14 - DNA Repair
1/4
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
5 Terms
List and explain the key DNA repair mechanisms, including direct repair, base excision repair (BER), nucleotide excision repair (NER), mismatch repair (MMR), and double-strand break repair (NHEJ & HRR).
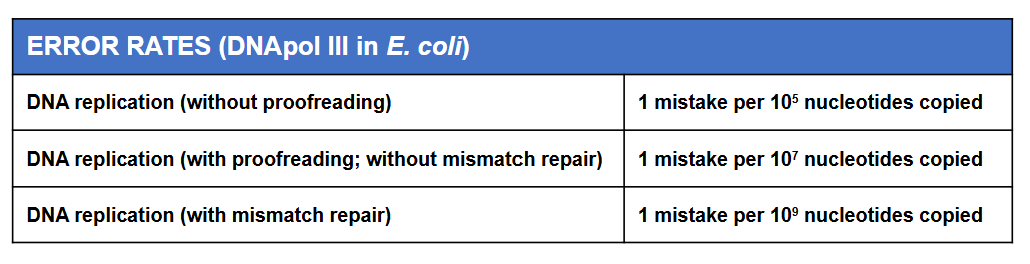
DNA repair systems are essential to the maintenance of the genetic integrity of organisms
Observed mutation rates result from the balance between mutations and their repair
Can prevent cellular malfunction, disease and cancers
Proofreading: DNApol III has a 3’ - 5’ exonuclease activity, cutting out the incorrect nucleotide just added
Direct Repair
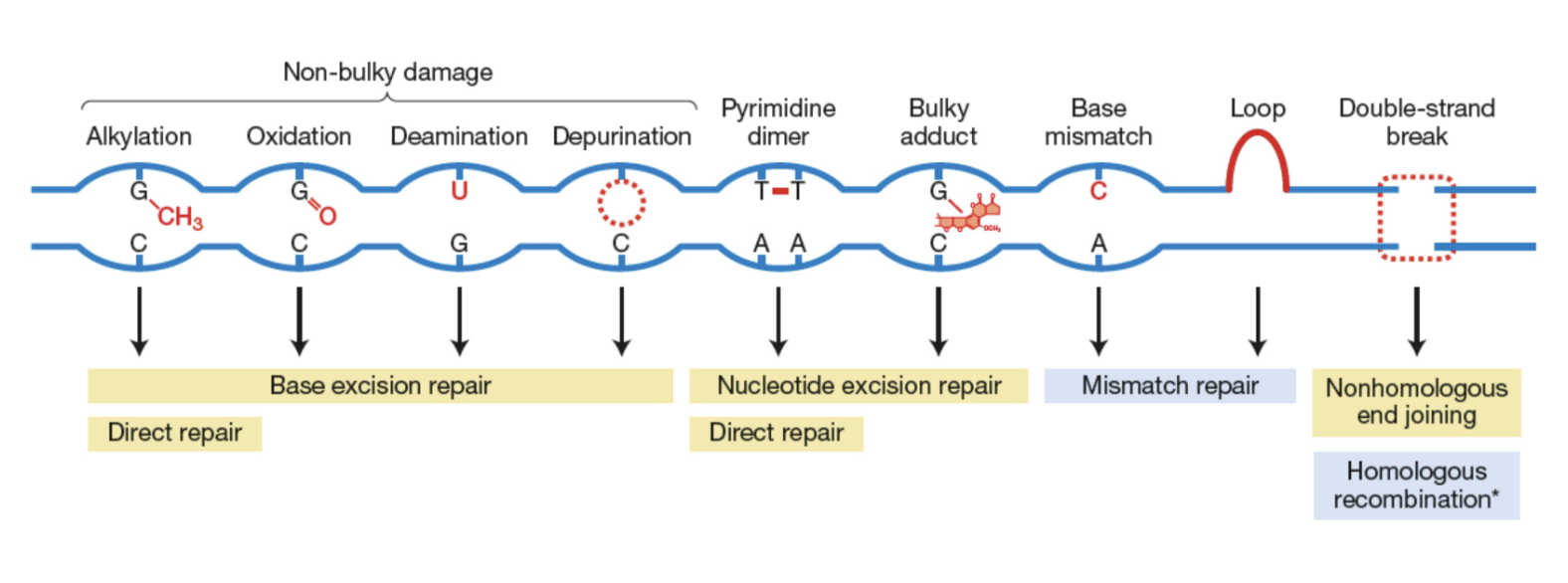
Direct repairs: fix “non-bulky” DNA damages without removing the affected nucleotide or disrupting the DNA backbone
Convert the damaged DNA back to the correct form
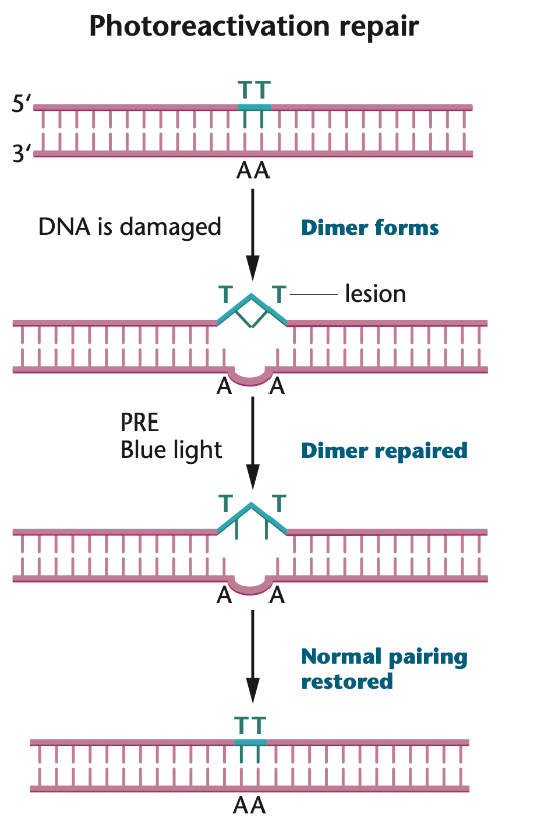
E.g. photoreactivation direct repair
Following UV radiation and the formation of a thymine dimer
A photoreactivation enzyme (PRE) or photolyase cleaves the cross-linking bond under visible blue light
The thymine binds back to their complement nucleotide
Base Excision Repair (BER)
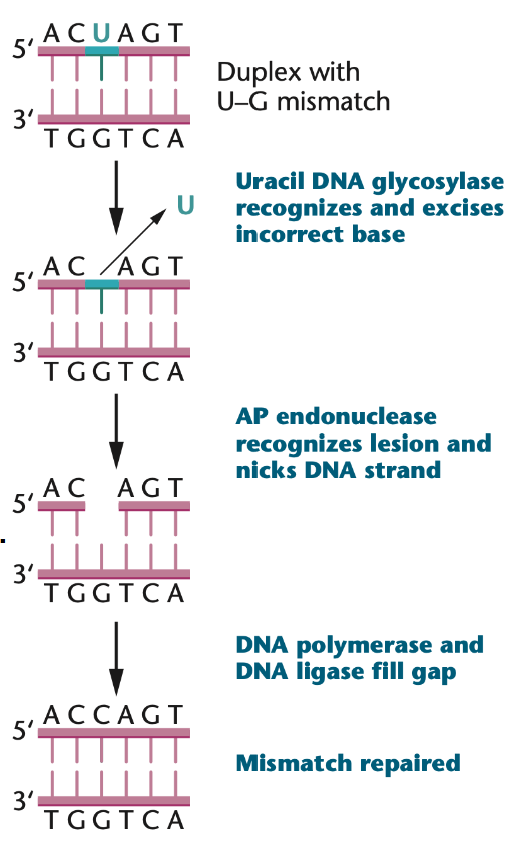
Base excision repair (BER) eliminates “non-bulky” damages that don’t distort the double helix and affect individual abnormal bases (e.g. uracil, 3-methyladenine)
During replication, a mismatch is recognized by a DNA glycosylase
The DNA glycosylase breaks the base-sugar bond, freeing the base
An AP endonuclease (apyrimidinic/apurinic endonuclease) cuts the phosphodiester bond, creating a nick
a DNA polymerase β excises the incorrect nucleotide and adds the correct one
A DNA ligase links the new nucleotide by forming phosphodiester bonds
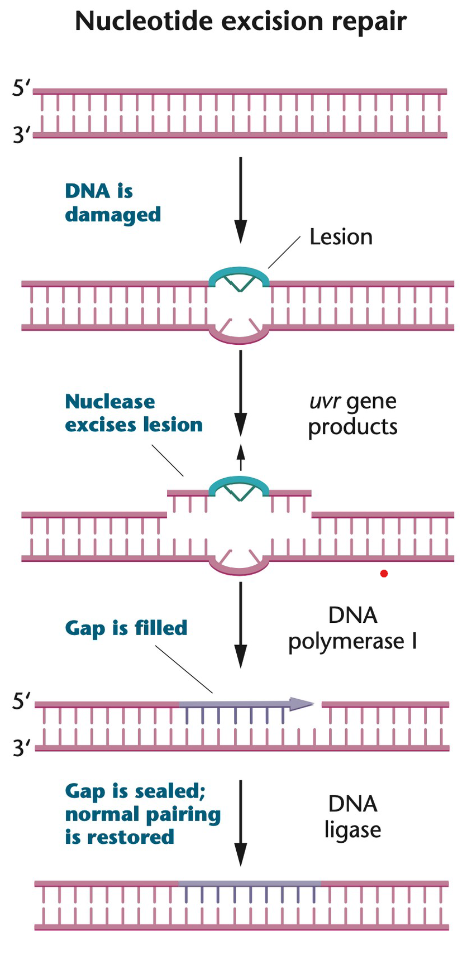
Nucleotide Excision Repair (NER)
Nucleotide excision repair (NER): mechanism that repairs many “bulky” damages in DNA that distort the double helix (e.g. thymine dimers, chemically modified bases, missing bases, and crosslinks)
In NER, several nucleotides in the damaged strand are removed the DNA, and the intact strand is used as a template
A nuclease excises the lesion (larger than the lesion itself)
A DNApol I synthesizes the missing nucleotides
A DNA ligase ligates the newly synthesized fragment by forming the phosphodiester bonds
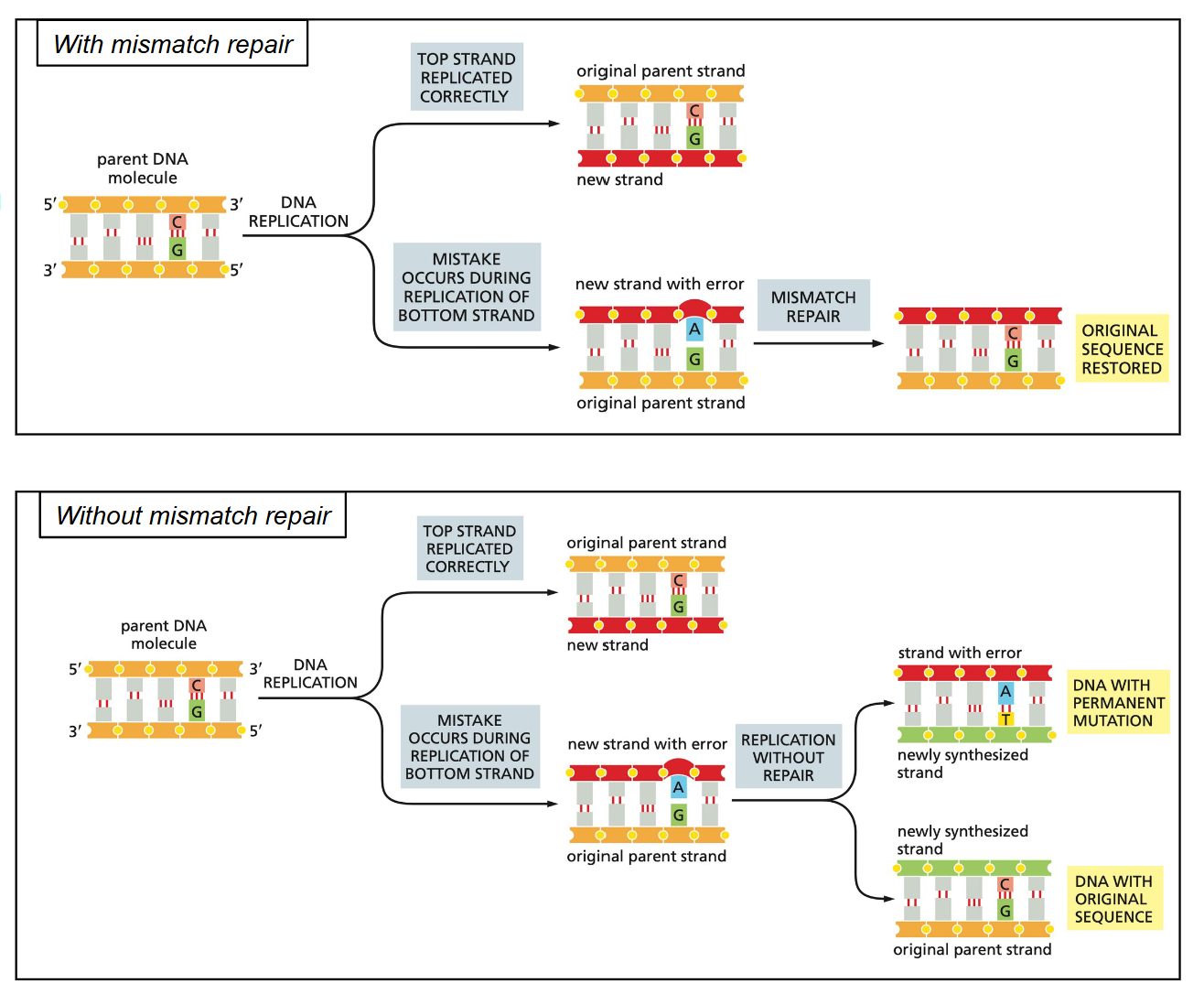
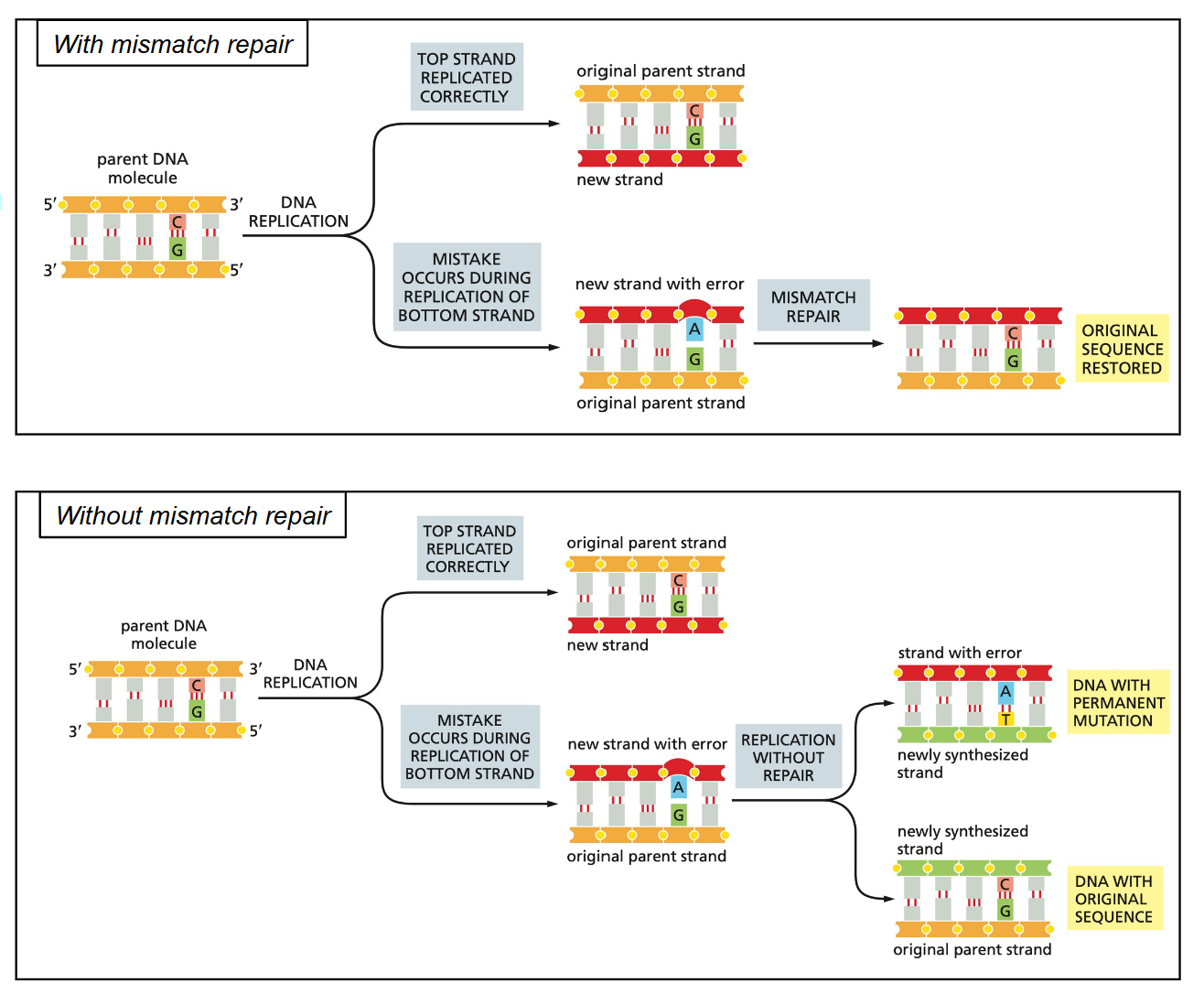
Mismatch repair
The mismatch repair (MMR) system must repairs the newly synthesized strand… not the parental strand
MMR fixes a base-pair mismatch, not an abnormal nucleotide (as in BER or NER)
1. Methylation of the adenine (in 5’-GATC-3’) on the old strand soon after the new strand is synthesized
2. The mismatch is recognized by (MutS)
3. A bridge is formed with MutL and MutH
4. An exonuclease removes a few nucleotides within the bridge
5. A DNA polymerase synthesizes a normal daughter strand using the parental strand as a template
Double-strand break (DSB) Repair
Non-homologous end joining (NHEJ) fixes double-strand breaks (DSB) by simply attaching the two broken strands back together
The DSB is recognized by end-binding proteins kept close together thanks to proteins that form a crossbridge
Additional proteins digest one of the two DNA strands (can result in the few deletion of a few nucleotides)
Gaps are filled in by a DNA polymerase
DNA ends are ligated together by a ligase
NHEJ doesn’t require a sister chromatid - can occur at any phase of cell cycle
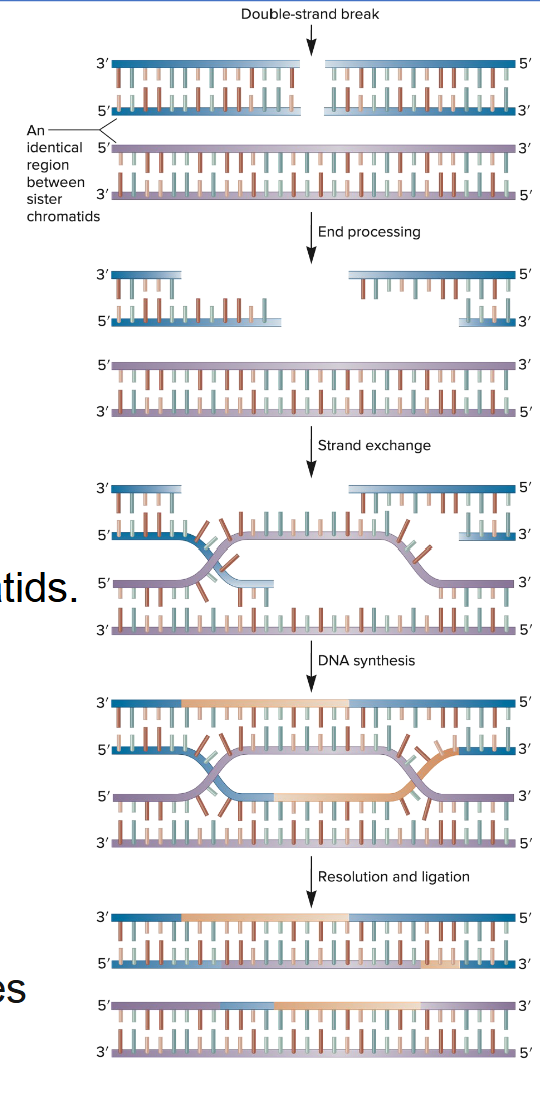
Homologous recombination repair (HRR)
Homologous recombination repair (HRR) fixes double-strand breaks (DSB) using a sister chromatid → also called Homology Directed Repair (HDR)
Needs a non-damaged DNA fragment
Digestion of short segments of both DNA strands at the break site
Invasion and exchange of strands between the broken/unbroken sister chromatids
The unbroken strands are then used as templates to synthesize DNA
Finally, the crisscrossed strands (forming Holliday junctions) are resolved (i.e. broken and rejoined)
The sister chromatid used a template is available only during the S and G2 phases of the cell cycle
Contrast the molecular steps involved in base excision repair (BER) and nucleotide excision repair (NER)
1. Base Excision Repair (BER)
Purpose: Repairs small, non-bulky lesions (e.g., oxidation, alkylation, deamination).
Damage recognition by a DNA glycosylase, which removes the damaged base.
Creation of an AP site (apurinic/apyrimidinic).
AP endonuclease cuts the DNA backbone at the AP site.
DNA polymerase adds the correct nucleotide.
DNA ligase seals the nick.
2. Nucleotide Excision Repair (NER)
Purpose: Repairs bulky, helix-distorting lesions (e.g., thymine dimers, chemical adducts).
Damage recognition by NER complex.
Local DNA unwinding near the lesion.
Dual incision: Excision of a short oligonucleotide segment (~24–32 nucleotides) containing the lesion.
DNA polymerase synthesizes the replacement strand.
DNA ligase seals the final nick.
Summary of Differences
Feature | BER | NER |
|---|---|---|
Type of damage | Small, non-helix-distorting lesions | Bulky, helix-distorting lesions |
Initial enzyme | DNA glycosylase | NER damage-recognition complex |
Excised region size | Single base | 24–32 nucleotides |
Cleavage enzyme | AP endonuclease | Dual endonuclease complex |
Common outcomes | Repair of oxidized or deaminated bases | Repair of UV-induced dimers, bulky adducts |
Contrast the mechanisms of Non-Homologous End Joining (NHEJ) and Homologous Recombination Repair (HRR)
1. Non-Homologous End Joining (NHEJ)
Definition: A repair mechanism that directly joins the ends of a double-strand break (DSB) without using a homologous template.
Steps:
Ku proteins recognize and bind to DNA ends.
The DNA ends are processed (can involve trimming or filling).
The break is ligated by a ligase complex.
Characteristics:
Fast but error-prone
Often results in mutations, such as small insertions or deletions (indels)
Functions in all cell cycle phases
2. Homologous Recombination Repair (HRR)
Definition: A repair mechanism that uses a homologous DNA sequence (usually a sister chromatid) as a template to accurately repair a DSB.
Steps:
DSB is resected to produce single-stranded overhangs.
RAD51 mediates strand invasion into the homologous template.
DNA synthesis occurs using the homologous strand as a template.
The repaired strands are resolved and ligated.
Characteristics:
Error-free
Requires the presence of a homologous sequence
Active mainly in the S and G2 phases of the cell cycle
Summary Table
Feature | NHEJ | HRR |
|---|---|---|
Template used | None | Sister chromatid (homologous DNA) |
Accuracy | Error-prone | Error-free |
Cell cycle phase | All phases | S and G2 phases only |
Key proteins | Ku proteins, ligase | RAD51, recombination machinery |
Typical outcome | Indels or mutations possible | Precise restoration of sequence |
Predict the consequences of base pair mismatch in the presence or absence of mismatch repair (MMR)
1. In the Presence of Mismatch Repair (MMR)
MMR detects and corrects mismatched base pairs that escape proofreading during DNA replication.
The system identifies the newly synthesized strand, removes the segment containing the mismatch, and fills in the correct bases using DNA polymerase and ligase.
Outcome:
Genome stability is maintained.
Mutations are prevented.
No lasting consequences if repair is successful.
2. In the Absence of Mismatch Repair (MMR)
Mismatches remain uncorrected in the DNA after replication.
These mismatches can lead to permanent mutations in the next round of replication when the DNA is copied again.
Potential consequences:
Point mutations (e.g., A–C becomes A–T after replication).
Increased mutation rate (mutator phenotype).
Genetic disorders or cancer, such as hereditary non-polyposis colorectal cancer (HNPCC) linked to defective MMR genes.
Key Takeaways
Condition | Consequence |
|---|---|
MMR active | Mismatch is repaired → no mutation |
MMR inactive | Mismatch persists → fixed as a mutation |
Leads to increased mutation rate and instability |
Illustrate how homologous repair can occur and how Holliday Junctions can be resolved.
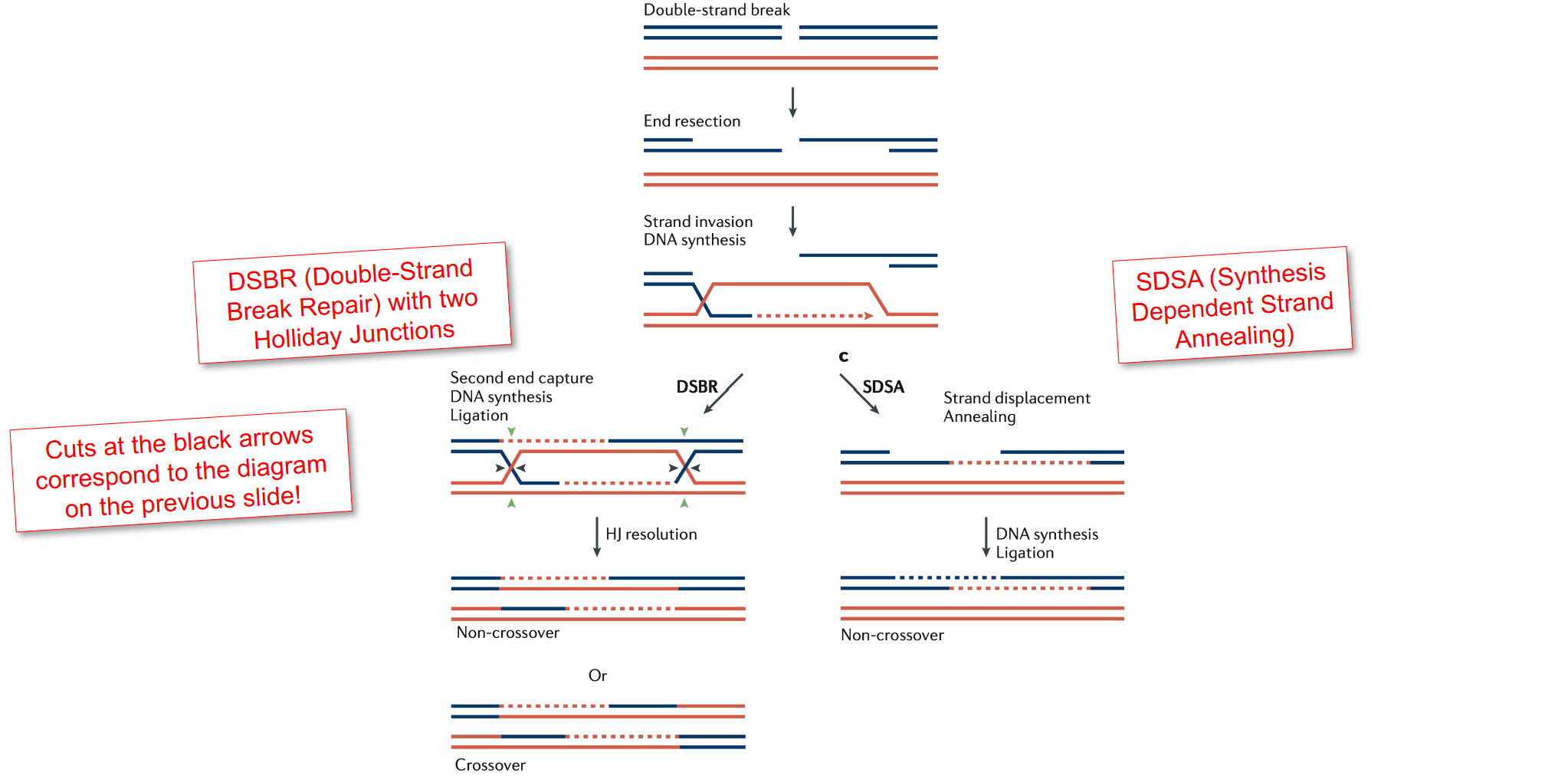
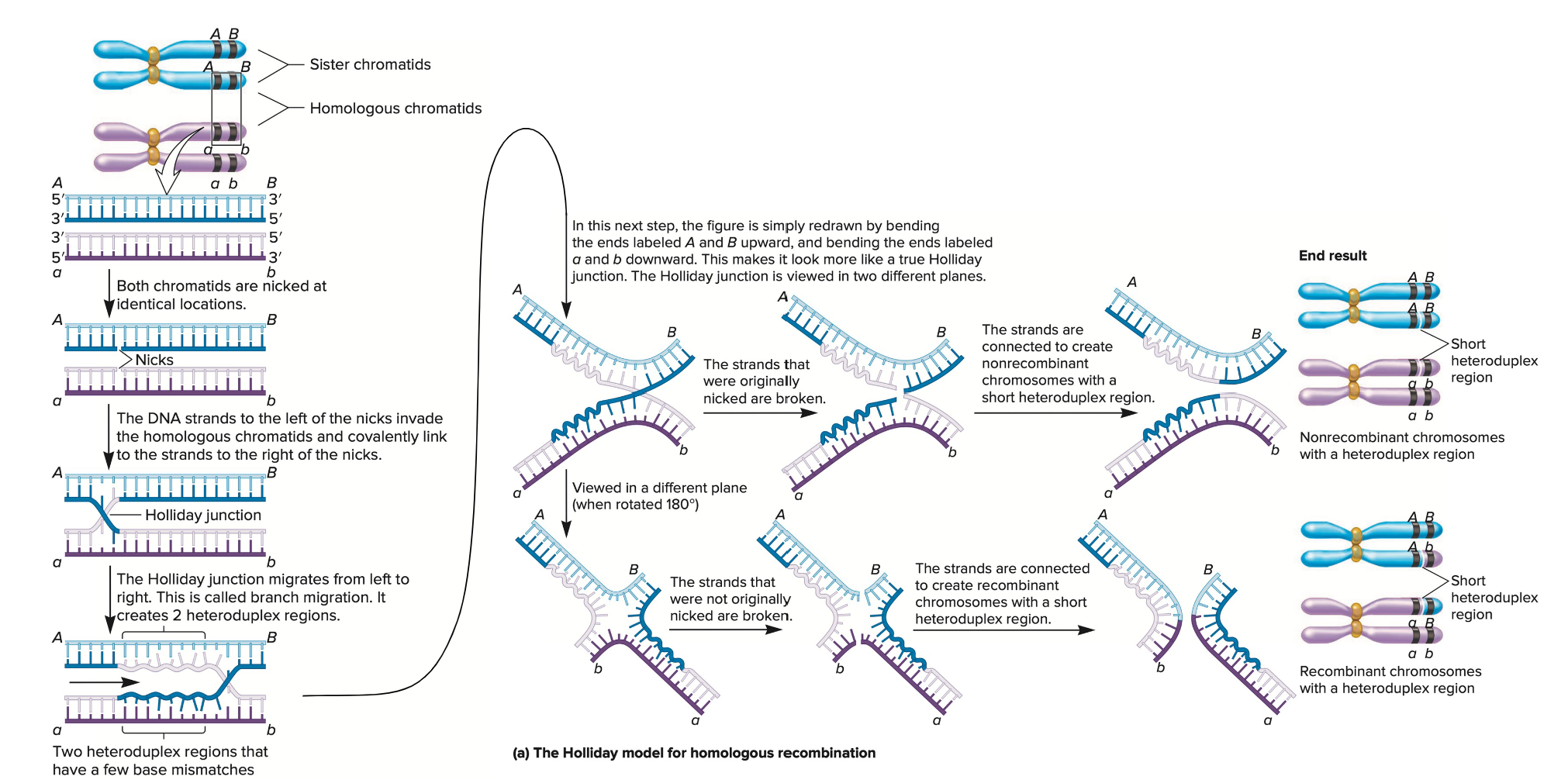
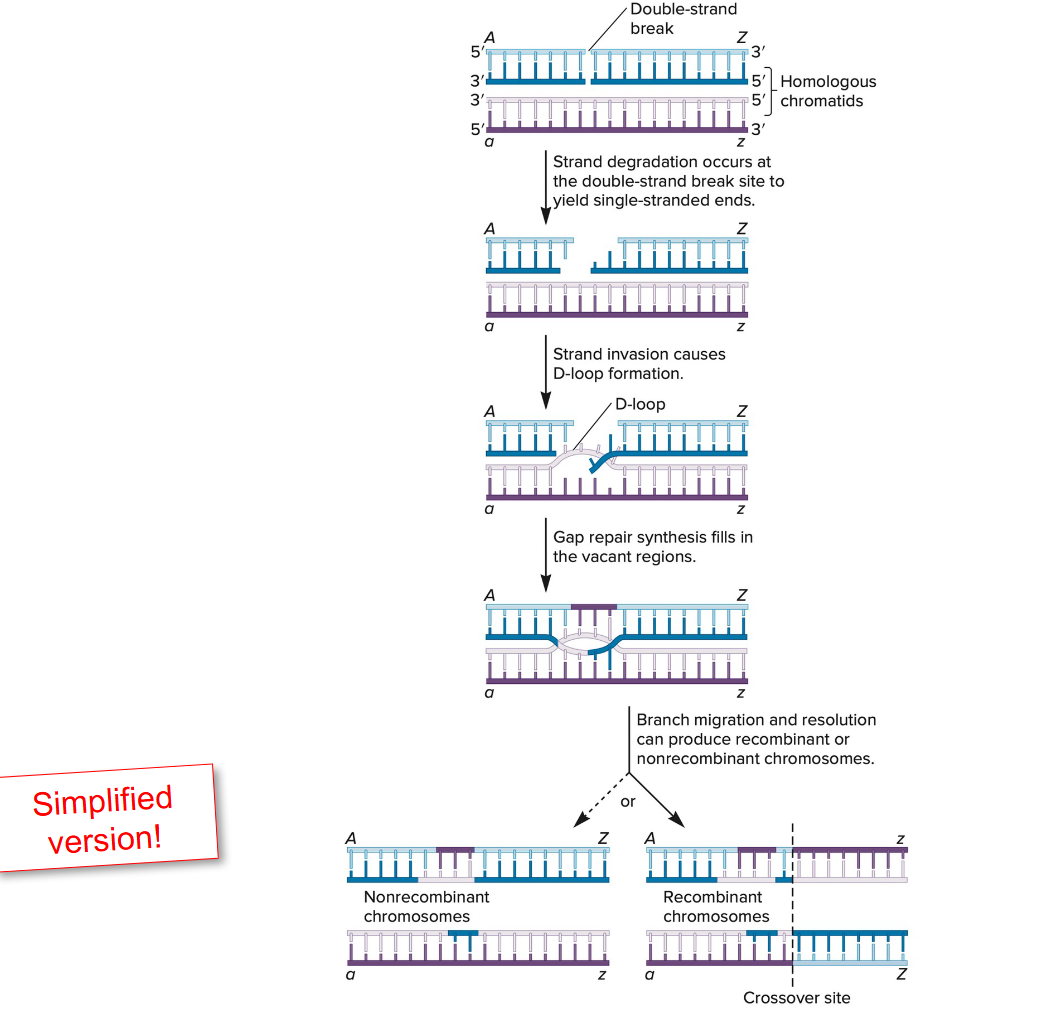
Homologous Recombination Repair (HRR) Overview
HRR repairs double-strand breaks (DSBs) using a homologous DNA template, typically a sister chromatid.
It is an error-free mechanism, primarily active during the S and G2 phases of the cell cycle.
Steps of Homologous Repair
DSB Recognition and Resection
The DSB is detected and the ends are resected, creating 3′ single-stranded overhangs.
Strand Invasion
A RAD51-coated 3′ overhang invades the homologous duplex DNA (usually from the sister chromatid).
DNA Synthesis
DNA polymerase extends the invading strand using the homologous template.
Holliday Junction Formation
A Holliday junction forms — a cross-shaped structure where strands from homologous DNA molecules are exchanged.
Branch Migration
The junction can move along the DNA, extending the region of heteroduplex DNA.
Resolution of Holliday Junctions
The junction is cleaved by resolvase enzymes in two possible ways:
Horizontal resolution: Produces non-crossover products.
Vertical resolution: Produces crossover products.