1404 Biochemistry- Nitrogen Metabolism-Protein Turnover
1/20
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
21 Terms
What are the 9 essential amino acids?
histidine
isoleucine
leucine
lysine
methionine
phenylalanine
threonine
tryptophan
valine
what is protein turnover?
it is the continual renewal or replacement of protein
The rates of turnover vary from tissue to tissue, and
the relative contributions of different tissues to total
protein turnover change with age and adaptation
to various levels of protein intake
Proteostasis Network
The proteostasis network (PN) is a complex system of molecular pathways that maintains protein homeostasis (proteostasis)—ensuring that proteins are correctly synthesized, folded, trafficked, and degraded in the cell. It is essential for cellular function and preventing diseases related to protein misfolding or aggregation.
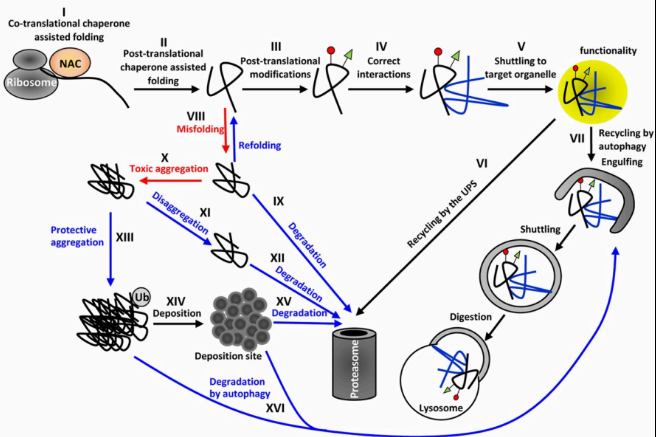
What are the protein degradation pathways?
Ups
Macroautophagy
CMA
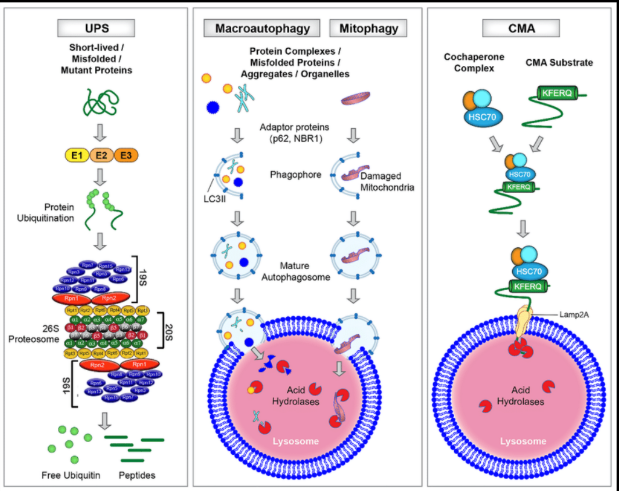
What is ubiquitin/Proteasome system (UPS)?
The major pathway of selective protein degradation in eukaryotic
cells
Uses ubiquitin as a marker that targets cytosolic and nuclear proteins
for rapid proteolysis
The first ubiquitin attaches the amino group of the side chain of a
lysine residue on the protein substrate (isopeptide bond). Additional
ubiquitins are then added to form a multiubiquitin chain.
-Mono-ubiquitylation
-Multiple mono-ubiquitylation
Polyubiquinated proteins are recognized and degraded by a large, multisubunit protease complex; aka the proteasome Chains of 4 or more ubiquitin molecules target protein for destruction
Ubiquitin is released in the process, so it can be reused in another
cycle
Process requires ATP (attachment and degradation)
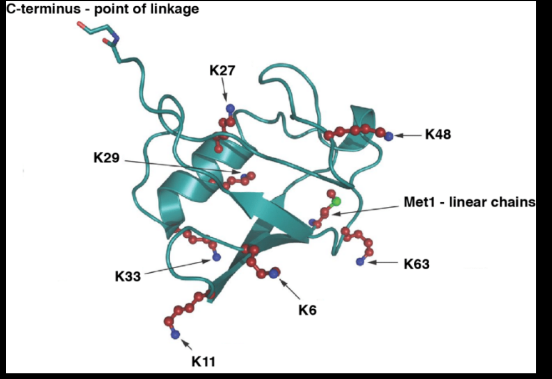
Ubiquitin
A 76-amino-acid polypeptide that is highly
conserved in all eukaryotes
Adopts a compact globular fold, the β-grasp
or ubiquitin-like fold
Covalent attachment of ubiquitin to target
proteins is mediated by an elaborate
enzymatic conjugation cascade; in most cases,
lysine residues serve as acceptors for ubiquitin
Can also be attached to the α-amino group
of the N-terminal residue of a protein or to
serine or threonine residues via the formation
of a peptide bond or ester bond, respectively
Can serve as its own substrate resulting in the
formation of ubiquitin chains on target proteins (poly- ubiquitylation)
Contains 7 lysine residues and each of these as well as
its N-terminal residue can serve as an attachment site
for ubiquitin resulting in 8 homotypic, numerous
heterotypic chains and branched chains
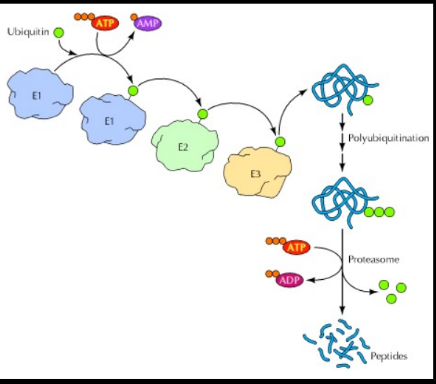
What happnes in the UPS?
1. Ub activation – A thioester is formed between the
carboxyl of the terminal glycine residue of Ub and a
cysteine residue of the activating enzyme, or E1. The
energy that fuels this reaction is provided by
hydrolysis of ATP.
2. Conjugation to E2 – Activated Ub is transferred to
a cysteine residue of a conjugating enzyme, or E2.
3. Binding of Ub to E3 – A new transesterification
attaches Ub to the enzyme ubiquitin ligase, or E3. Most cells have one type of E1, but different forms of
E2 and E3. Different kinds of E2 and E3 recognize
different proteins as substrate.
what is proteasome?
The proteasome is a large protein
complex responsible for degradation of
intracellular proteins
requires metabolic energy
what is proteasome made up of?
The proteasome is made up of two subcomplexes: a catalytic core particle (CP; also
known as the 20S proteasome) and one or two terminal 19S regulatory particle(s)
(RP) that serves as a proteasome activator.
What is autophagy?
Autophagy is a cellular process in which cytoplasmic contents are degraded within the
lysosome/vacuole, and the resulting macromolecular constituents are recycled
Autophagy complements the ubiquitin-proteasome system in mediating protein turnover
Whereas the proteasome degrades individual proteins modified with ubiquitin chains,
autophagy degrades many proteins and organelles en masse
How is macromolecules destined for autophagic degradation selected?
Through sequestration
within a specialized double-membrane compartment termed the phagophore, the precursor to
an autophagosome, and then are hydrolyzed in a lysosome- or vacuole-dependent manner
A pair of distinctive ubiquitin-like proteins (UBLs), Atg8 and Atg12, regulate degradation by
autophagy in unique ways by controlling autophagosome biogenesis and recruitment of
specific cargos during selective autophagy
what are the proteins in cells that are degraded by autophagy?
- Cytoplasmic proteins
- Misfolded proteins
- Insoluble misfolded protein complexes
(polymers, aggregates and
aggresomes)
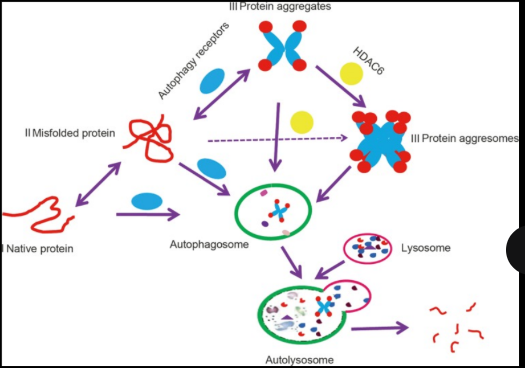
what is chaperone-mediated autophagy (CMA)?
Degrades soluble or unfoldable proteins in
a molecule-by-molecule fashion.
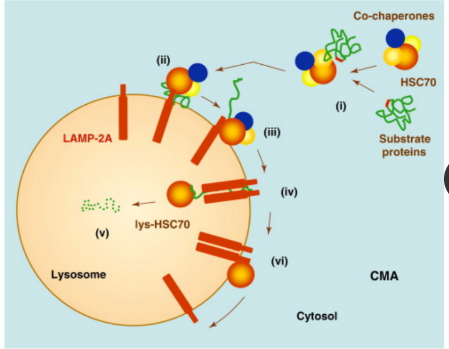
How can chaperone-mediated autophagy be activated?
Can be activated by prolonged starvation
to provide amino acids for essential protein
synthesis
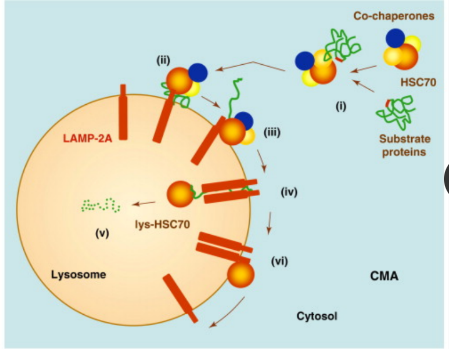
What are the key factors involved in the CMA process?
Heat shock protein 70 (HSC70) and
lysosome membrane protein type 2A
(LAMP2A) are two key factors that are
involved in this process.
Proteasome network and disease
Changes in hormone secretion, tissue perfusion, oxygen availability, energy-protein
intake, free amino acid pattern, hydration state, acid-base balance as well as
activation of the systemic inflammatory response may affect protein synthesis and
degradation.
What diseases are associated with the disruption of protien turnover?
- Angelman syndrome is caused by maternal deficiency of the E6-AP ubiquitin E3 ligase (UBE3A)
- Other disorders of E3 ligases have been identified, including autosomal recessive juvenile Parkinson
disease, the APECED form of autoimmune polyendocrinopathy syndrome, von Hippel-Lindau syndrome,
and congenital polycythemia
- Disorders that disturb ubiquitin regulatory signaling include at least two subtypes of Fanconi anemia,
the BRCA1 and BRCA2 forms of breast and ovarian cancer susceptibility, incontinentia pigmenti, and
cylindromatosis
- Many disorders affect ubiquitin pathways secondarily
What is nitrogen balance?
Reflects the equilibrium between protein intake and losses
Occurs when nitrogen intake equals nitrogen output (NB = 0), and is also referred to
as nitrogen equilibrium
A positive NB or anabolic state exists when nitrogen intake exceeds nitrogen output
-A net 24-hour positive NB of 2 to 4g is optimal for anabolism
When nitrogen excretion is greater than nitrogen intake,
a negative NB or catabolic state exists
How to calculate NB?
Subtract the total nitrogen output from total nitrogen intake
The total nitrogen intake is determined by dividing the daily protein intake (grams)
from both enteral and parenteral sources by 6.25
Nitrogen output consists primarily as urine urea nitrogen (UUN)
An aliquot of a 24-hour urine collection is assayed for its urea nitrogen content by a standard
enzymatic laboratory technique
The above value, plus 4 (the constant used for nitrogen losses from the skin and feces),
is subtracted from the grams of nitrogen intake during the same 24-hour period to
calculate the NB

What are the factors affecting NB?
Dietary intake
The recommended protein intake is 0.8g/kg body wt/day
. This amounts to about 58 g protein/day for a 72-kg
(160-lb) man and about 44 g/day for a 55-kg (120-lb)
woman
Dietary Content of Carbohydrate and Fat
Growth
Pregnancy
Illness and major trauma
What is the clinical significance of NB?
Protein-Energy Malnutrition (PEM)
The most common form of malnutrition in the world
While the symptoms vary widely from case to case, it is common to classify most cases
as either marasmus or kwashiorkor
Marasmus
Caused by inadequate intake of both protein and energy
Kwashiorkor
Caused by inadequate intake of protein with adequate energy intake