Week 8: Chromatin + Nuclear Organization
1/20
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
21 Terms
DNA organization
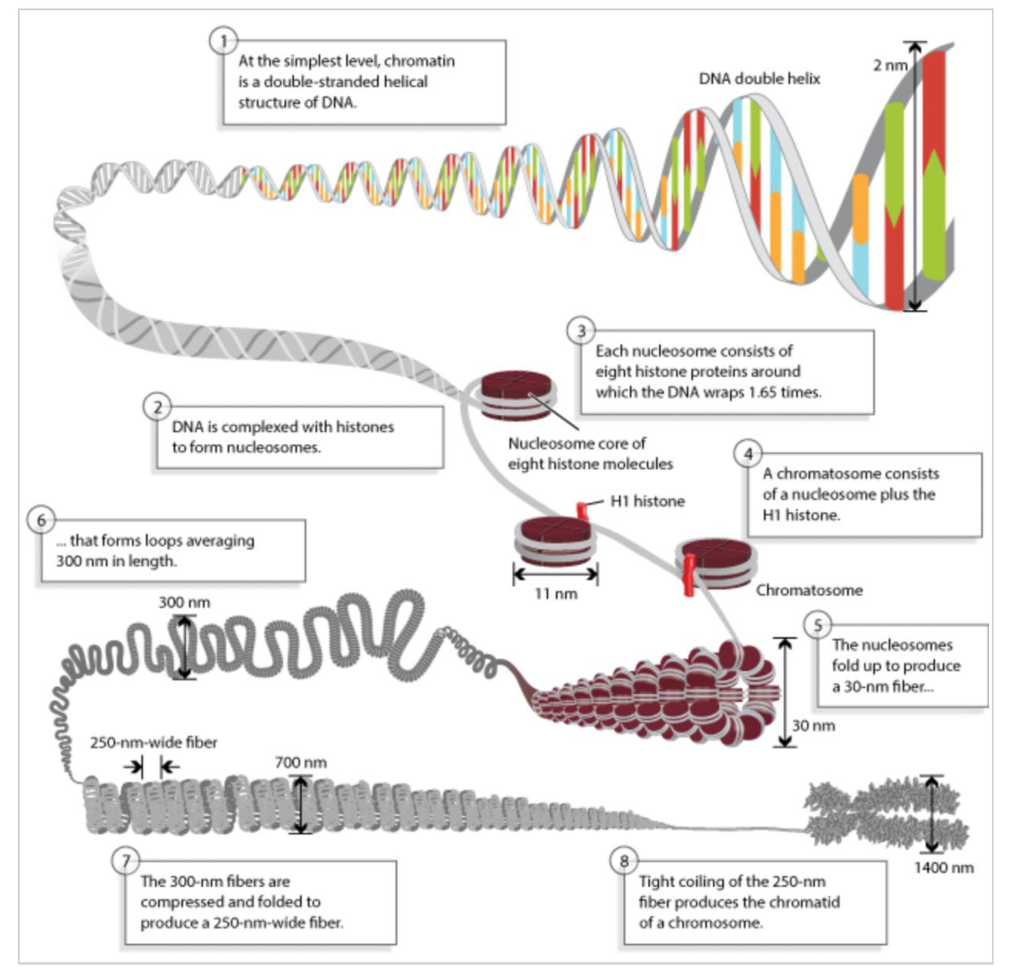
2m of DNA compacted to fit into nucleus
1st order = 10nm chromatin fibre
in vivo → all 10nm
packaged w/o 30nm fibres into mitotic chromosomes
regulated w/o changes b/w 10-30nm fibres
30nm fibres + higher-order fibres = artifact of in vitro purification
DNA wraps around core histone octamer protein complex → forms nucleosome
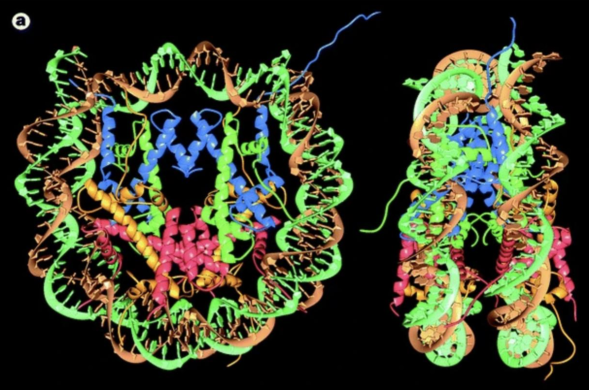
nucleosome
fundamental structural unit of chromatin
DNA-histone complex
DNA wraps ~1.6 turns around histone complex
histone proteins = H2A/H2B/H3/H4 → 2x of each form octamer
mostly a-helices
tails sticking out can be modified → recognized by other proteins
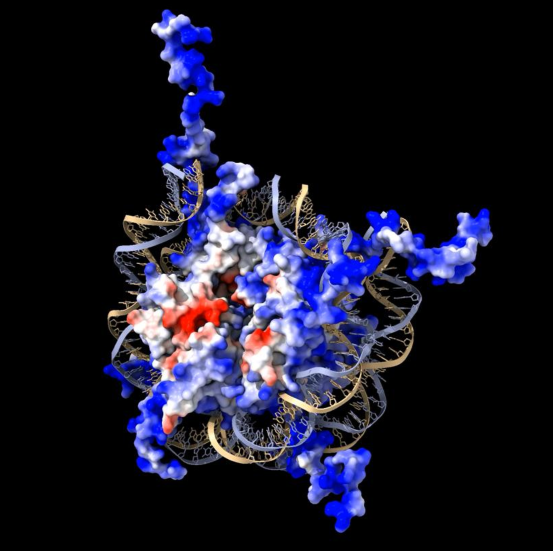
histone
(+) charged protein core of nucleosome + wrapped by DNA
make tight but not sequence specific contact b/w DNA + (+) charged proteins
has acidic (-) patch → no interactions w/ DNA
drives orientation
tails are flexible unstructured domains
stick out from nucleosome
can be post-translationally modified to alter gene expression
basic (+ charged) properties help neutralize (-) charge of DNA phosphate backbone
chromatin properties
(-) charge of phosphate-sugar backbone stabilized by (+) charges of histone proteins
reduce steric repulsion + enables bending + flexible polymer properties
naked DNA properties
periodic structure w/ regularly spaced (-) charge from phosphate backbone
requires metal ions + small amino-rich molecules to neutralize charge + enable flexibility
post-translational modifications
contributes to gene regulation
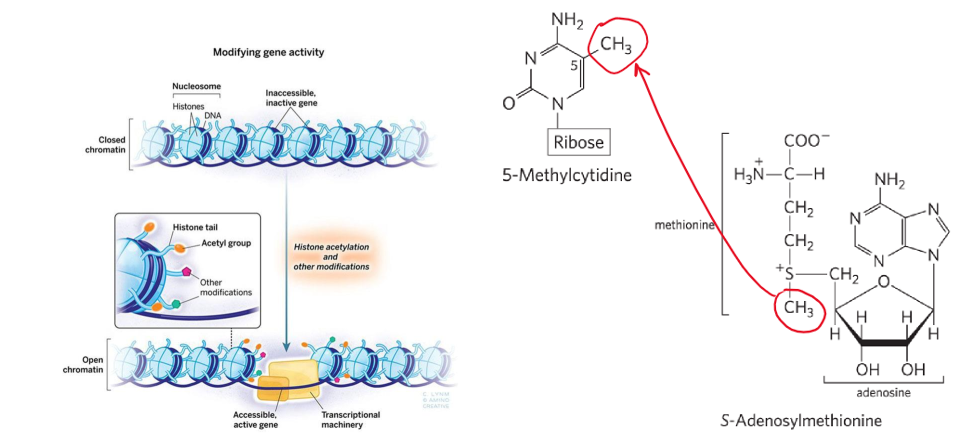
DNA methylation = changes TF recruitment → changes gene expression + DNA metabolism
A + C methylated more than G + T
eukaryote → 5% of cytidine residues methylated → common at CpG sites
prokaryotes → adenine commonly methylated
DNA methylases use S-adenosylmethionine (SAM) as methyl donor
modification to histone tails = changes nucleosome packaging = changes gene expression
nucleosomes moved out of way so polymerase can go to site
DNA needs to be bent at promoter site
open = active chromatin
closed = inactive chromatin
provides platform for recruiting protein complexes → activate/silence particular region of genome
in combo w/ DNA methylation → mark specific genes for activation/repression
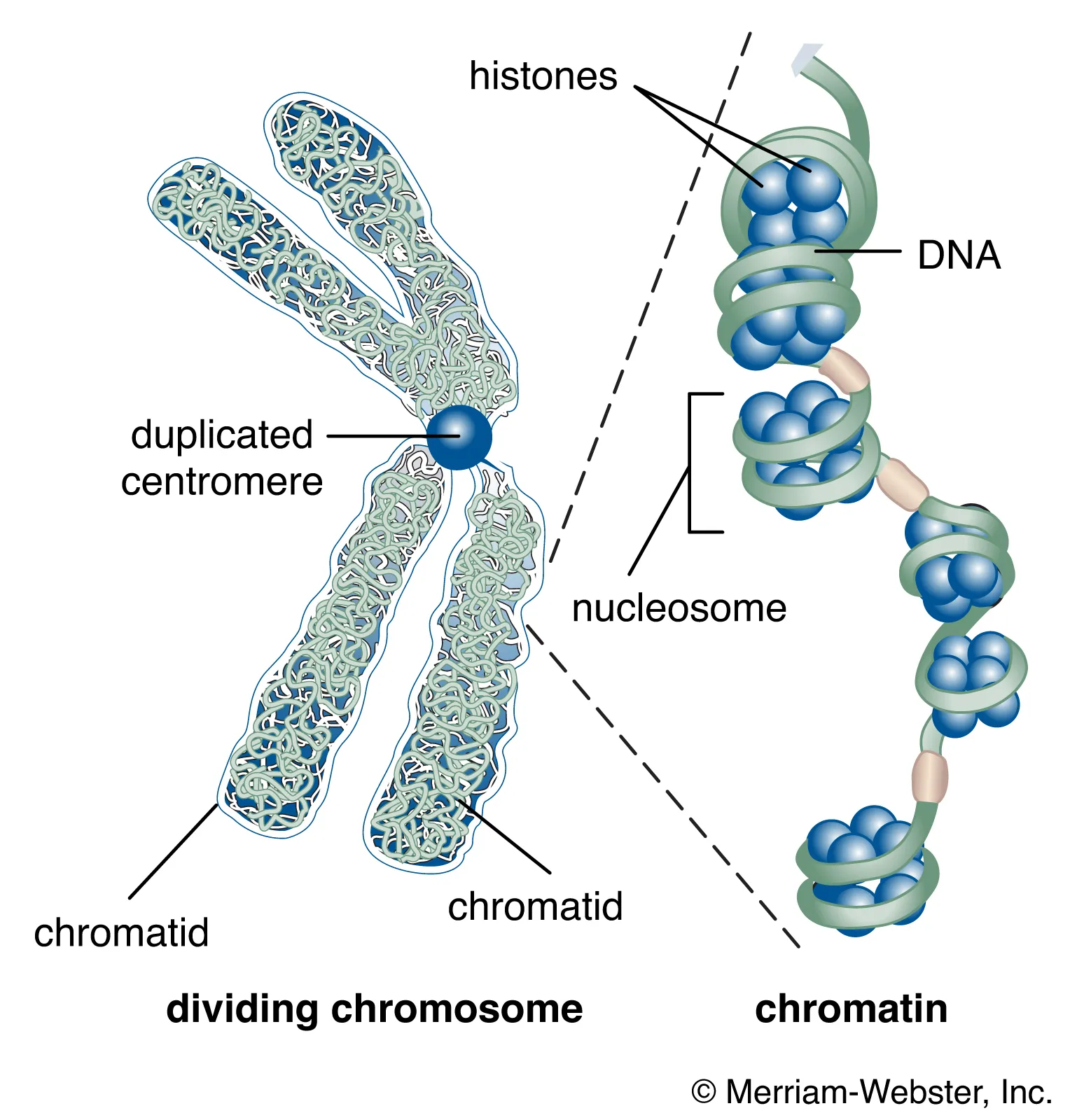
chromatin
complex of DNA and proteins found in eukaryotic cells
package long DNA molecules into more compact, denser structures
made up of string of nucleosomes attached by linker DNA
10nm = in vivo
30nm+ = in vitro artifacts
needs to be “open” for proteins to reach genetic material
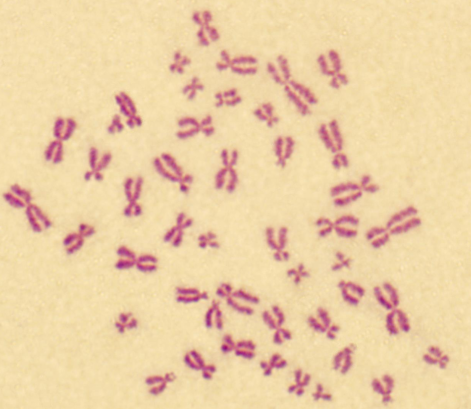
karyotyping
technique that visually assesses # and structure of chromosomes in cell, identifying abnormalities related to genetic disorders
length + staining density of genomic regions assessed w/ light microscopy
arrest cell in cell cycle
isolate DNA from cells
add methanol + other things
take mixture + drop onto slide
mitotic chromosomes spread out into little splats
different stains used to look at them
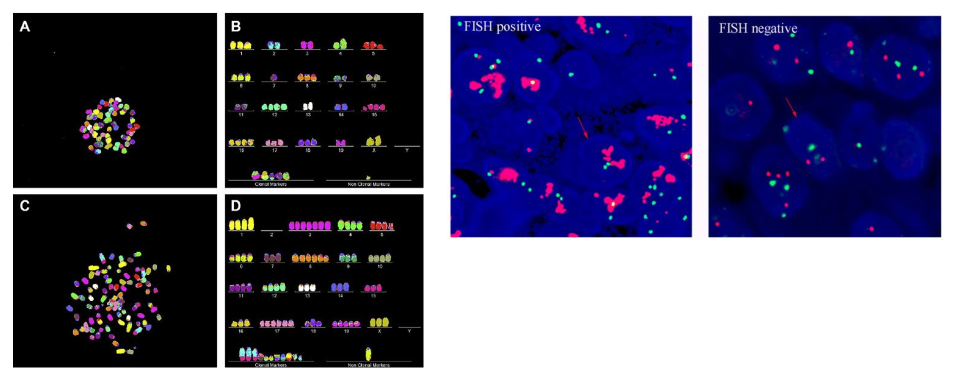
FISH (fluorescence in situ hybridization)
method to visualize/detect specific sequences of DNA → where segments of genome are
fluorescent probes made from specific DNA + cut into small fragments
cells fixed + chromatin chemically denatured → heated to melt DNA
fluorescent probes annealed + non-specific probes washed away
detection via fluorescence microscope
interphase chromosomes
chromatin from individual chromosomes form distinct domains
fibres within territories are 10-nm
strings of nucleosomes attached by linker sequences
epigenetic modifications may change local chromatin environment
change space b/w nucleosomes
provide platform for protein/protein-RNA complexes to bind + regulate gene expression
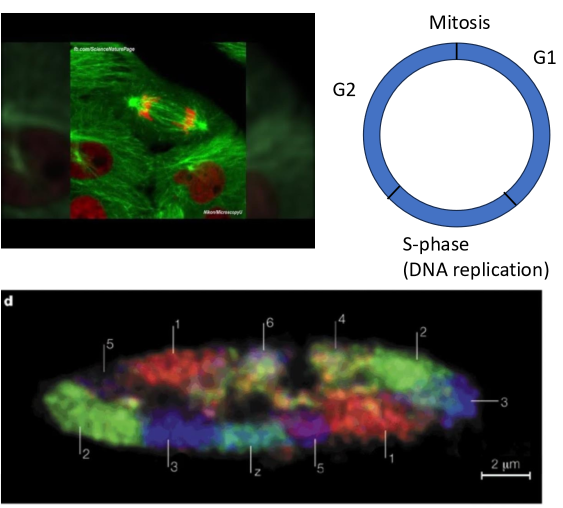
chromatin in situ
interphase
chromatin organized into compact discrete structures → chromosome territory
fibers in territories = 10-nm chromatin
strings of nucleosomes attached by linker sequences
epigenetic modifications may change local chromatin environment
change nucleosome spacing
provide platform for proteins/protein-RNA complexes
cell cycle chromatin organization
interphase (G1/S/G2) = cell grows + DNA replicates
genes packaged in looser, more accessible structure
non-transcribed regions more condensed, less accessible
DNA + histone modifications
chromatin form chromosome territories
mitosis = cell division
chromatin condenses → chromosomes
organized for efficient segregation to separate
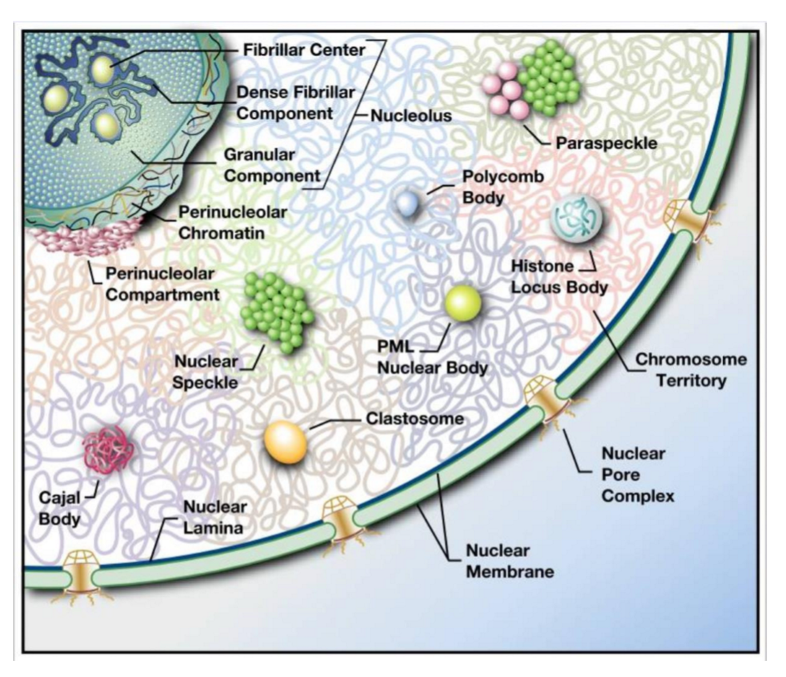
nuclear bodies
range in size + function → large nucleolus to small polycomb bodies
non-membrane bound sub-nuclear compartments w/ specific function
eg. nucleolus = ribosome biogenesis
spatial + temporal regulation of nuclear processes
sequestration of molecules improves reaction efficiency
active genomic regions → clustered around transcription factories w/ active RNA polymerase
can be visualized w/ fluorescent microscopy
protein phase separation
process where proteins dynamically aggregate to form membrane-less compartments, influencing various cellular functions and processes
some proteins + RNA under certain conditions self-solubilize → form phase-separated compartment
eg. concentration, post-translational modification
interlocking proteins via multivalent domains
proteins post-translationally modified → drives association or dissociation
[sufficient] → spontaneous formation of membrane-less compartments
proteins can interlock
pi-stacking via disordered/low-complexity regions
phenylalanine rings + glycine build up force → F-G repeats
need to be on flexible part of protein to stack
![<p>process where proteins dynamically aggregate to form membrane-less compartments, influencing various cellular functions and processes</p><ul><li><p>some proteins + RNA under certain conditions self-solubilize → form phase-separated compartment</p><ul><li><p>eg. concentration, post-translational modification</p></li></ul></li></ul><ol><li><p>interlocking proteins via multivalent domains </p><ol><li><p>proteins post-translationally modified → drives association or dissociation</p></li><li><p>[sufficient] → spontaneous formation of membrane-less compartments</p></li><li><p>proteins can interlock</p></li></ol></li><li><p>pi-stacking via disordered/low-complexity regions</p><ol><li><p>phenylalanine rings + glycine build up force → F-G repeats</p><ol><li><p>need to be on flexible part of protein to stack</p></li></ol></li></ol></li></ol><p></p>](https://knowt-user-attachments.s3.amazonaws.com/e86423a7-7519-4fd4-8d73-cd66c487d817.png)
fluorescence microscopy
way to visualize/detect proteins
antibodies raised to detect protein of interest or protein of interest tagged w/ fluorescent protein, eg. GFP
cells fixed + permeabilized → antibodies can fit into cellular compartments
fluorescent-tagged secondary antibody often used
DNA counterstains, ie. DAPI + Hoescht, define nucleus
give sense of regions of more or less DNA → compaction/heterochromatin
progeria
rare genetic disorder resulting in rapid againg
caused by point mutation in lamin A
de novo (meiotic polymerase replication) error causes C → T mutation in exon 11
generates cryptic splice site
1 allele expresses truncated form of lamin A
50 AA shorter than WT
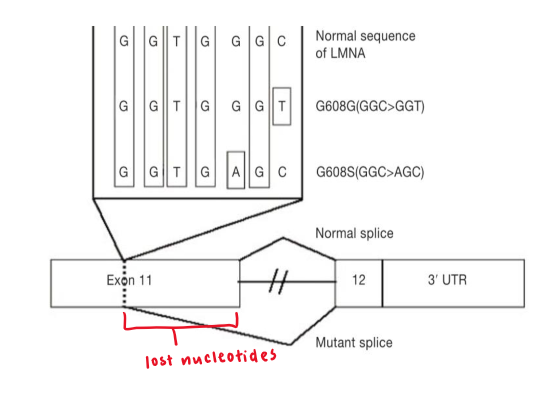
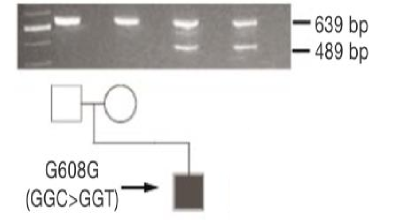
lamin A RNA
affected individual has both full-length + truncated version of lamin A mRNA
WT = 639 bp
mutant = 489 bp
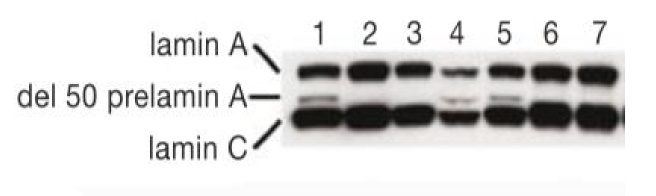
lamin A protein
lamin C reacts w/ antibody for lamin A
affected individual has small amount of alternatively spliced version
WT (2/3/6/7)
mutant (1/4/5) = del 50 prelamin A variant band smaller than WT
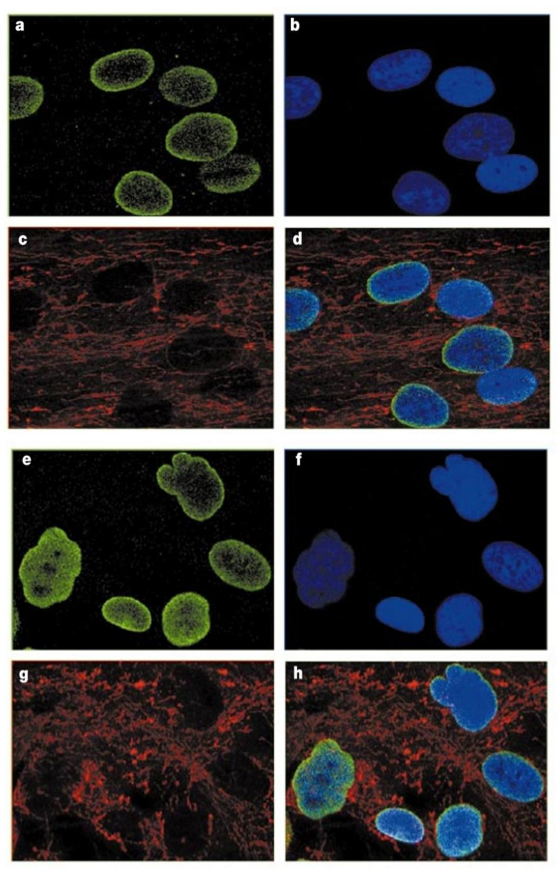
progeria cells
control (A-D)
nucleus = smooth, oval shape
lamin localizes to periphery, some in nucleoplasm
mutant (E-H)
nucleus = reticulated + irregular
lamin localizes throughout nucleoplasm
green LaminA; blue DAPI; red mitochondria
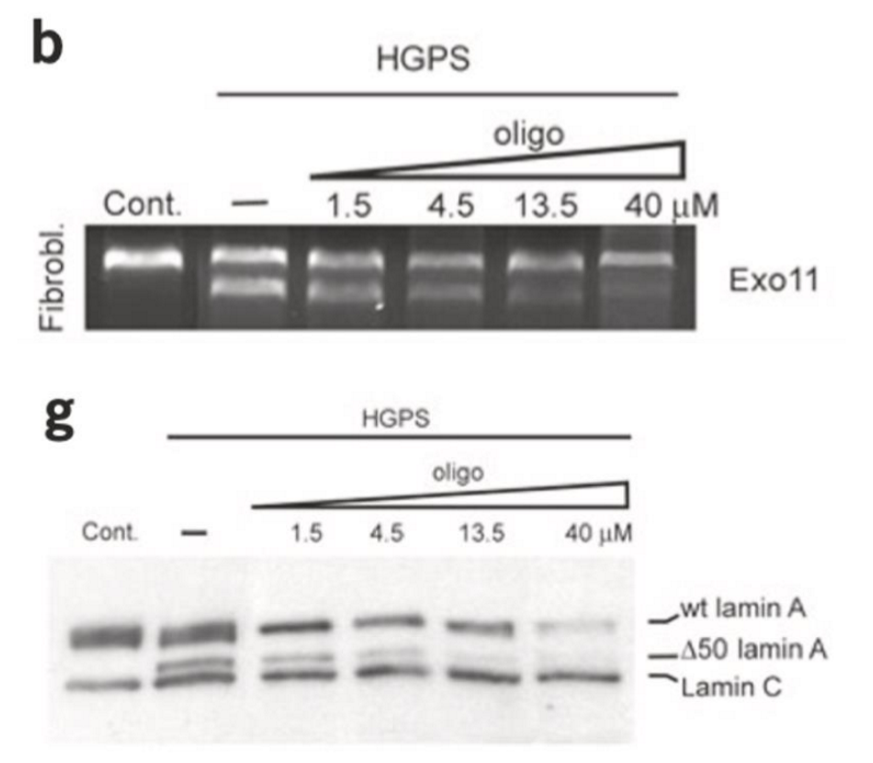
progeria cell treatment
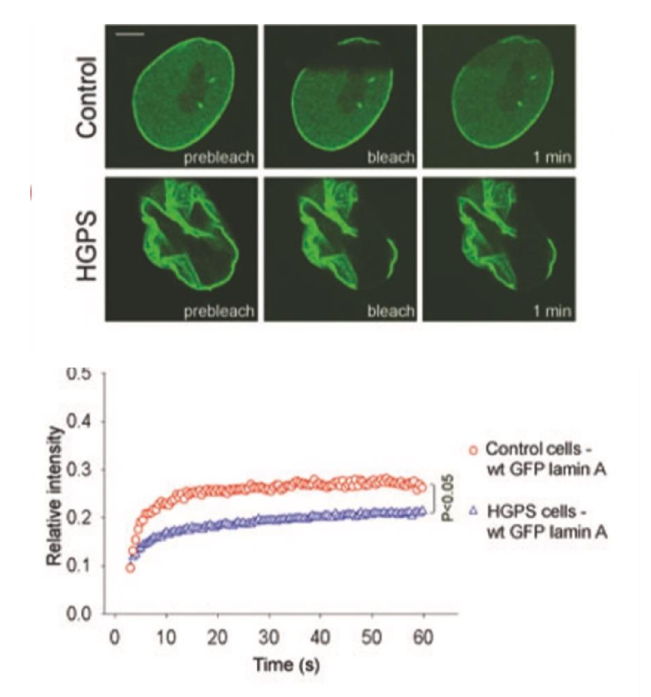
progeria Lamin slow to recover compared to WT
⬇ mobility of Lamin A protein
WT dynamics restores by ⬆ oligo (nucleic acid = small RNA molecule) to progeria cells
⬇ amount of alternatively spliced Lamin A mRNA
oligo targets cryptic exon 11/12 boundary → sends RNA for degradation
FRAP (fluorescence recovery after photobleaching)
measures recovery of fluorescence after being bleached
determines mobility + dynamics of tagged proteins in live cells
often tagged w/ GFP
recovery time ⬆ = dynamic ⬆