the tracks in a specific direction
Types:
Kinesin:
walk along microtubules and involved in vesicle transport
plus-end directed ( outward transport- away from cell body)
movement from “-” to “+” end of microtubule using energy fro. hydrolysis of ATP
Dyenin:
minus end directed ( inward transport)
movement from “+” to “-” end of microtubule filament towards cell center
It converts chemical energy from ATP hydrolysis into mechanical energy of movement to walk along microtubule while carrying a vesicle
What are the characteristics of Actin/ microfilaments?
The thinnest type of cytoskeleton
Involved in cell crawling
Associated with motor proteins
made of actin monomers
Involved in vesicle trafficking/ movement
Describe Actin/ microfilaments and examples
One of the most abundant proteins in eukaryotic cells
5% of total cellular protein
Polymers twist into helix
Actin monomers can add to plus or minus end, but add to plus end faster
Actin:
ATP →tightly bind to filament
ADP → not as tightly bound
Dynamic instability:
rapid extension and shrinking of filaments
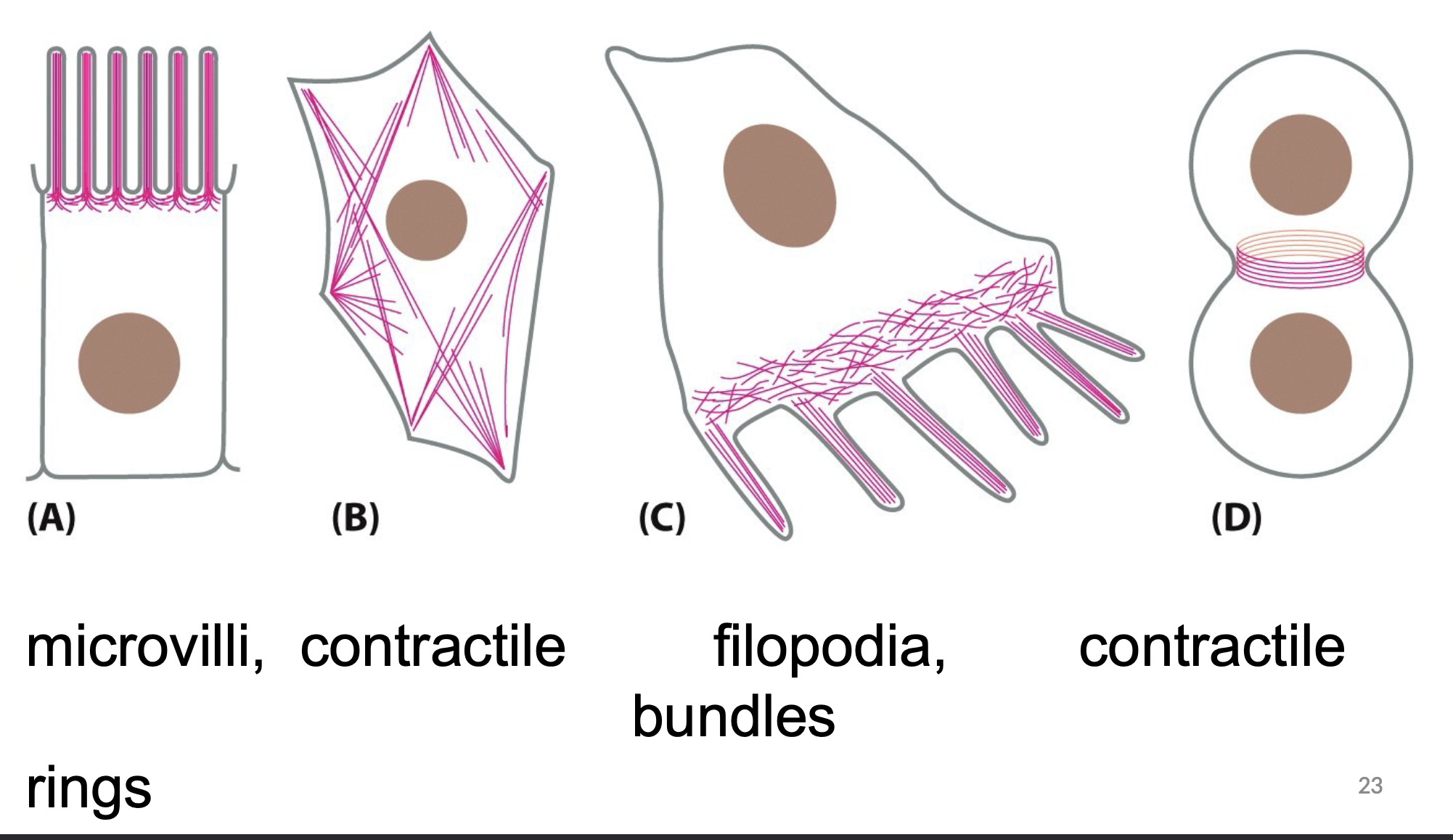
Describe the type of motor protein in actin/ microfilaments
Myosin:
Found in most cells and used in vesicle transport
used to move actin filaments relative to plasma membrane (cell shape change)
ATP dependent
Plus-end directed motor protein (- to + end)
movement uses energy of ATP hydrolysis to provide energy for changes needed for movement
Do all cells have the same DNA, and how do we get different cells?
Yes, most cells have the same DNA, but they become different through gene expression, where cells turn certain genes on or off
List the 4 nucleotides and how they interact with each other in the double helix
Adenine, Thymine, Guanine, Cytosine
A → T ; G → C
Define a gene
Linear sequence of nucleotides along a segment of DNA that provides coded instructions for synthesis of RNA, then translated into protein, leading to the expression of hereditary character
Describe gene expression
The process of turning on a gene to code for mRNA to produce protein
It must be regulated, because not al genes can be turned on or off at the same time
Regulation of gene expression conserves energy
Describe the coding region of DNA
The genetic doe of how DNA codes for an amino acid is nor obvious from a base pair sequence
There are protein coding region soft DNA where the DNA is used to make RNA and then protein
Describe the non-coding region of DNA
~2-5% of the human genome contains genes
Remaining portion= non coding DNA (do not code for protein)
Role: not fully understood, but may be important in regulation of gene expression
Genome: all DNA/ genetic information
Compare prokaryotic and eukaryotic genomes
Prokaryotic:
genomes are circular and contain plasma membrane and chromosome
Eukaryotic:
Linear and organized into multiple chromosomes
Chromatin:
DNA complexed with proteins
Chromosomes:
linear DNA molecules and associated proteins that is folded into a compact structure
Chromatin condenses to form chromosomes
Describe eukaryotic chromosomes
Except germ cells (sperm and eggs) and RBCs, human cognation two copies of each chromosome
Maternal and paternal pairs —> homologous chromosome
only non-homologous pairs are sex chromosomes
Define karyotype
Ordered display of the full set of 46 chromosomes (23 pairs)
Describe the types of chromatin
Euchromatin:
more expanded DNA, less condensed
Heterochromatin:
Highly condensed DNA, more DNA packaging
more likely to see in dividing cells
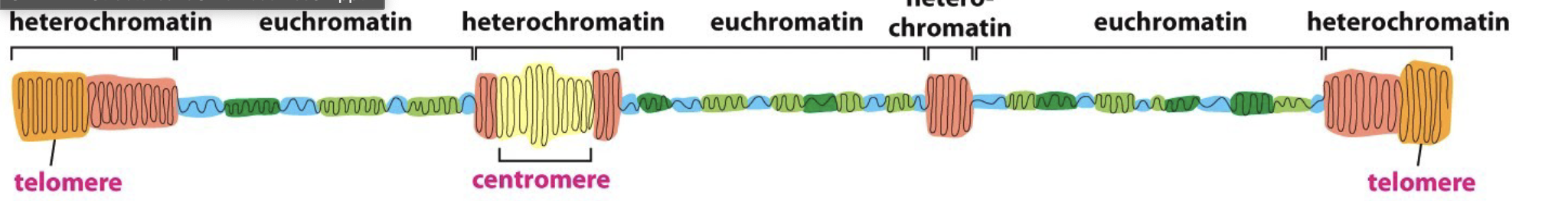
For gene expression differences, do you think that it is all or nothing?
No, generally a mixture of euchromatin and heterochromatin
When does DNA replication occur?
Occurs during interphase
duplicated in preparation for mitosis
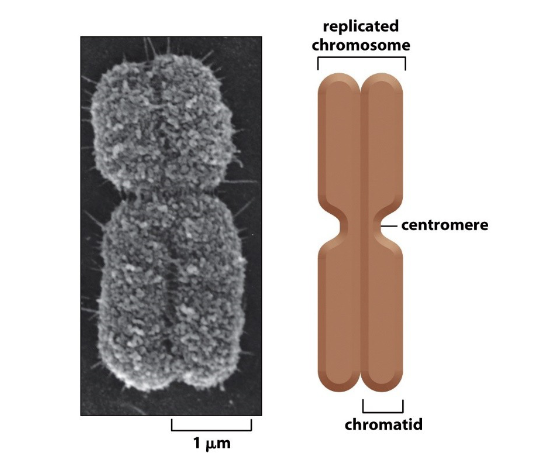
Describe the anatomy of a chromosome
Duplicated mitotic chromosome is highly condensed
Contains two identical daughter DNA molecules
Each one chromatid (sister chromatids)
Centromere:
specific sequence that allows duplicated chromosomes to be separated
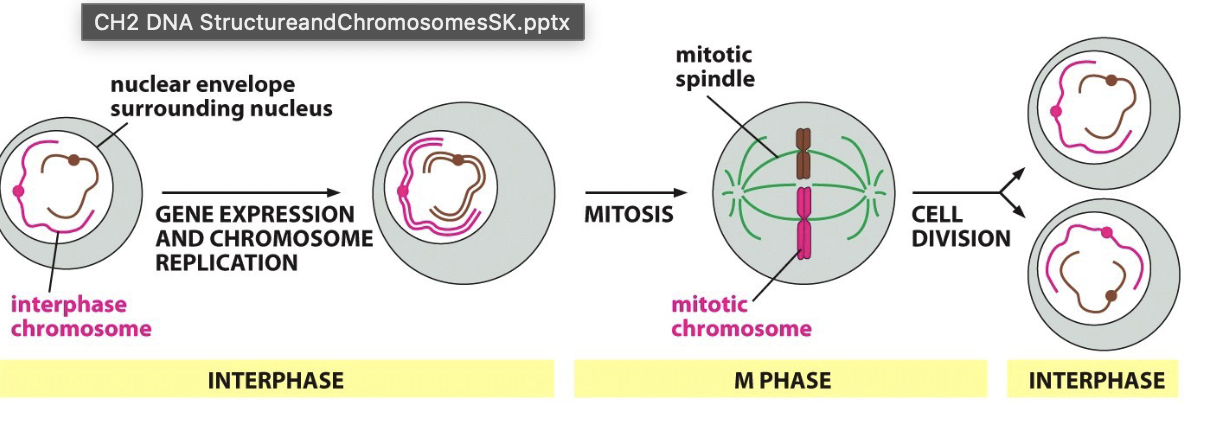
Describe interphase vs m-phase chromosomes
Interphase:
when chromosomes are duplicated
chromatin condenses to prepare for distribution
M-phase:
mitosis leading to distributed 2 daughter cells
Define a nucleosome
Structure when DNA wraps around proteins called histones
Define a histone
Proteins that bind DNA for packaging
Describe the anatomy of a nucleosome
Beads on a string
String = linker DNA
Beads = nucleosome core:
DNA wrapped around proteins called histones
What properties would histones have to bind DNA?
Positively charged histone subunits
Describe the process of nucleosome anatomy
Histones from an octamer (8 monomers) w/ 2 of each histone: H2A, H2B, H3, and H4 all joined together in a cylindrical structure with a positively charged amino acid to interact w/ DNA and form a nucleosome
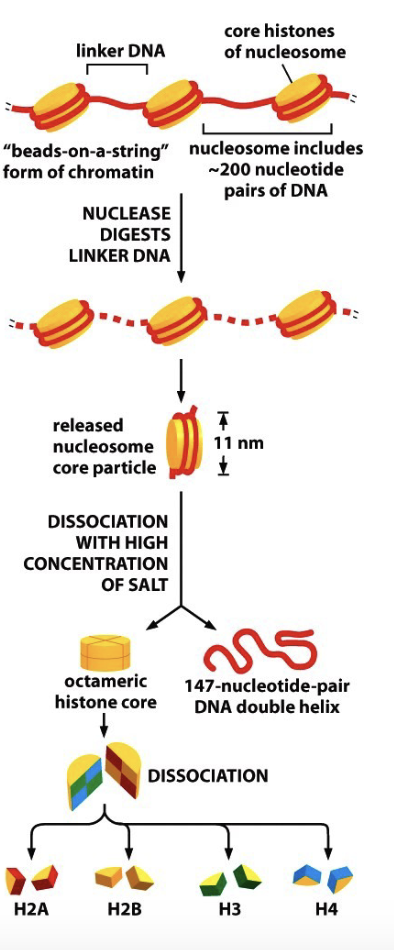
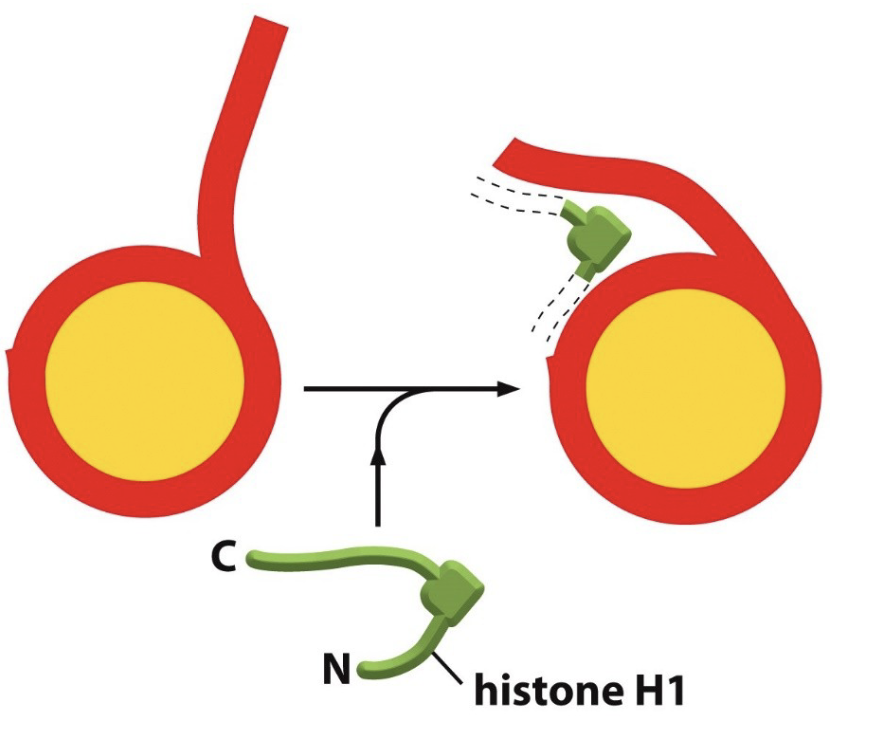
What is the importance of H1?
Histone H1 binds to the linker DNA and bends it, so that those nucleosomes can twist into a helix or spiral
has a long C-terminal that helps bind to chromatin
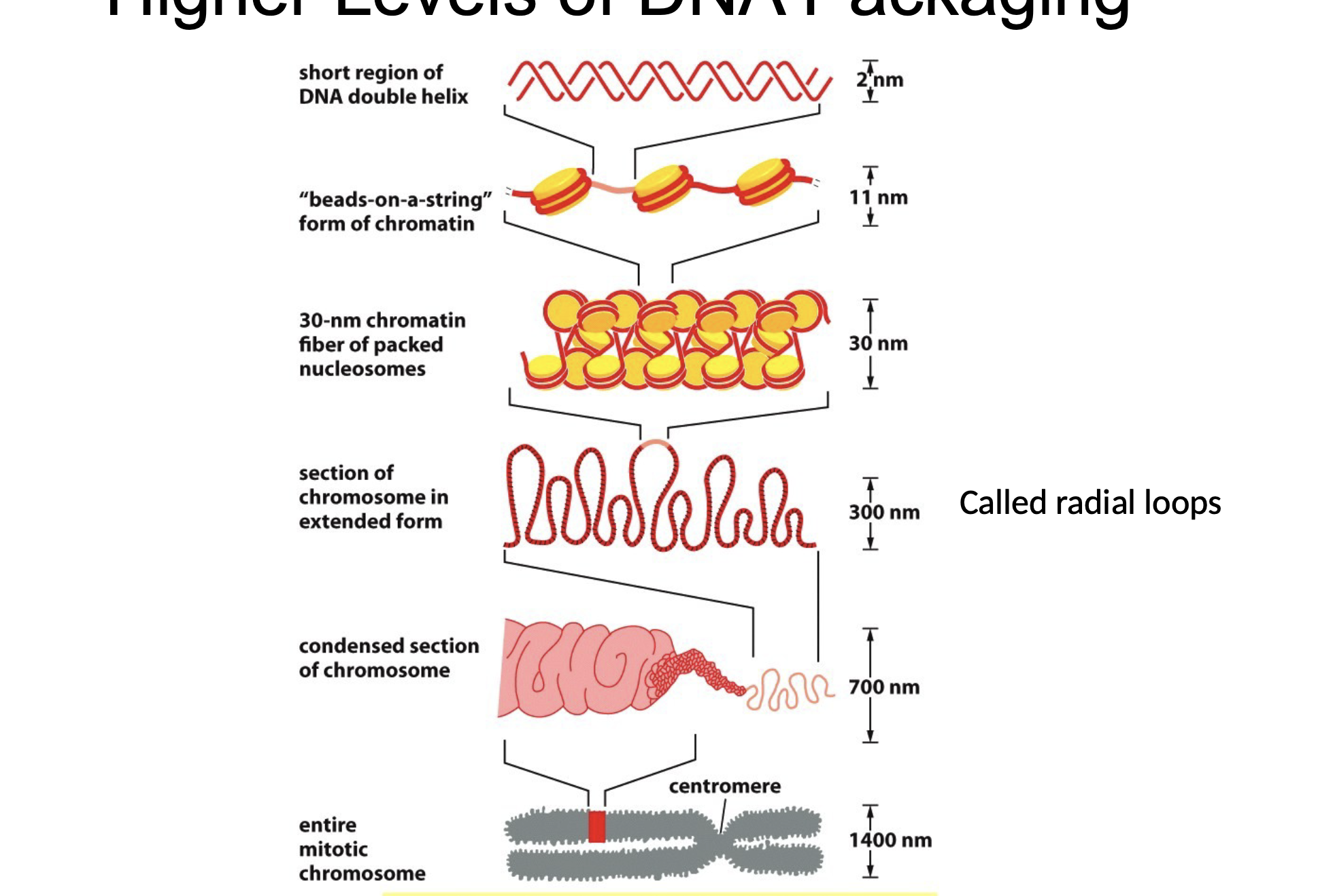
Describe DNA packaging from least to most condensed
Naked DNA/DNA double helix (least)
Beads on string (nucleosome)
30-nm filament
Radical loops
Condensed section of chromosomes
Mitotic chromosome (most)
Why would we need to access tightly packaged DNA?
DNA replication
DNA repair
Gene expression ( transcription and translation)
What is the role of the H1 linker protein?
Pulls nucleosomes together and pack them tightly to help bind the linker DNA, which aids in chromatin condensation
Describe the role of histones H2A, H2B, H3, and H4
H2A and H2B provide structural, disk shaped core that is the fundamental unit of chromatin
H3 and H4 are essential for initial tetramer formation
Describe whether euchromatin or heterochromatin can result in increased gene expression and why?
Euchromatin can result in increased gene expression b/c its less condensed and loosely coiled, meaning more accessible for transcription
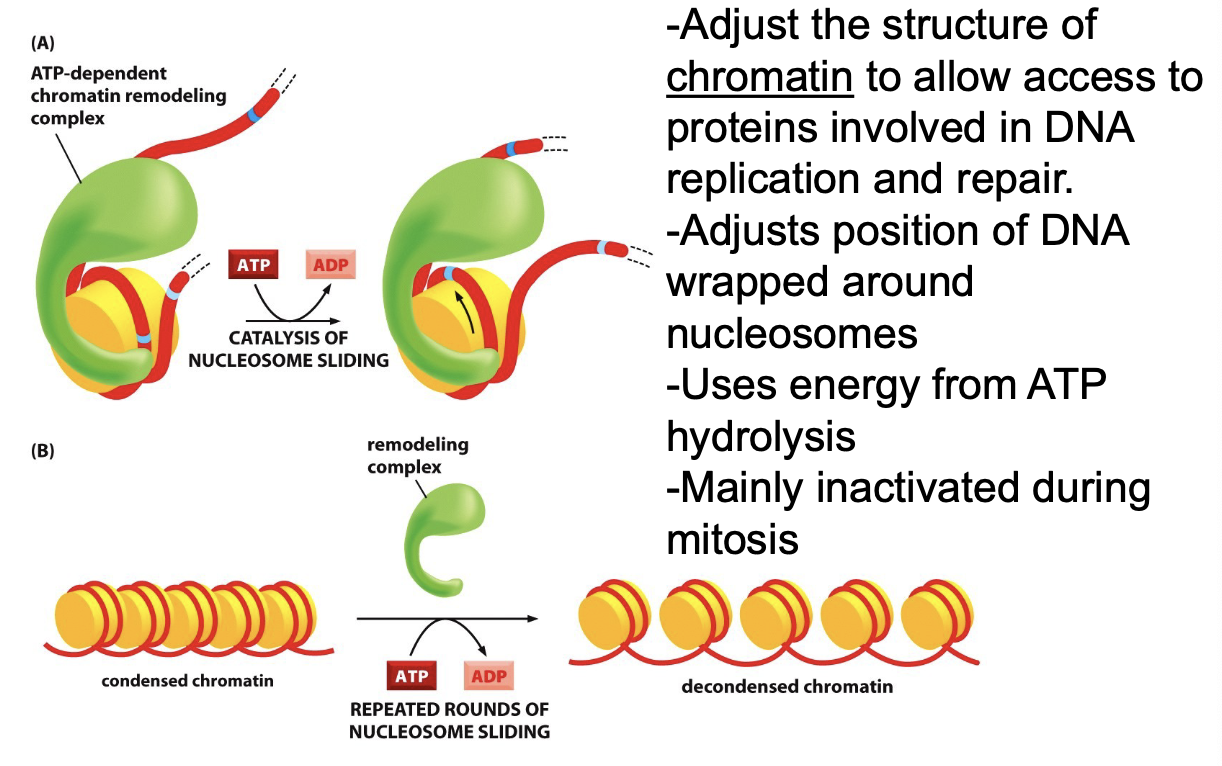
Describe ATP-Dependent Chromatin Remodeling:
Regulates DNA condensation
Adjust the structure of chromatin to allow access to proteins involved in DNA replication and repair
Adjusts position of DNA wrapped around nucleosomes
Uses energy from ATP hydrolysis and inactivated during mitosis
What are the effects of post transitional modifications to histones?
Addition of these chemical groups to stones cause changes in gene expression
Alter chromatin structure based on chemical modification of histones (could reduce strength of interactions for histones)
Can serve as binding or docking sites for proteins that alter condensation of DNA
Describe the Central Dogma of Biology
DNA is transcribed to RNA, then translated to proteins
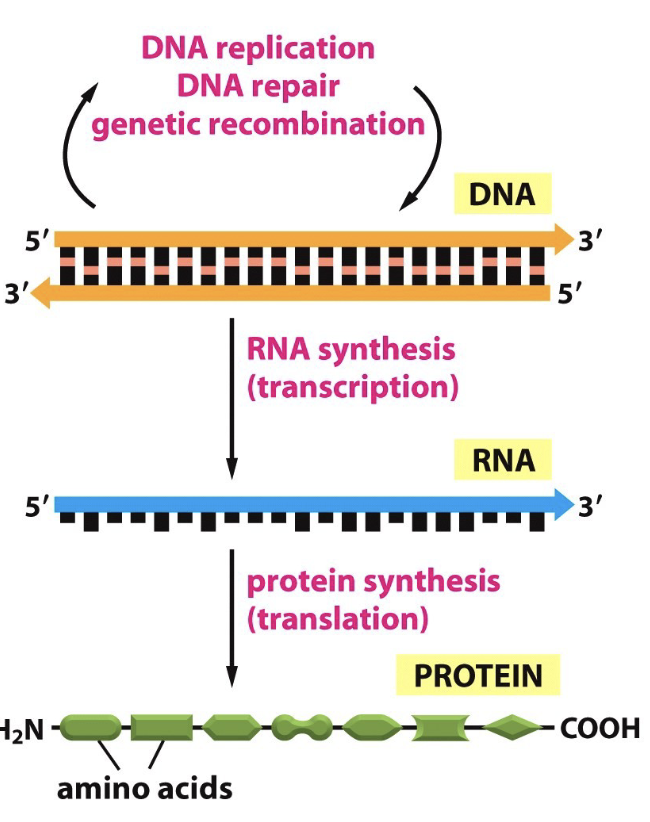
Does this process of Central Dogma ever of backwards?
No, but retroviruses like HIV are an exception b/c it has a reverse transcriptase enzyme that allows this reverse
Describe Transcription
One strand of DNA serves template (3’ to 5’)
Coding strand: matches RNA
RNA added in 5’ to 3’ direction
Describe Huntington’s Disease (HD):
Fatal genetic dominant disorder that causes progressive breakdown of nerve cells in brain cell causing loss of motor and memory
Known as the quintessential disease where every child of a parent w/ HD has 50/50 chance of carrying gene
Happens when patients have a mutation in the Huntington gene, resulting in production of rom of Huntington protein that attacks neuron
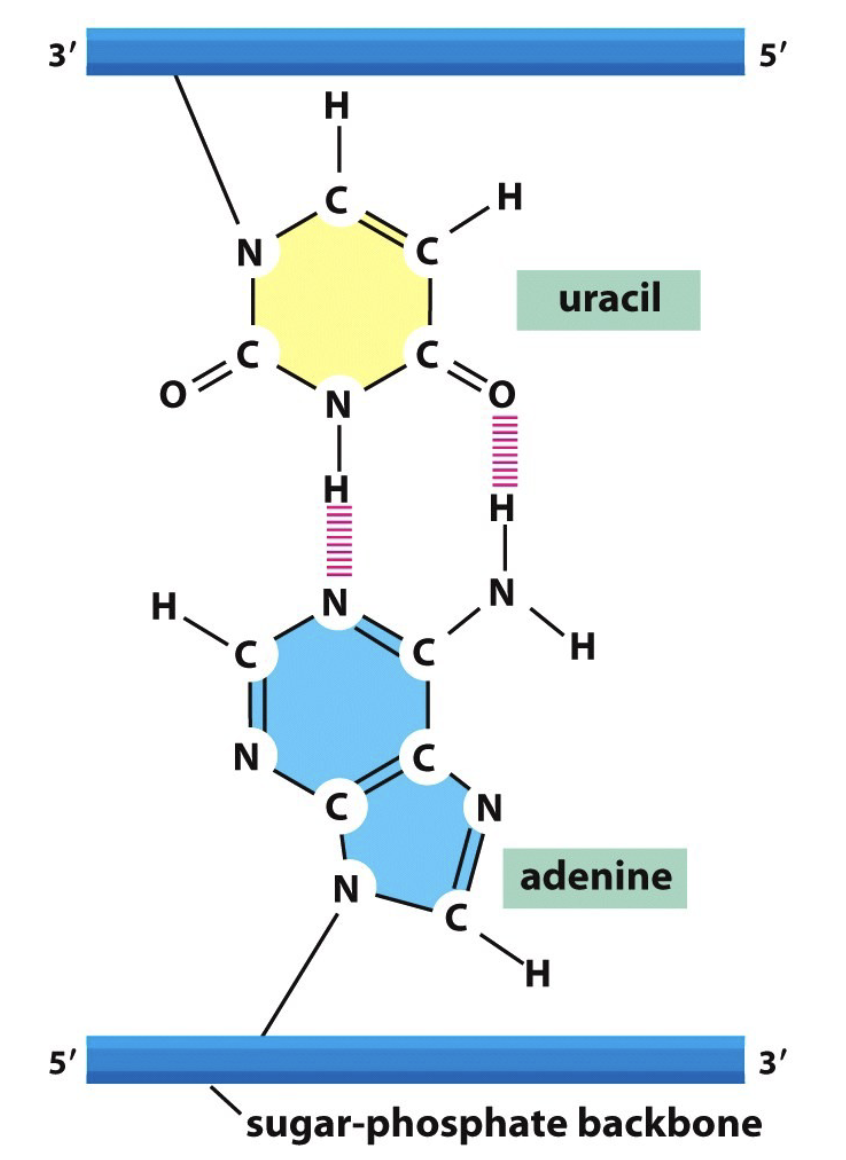
Compare RNA and DNA
RNA:
single stranded
ribose nucleic acid w/ 2 OHs attached
uses Uracil instead of Thymine
RNA nucleotides:
purines → A and G
pyrimidines → C and U
DNA:
Double stranded
deoxyribose nucleic acid with 1 OH
uses Thymine
Describe the roles of the types of RNA
mRNA:
code for proteins
rRNA:
form the core of the ribosome and catalyze protein synthesis
miRNA:
regulat gene expression
tRNA:
serve as adaptors between mRNA and amino acids during protein synthesis
What can all cells do and name an example
can express different genes at different rates
The same cell can make a lot of gene A and at the same time make a smaller amount of gene B
Ex.)
Skin cells need some DNA polymerase, but not all the time
Insulin secretion in the pancreas
What is needed for Transcription?
Template:
region of DNA to be transcribed into RNA
Monomers for new RNA strand:
ribonucleotides (ATP, UTP, CTP, GTP)
Enzymes to polymerize monomers:
RNA polymerase
Key differences to DNA replication:
only one strand of RNA made and no primers needed
Why is the term “pre-mRNA “ used when genes are transcribed here?
Because splicing addition of 5’ caping, and adding of poly- A - tail needed for mature RNA
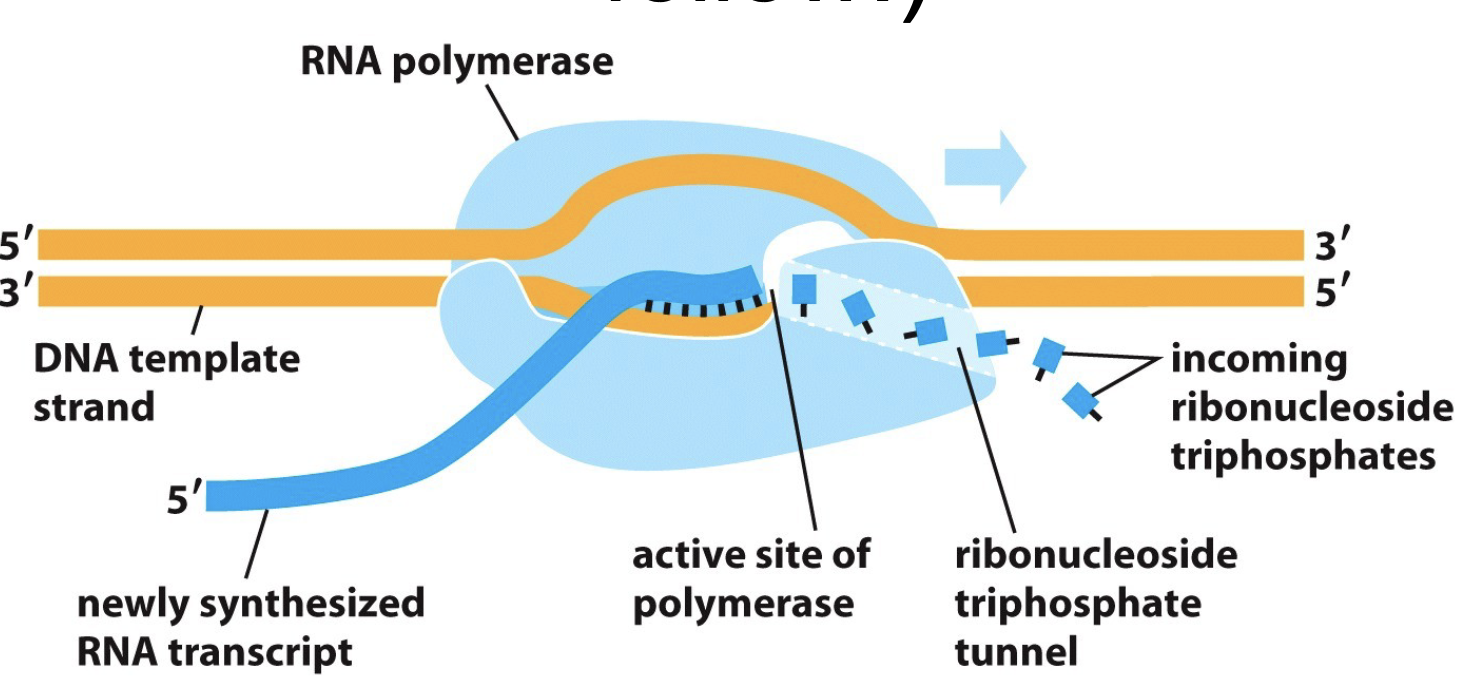
What are the basics of Transcription?
RNA polymerase moves along DNA by unwinding the DNA helix in front
Add ribonucleotides one by one to RNA chain and uses DNA chain as template
Resulting RNA transcript is complementary to template
Polymerase moves in 3’ to 5’ direction and displaces newly formed RNA strand along DNA template
Describe the type of RNA polymerase in Eukaryotic cells
RNA polymerase I:
makes most rRNA genes
RNA polymerase II:
makes mRNA, miRNA, and small RNBAs in spliceosomes
RNA polymerase III:
makes tRNA, Ss rRNA, and other small RNAs
Describe the starting and stopping transcription process
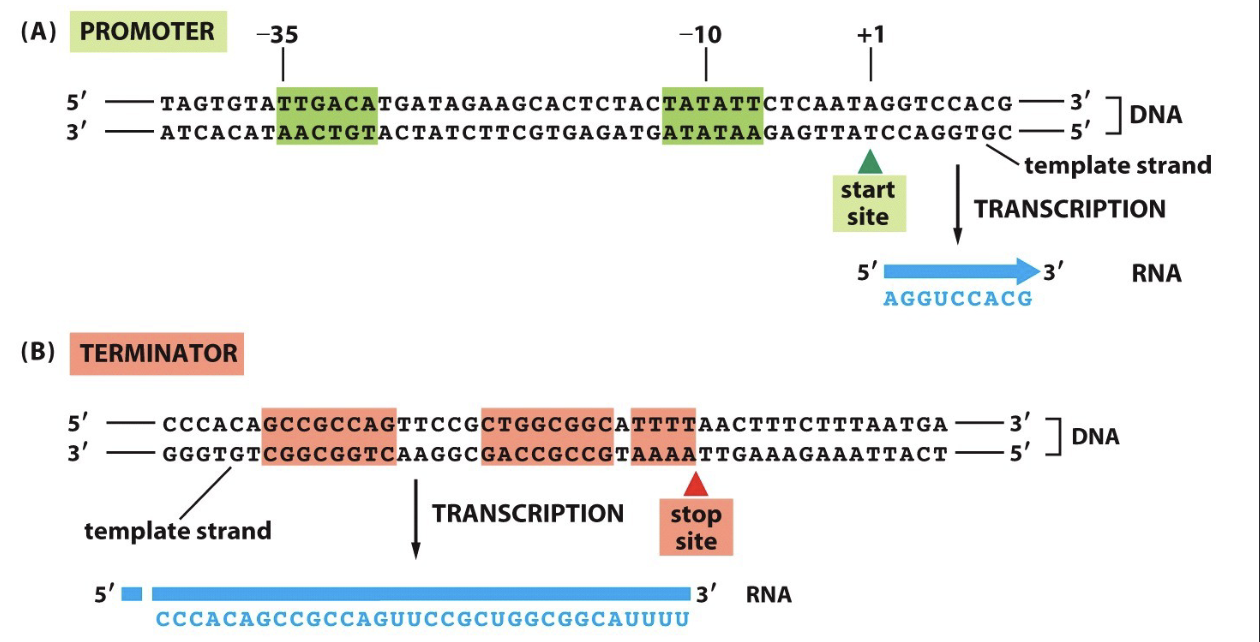
Promoter:
region of DNA that signals the start of RNA synthesis
Terminator:
region of DNA that signals the end of RNA synthesis
Process:
RNA polymerase encounters DNA nd slides down the double helix and latches on tightly to promoter
Goes on to open up double helix to allow the process to continue until reaching the terminator region
the polymerase halts and releases the DNA template band the new RNA transcript
Transcription moves in 5’ to 3’ direction of the new mRNA transcript (left to right)
What are the parts of Transcription?
Template strand:
DNA strand being transcribed
Promoter:
TATAA Box in eukaryotic cells. Transcription factors and RNA pol II assemble at promoter region on non-template strand
+1 or start site:
actual starting point of transcription located on template strand
Upstream:
sequence before start site
Downstream:
sequence after start site
Coding strand:
contains promoter region (TATAA Box) orientation of promoter determines which direction that gene is transcribed
Define Transcription factors and describe the factors necessary to promote initiation process of Transcription of agent in eukaryotes
Transcription factors:
proteins that bind to DNA to facilitate transcription
these factors assemble at promoter to initiate transcription
Process:
TATA binding protein (TBP) recognizes and binds the core promoter (TATA box) and bends DNA
TFIIA and TFIIB join and TFIIB determines start site
TFFIIF bonded to RNA pol II brings enzyme to promoter
TFIIE and TFIIH bind
TFIIH acts as helicase to allow polymerase to start transcribing and phosphorylates RNA pol II tail, which releases general transcription factors (except TFIID)
Now RNA pol II adds ribonucleotides (UTP, ATP, CTP, GTP) to growing RNA strand
RNA pol II is released to undergo elongation process, where it adds ribonucleotides to the growing RNA strand
Describe the Elongation process in Transcription
RNA pol II adds ribonucleotides to growing RNA strand, making it longer
Describe the Termination process in transcription
Termination sequence exists that releases mRNA
RNA pol II has no specific signals that terminate transcription
A protein complex will bind to two locations on the growing pre-mRNA once RNA polymerase transcribed
Complex binds common AAUAAA sequence and a UG sequence
Protein in complex (CPSF) will cleave pre-mRNA at site between two bound sequences
This releases the pre mRNA
Poly (A) polymerase is a part of the same complex and will begin to add a poly-A tail to pre-mRNA (mRNA processing)
Explain the purpose of the three important modifications to eukaryotic pre- mRNA
1) 5’capping:
essential for ribosome recognition and translation
2) Addition of poly A tail:
plays a role in nuclear export of the mRNA and potential role in the protection from nucleases
3) Splicing:
Remove introns (noncoding regions) and link exons (coding sequences) to create mature mRNA
Define Spliceosome
a large RNA/protein complex
consists of snRNPs (small nuclear ribonucleoprotein particles)
snRNPs recognize those specific sequences and catalyze the covalent linkage of exon sequences
A -branch point:
where the beginning of intron is attached to form lariat structure
Describe Alternative Splicing
1 gene DOES NOT equal 1 protein
can produce various mRNAs from pre-RNAs to produce various mRNAs and proteins
Alternate splicing → rearrangement of protein domains (combination of new exons)→ new “patchwork” proteins
Define Untranslated Region (UTR)
when not all mRNA transcript codes for proteins
What is a protein domain?
a segment of a gene codes for a section of a protein that does a specific function/ task
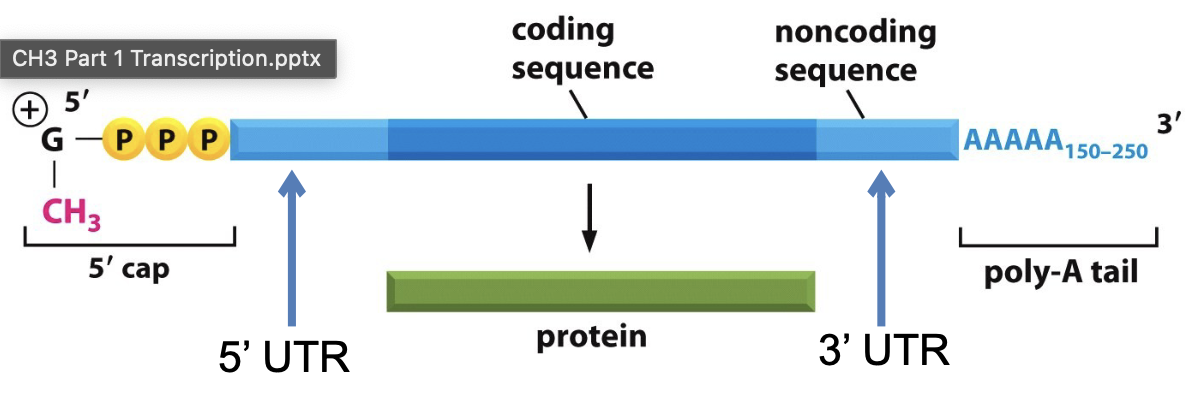
What are the components of a fully processed mRNA transcript ready for export?
5’ cap
5’ UTR
Coding sequence
3’UTR
poly-A tail
What are recognized by Nuclear Export proteins?
5’cap and poly A tail
binding both of these act as a signal to exit the nuclear core
then exchange that export factor for an initiation factor, so the ribosome starts translating
What therapy could treat HD at the level of RNA to prevent the production of toxic protein?
Gene silencing/ Antisense Therapy:
take known sequence of the HTT gene, generate an oligonucleotide that attaches to mRNA
Double stranded mRNA will be chopped up b/c it is not recognized/ doesn’t belong
What is translation?
Using three nucleotides (codon) to code for amino acids
What are some important codons to know?
Start codon:
AUG (Met)
Stop codons:
UAA, UAG, UGA
What are key characteristics for genetic code?
1) Degeneracy:
amino acids associated with more than one codon
Ex.) Proline (Pro)
2) Specificity:
each codon only codes for one amino acid
Ex.) UGU → always cysteine
What is a wobble nucleotide?
usually the third nucleotide, because it does not change the amino acid
What are the key parts of Translation?
mRNA:
carries nucleotide code for the protein to be made
tRNA:
molecules interpret nucleotide code
Ribosome:
made of rRNA and protein
catalyzes formation of peptide bond
Explain the transfer RNA (tRNA)
an adaptor molecule made of RNA that interprets the genetic code embedded in mRNA (L shaped)
Anticodon:
sequence on tRNA that is complementary on mRNA
must be charged with correct amino acids before use of protein synthesis
Explain Ribosomes
Facilitate large and small subunits for protein synthesis
Contains proteins and rRNA (ribosomal RNA)
called ribozymes b/c contains RNA w/ catalytic activity that does formation of peptide bond
Found in Rough ER and cytoplasm in eukaryotic
Describe the types of mutations
Mis-sense mutation:
one amino acid change (substitution)
Ex.) sickle cell anemia
Non-sense mutation:
mutations result in a premature stop codon, meaning protein never made and mRNA degraded
Silent mutation:
change in mRNA sequence that does not change amino acid sequence
Frameshift mutation:
deletion of one nucleotide of mRNA which results in a change in all future amino acids
Most likely a stop codon and truncated protein (never made) or a nonfunctional protein gets degraded
Describe the functions of tRNA binding sites
“translation sandwich” mRNA between large and small ribosomal subunit
Aminoacyl site (A-site):
binds incoming aminoacyl- tRNA, which carries next amino acid to added chain
new tRNAs bind to this site
Peptidyl site (P-site):
where the initiator tRNA first binds (found)
holds tRNA molecule attached to growing polypeptide chain
Exit site (E-site):
Binds empty tRNA after amino acid added to chain, then tRNA exits ribosome
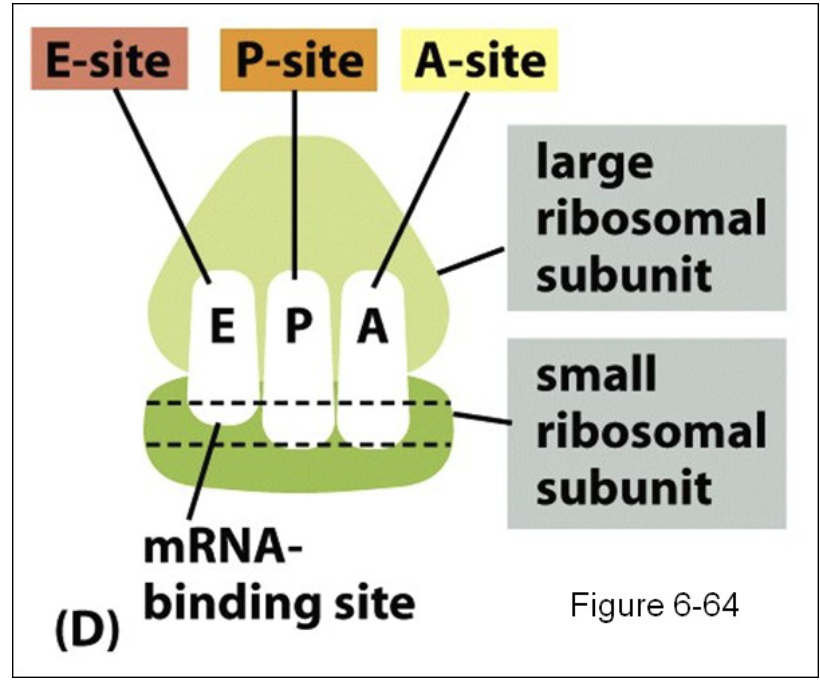
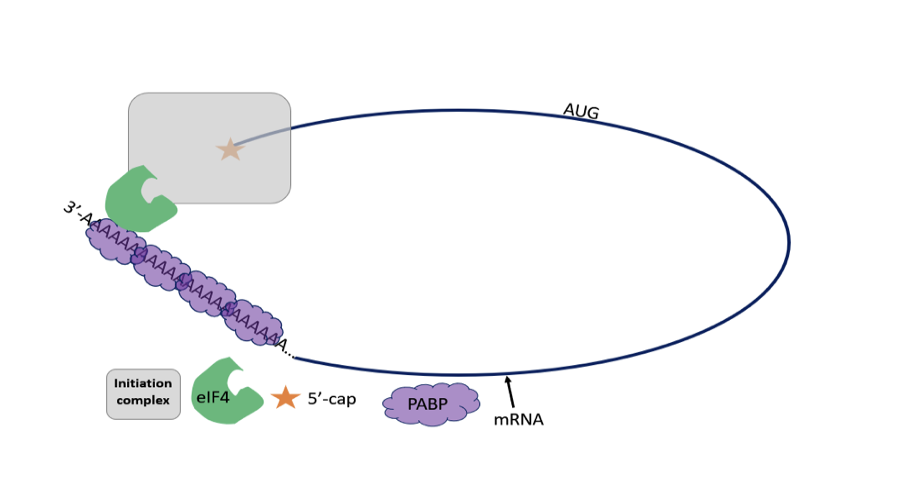
Describe the Initiation step of Translation
1) The initiator tRNA and initiation factors recognize 5’ cap and bind to small subunit of ribosome
2) The tRNA small ribosomal subunit bind to 5’ end of mRNA, moves along 5’ to 3’ in search of start codon (AUG)
3) When the start codon is found, initiation factors are related and the large ribosomal subunit binds
Describe the Elongation step of Translation
1) The charged tRNA carrying next amino acid binds to the A site creating new tRNA binding
2) A peptide bond is formed by the uncoupling of the tRNA on carboxyl end of the amino acid attached to the P-site, forming the peptide bond on the A site of tRNA
3) The large ribosomal subunit translocates three nucleotides (shifts down a codon)
4) The small ribosomal subunit moves 3 nucleotides to match the large subunit, and tRNA is ejected
This cycle is repeated until Stop codon is released
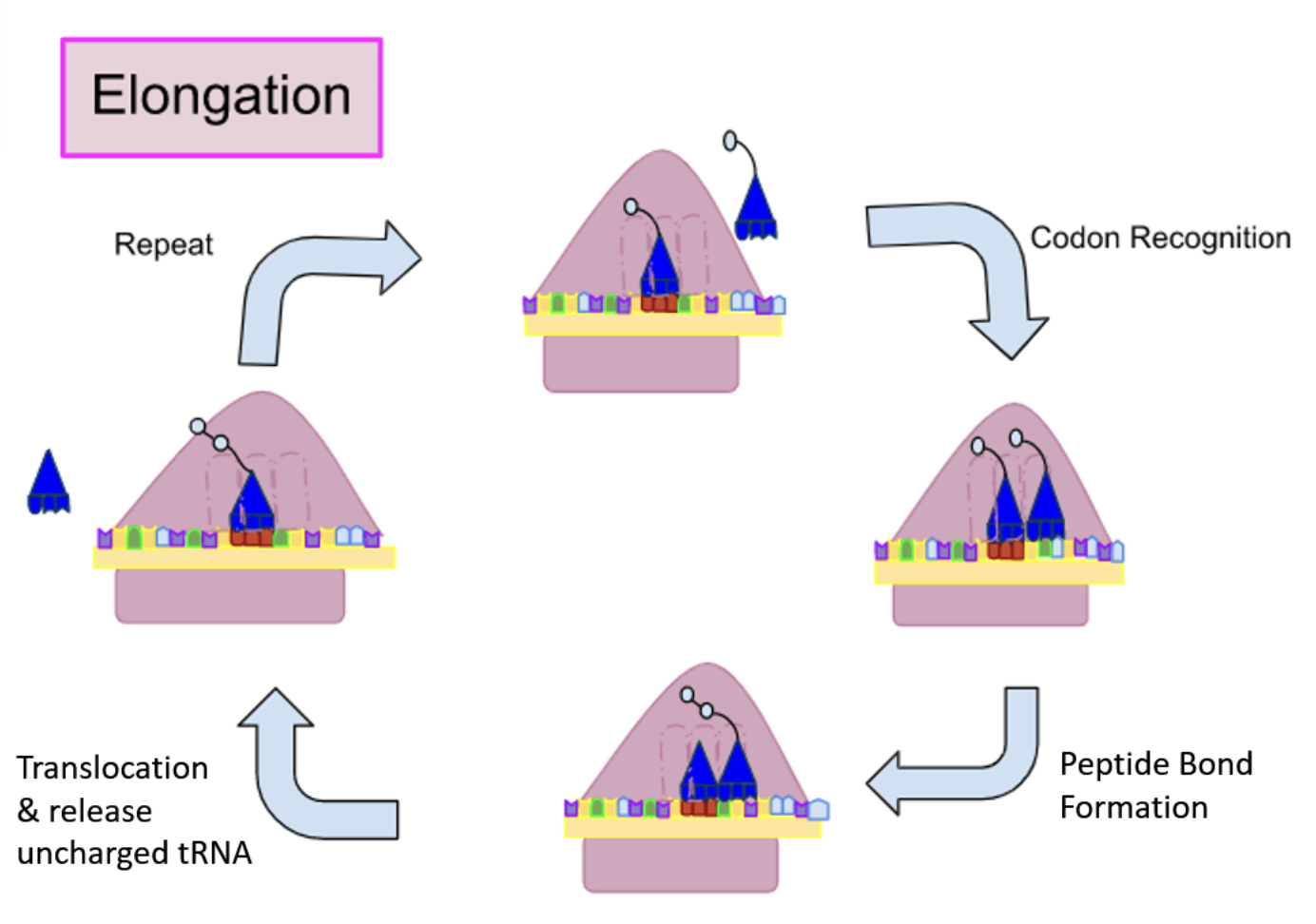
In elongation, how does the ribosome know which tRNA needs to be in the A-site?
Codon in the A-site is a complement for the anti-codon, so the anti- codon and codon match up to bring the amino acid that was attached to the tRNA
Describe what a “charged” tRNA is and describe what enzyme is responsible for the recycling of a tRNA that is “uncharged”
A charged tRNA means that the tRNA is covalently bonded to its corresponding amino acid
The aminoacyl tRNA synthetase enzyme recharges the empty tRNA, in order to make more proteins
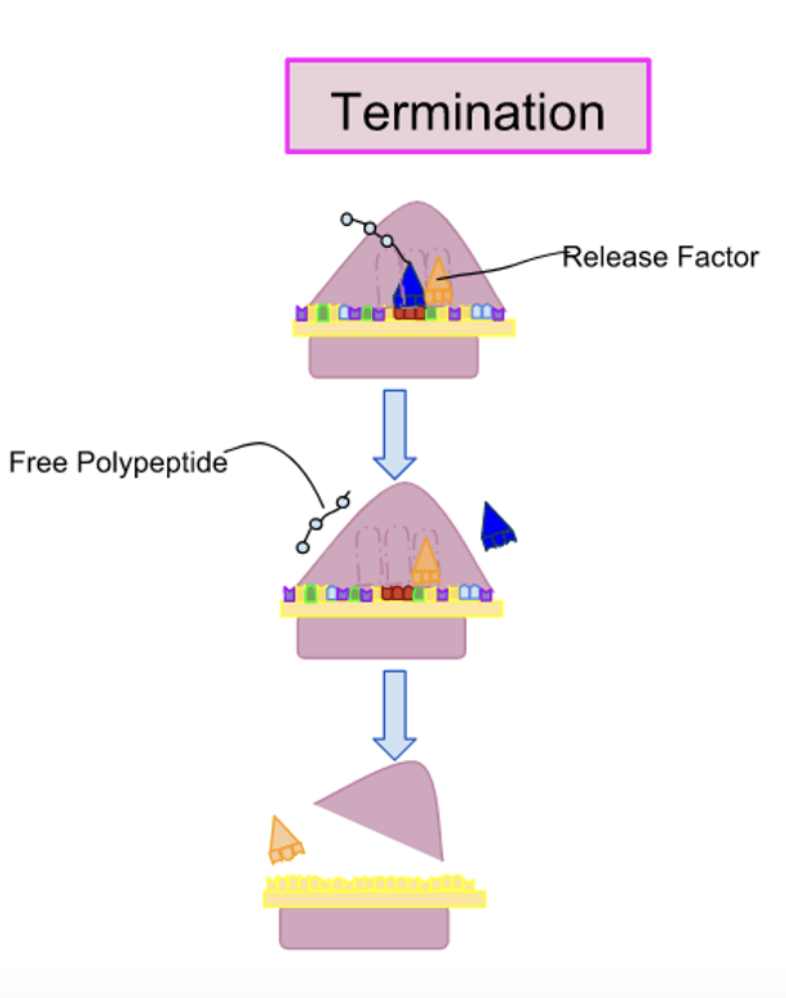
Describe the Termination step of Translation
1) The strip codons (UGA, UAA, UAG) are not recognized by a tRNA and do not specify an amino acid
2) The release factors ( made of proteins) bind to any stop codon that reaches A-site ( stimulates the release and dissociation of the ribosome)
3) The binding of release factors alter the enzyme activity in the ribosome, resulting in hydrolysis of polypeptide chain from the last tRNA
4) The disassembly of the entire complex occurs after the release of polypeptide chain
5) The ribosome also releases mRNA, that is later degraded, and dissociates into 2 subunits that can reassemble on another mRNA molecule to restart protein synthesis
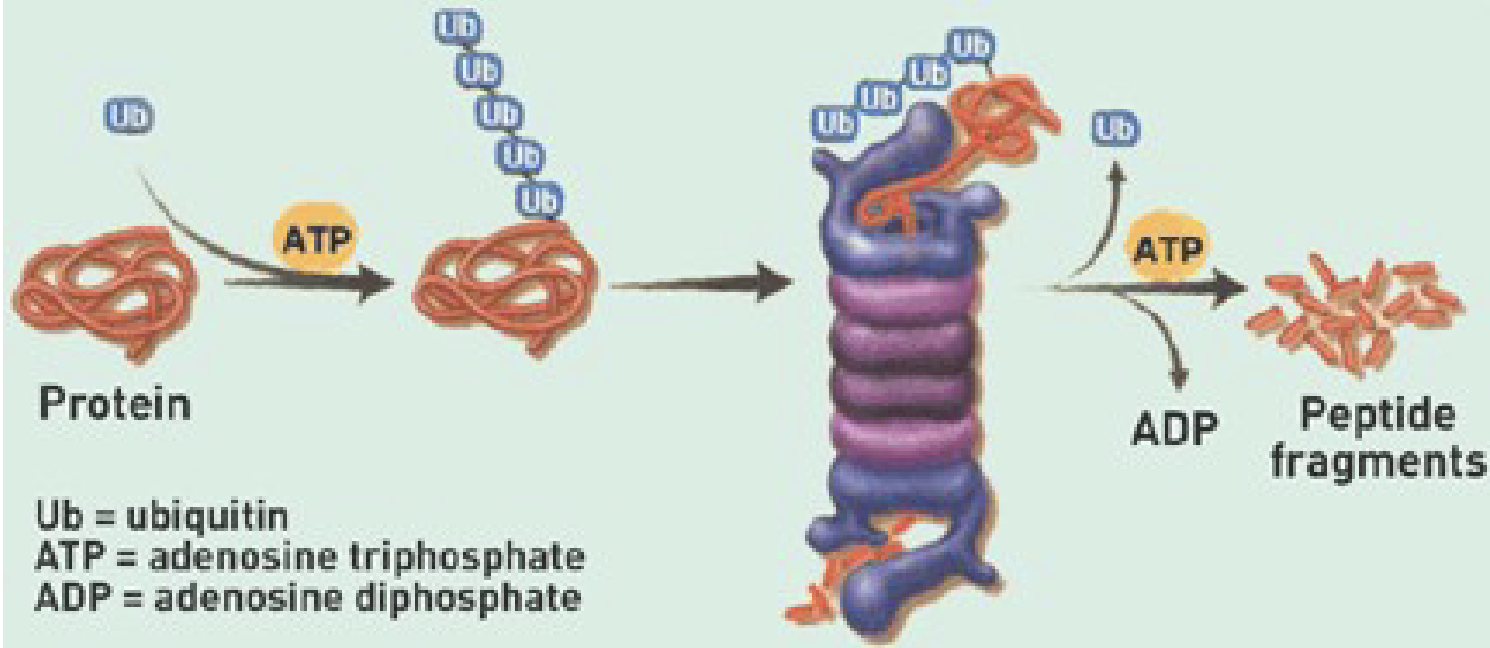
Describe the Ubiquitin Proteasome System
Ubiquitin:
small protein (peptide)
Proteasome:
Large cylindrical structure that degraded proteins via proteases that cleave peptide bonds
Process:
Polyubiquitination:
Ubiquitin (small protein) that is continuously added to protein to target it for proteasome
Proteasome:
Little tube w/ lid on either end
the ubiquitin chain targets the protein to the lid
the protein fed through tube ( filled with hydrolytic enzymes) to break a part peptide bonds
Results in peptide fragments that can be reused