DNA Replication and DNA Repair
1/76
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
77 Terms
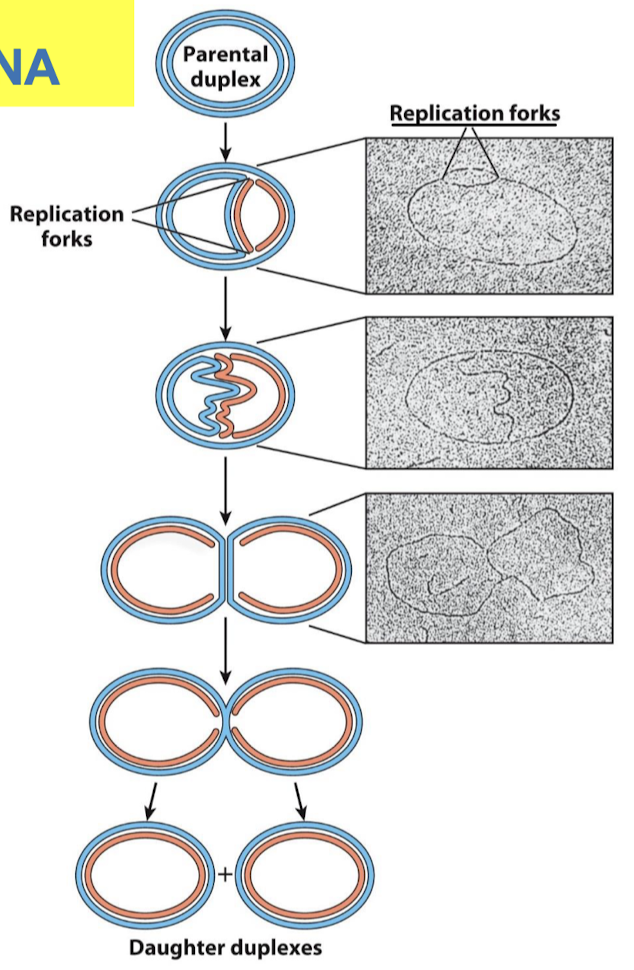
What are three fundamental principles of the replication of circular DNA in E. coli?
Both strands are replicated simultaneously (new strands are shown in red)
Two replication forks —> bidirectional replication
Loops always initiate at a unique origin (one origin of replication)
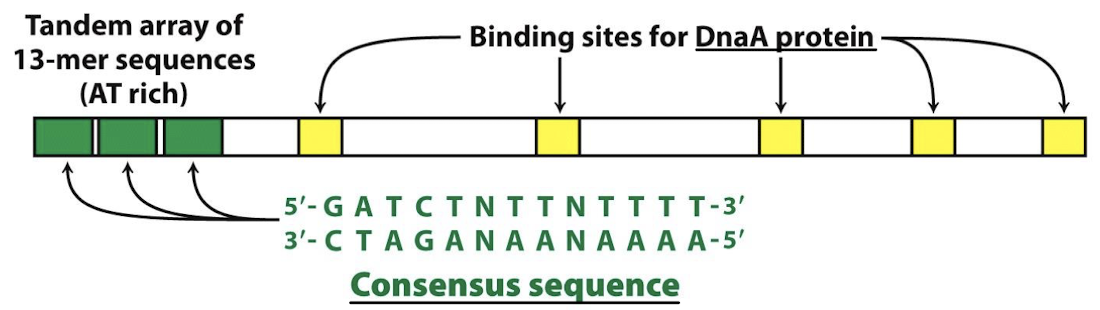
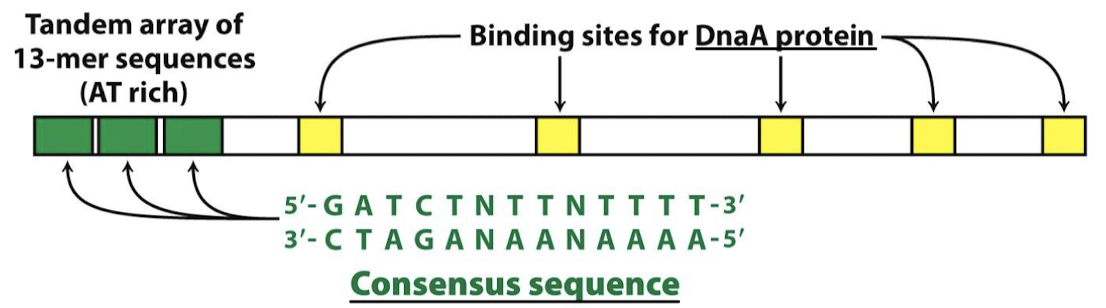
Is the origin of replication in E. coli highly conserved or not? What are the two elements/motifs that are common across E. coli origins of replication?
Highly conserved sequence elements:
Extremely A-T-rich DNA Unwinding Element (DUE) to facilitate DNA strand separation.
Five 9-bp motifs that are recognized by DnaA, known as DnaA boxes.
Why does it make sense for the DNA Unwinding Element (DUE) to be A-T-rich?
A/T base pairings only have two hydrogen bonds and are therefore weaker to promote DNA strand separation.
What kind of protein are DnaA proteins? Where do they bind? How many subunits?
DnaA proteins are ATPases. DnaAs bind to the DnaA boxes and additional sites. DnaA is a hexamer, so six subunits.
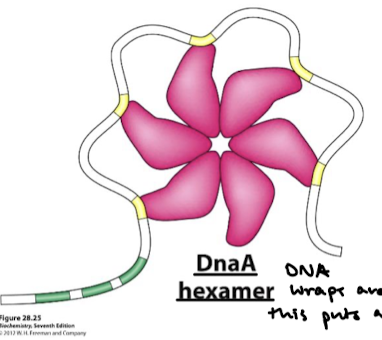
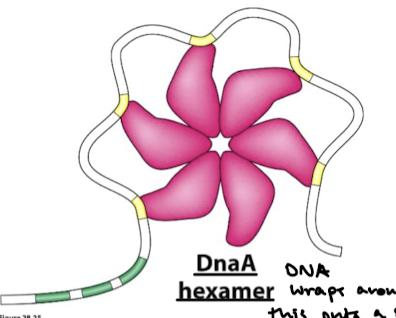
How does DNA wrap around DnaA? What does this supercoiling lead to? What proteins facilitate the DNA bending?
DNA wraps around the complex, resulting in positive supercoiling. Strain leads to the denaturation of nearby DUE sites. Associated proteins facilitate DNA bending.
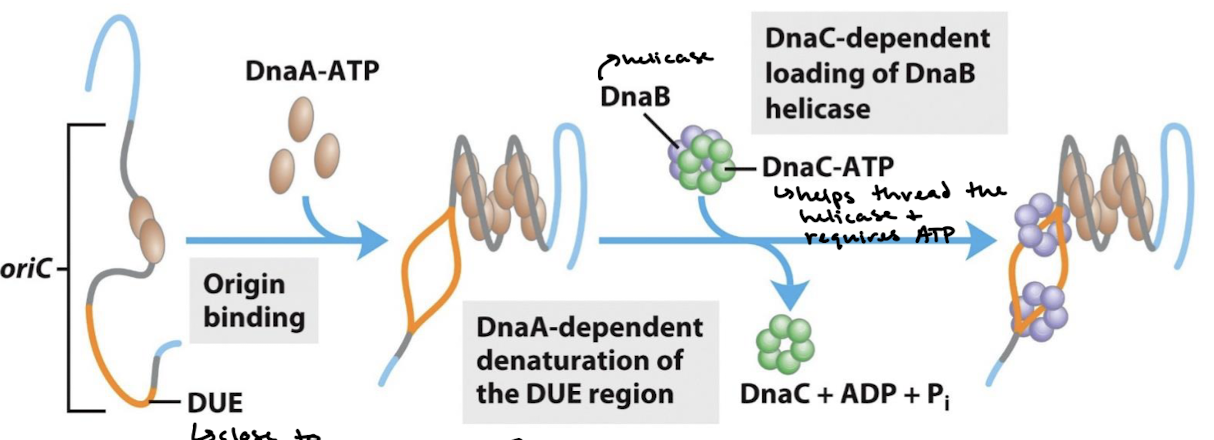
What are the steps in initiation of DNA replication in E. coli involving the proteins DnaA, DnaB, and DnaC?
DnaA-ATP binds the origin, inducing strain on the DNA.
DnaA-dependent denaturation of the DUE region.
DnaC-dependent loading of DnaB helicase (DnaC-ATP helps thread the helicase and requires ATP).
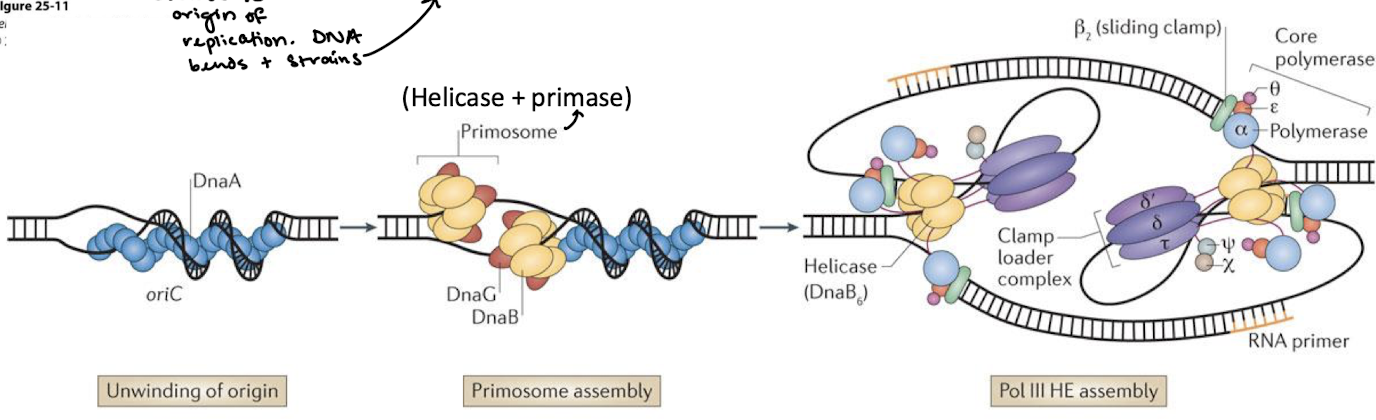
How is the E. coli primosome assembled in initiation of DNA replication?
The exact pathways for assembly of the E. coli primosome and Pol III HE complexes are not yet known.
What are the sites and proteins involved in termination of chromosome replication in E. coli?
Ter sites (termination sites; 23 bp sequences) are found near the opposite side of the initiation/origin and are in two different orientations (clockwise and counterclockwise fork traps).
Tus (terminus utilization sequence) proteins bind to Ter sites.
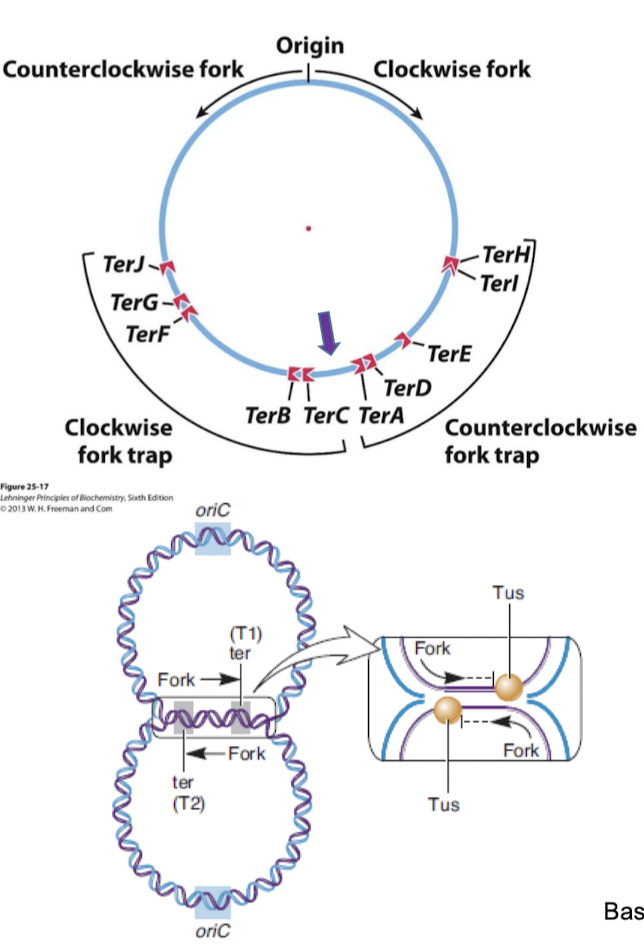
What is the “replisome trap” in termination of replication in E. coli? Why is this beneficial?
Tus-Ter sites form a replisome trap. They are directional traps and allow replication forks to cross them only when they going in the correct direction. If a replisome tries to cross in the non-permissive direction, the helicase becomes inhibited. This coordinates the two forks so that they will both terminate within the terminus region. Having this occur in a fixed region helps with activation of another mechanism to promote chromosomal segregation.
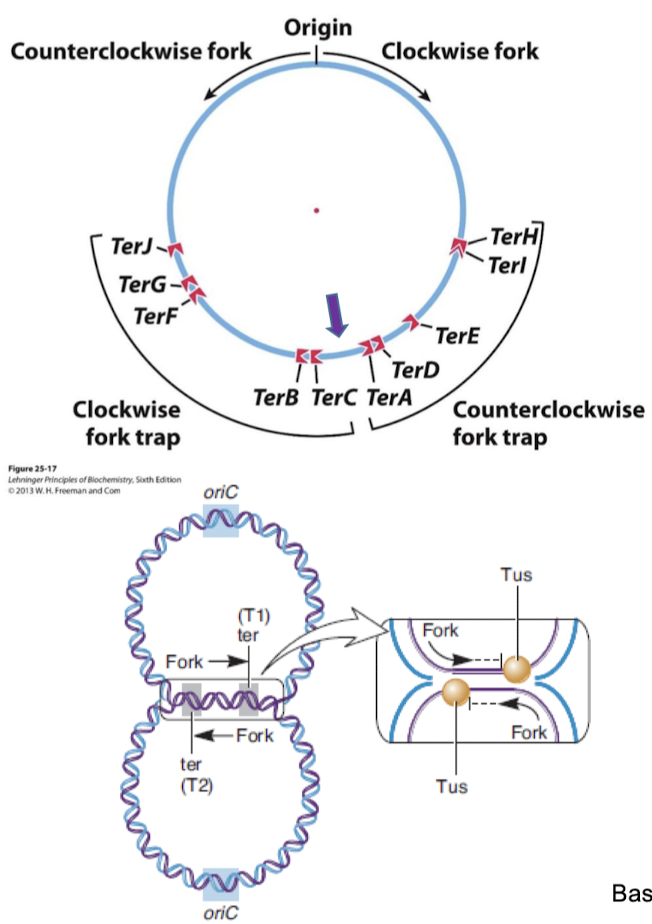
How many enzymes are required in E. coli for DNA replication? What are these enzymes called?
E. coli requires over 20 enzymes and proteins for DNA replication. The set is called the replisome,
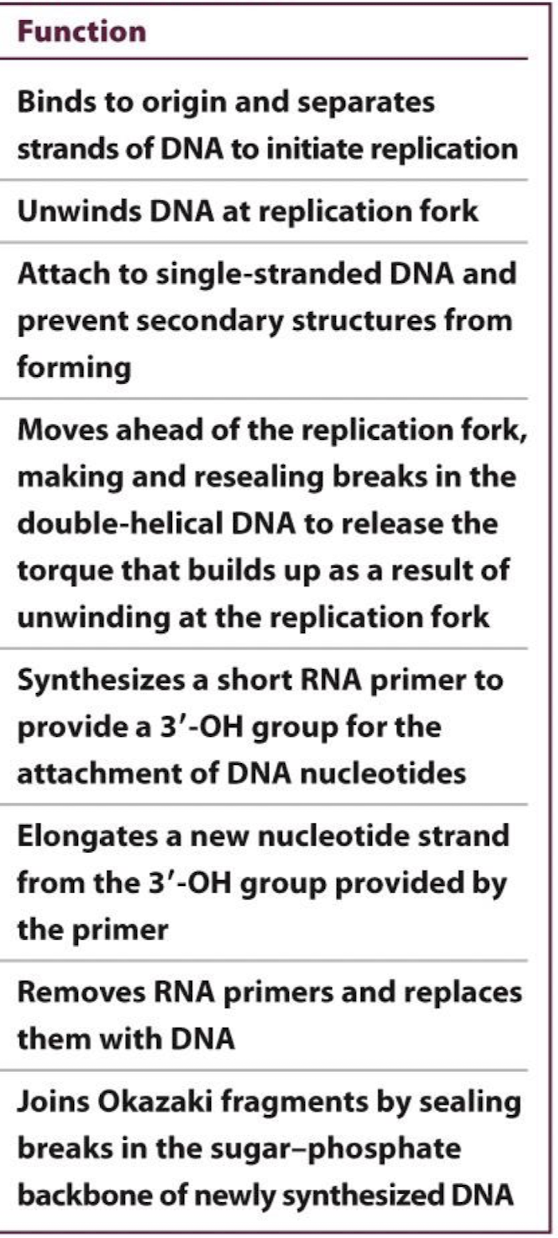
Using the function, identify the correct enzyme/component.

What are nine important differences between replication in prokaryotes versus replication in eukaryotes?
Location
Genome Size
Number of Chromosomes
Structure of Chromosomes
Number of Origins
Origin Defined Sequences
Presence of Origin of Replication Complexes (ORCs)
Distinctive Polymerases
Coordination with the Cell Cycle
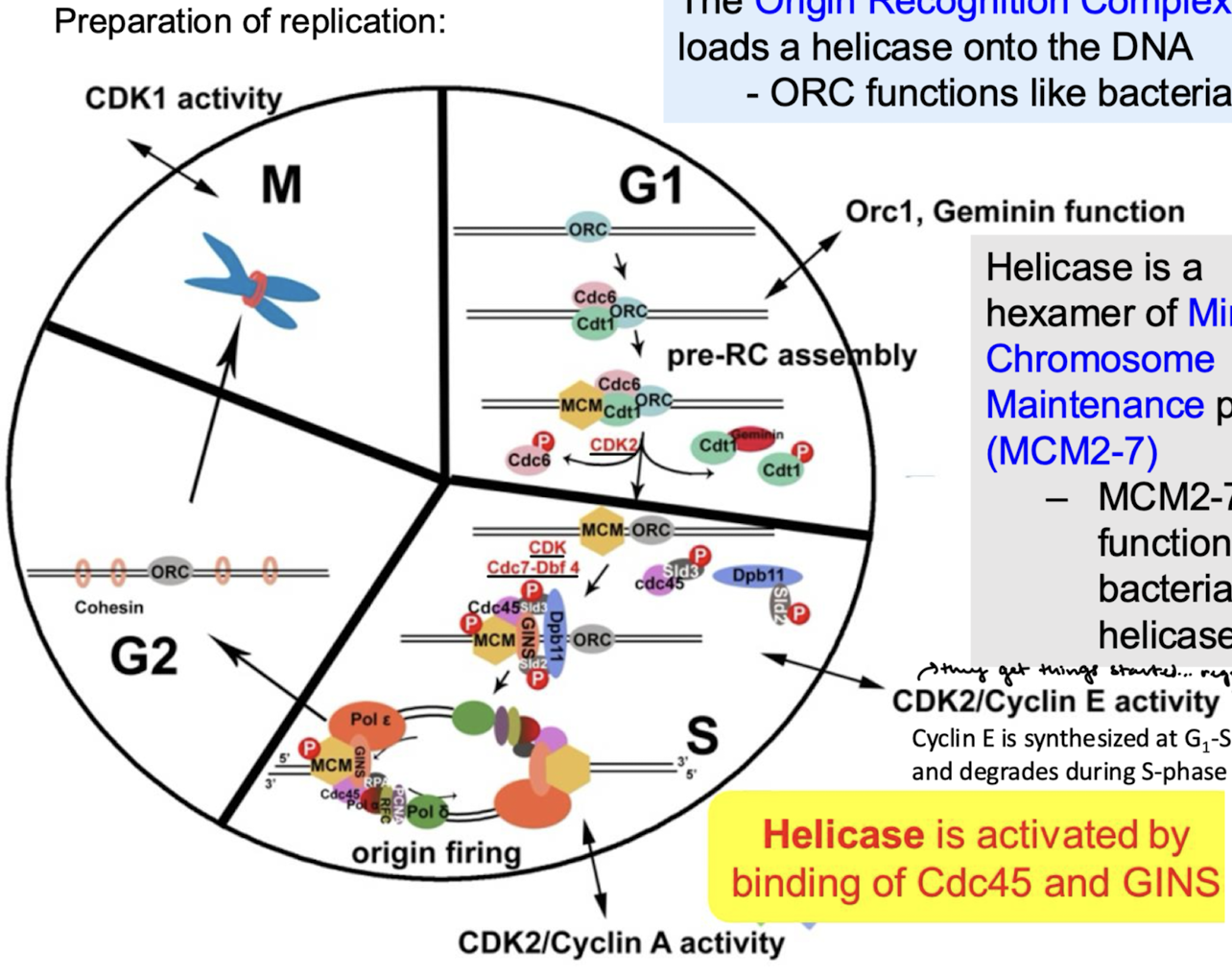
What is the commitment step of eukaryotic DNA replication initiation? When does the commitment step occur in the cell cycle?
Helicase activation. Helicases are activated only in the S-phase of the cell cycle.
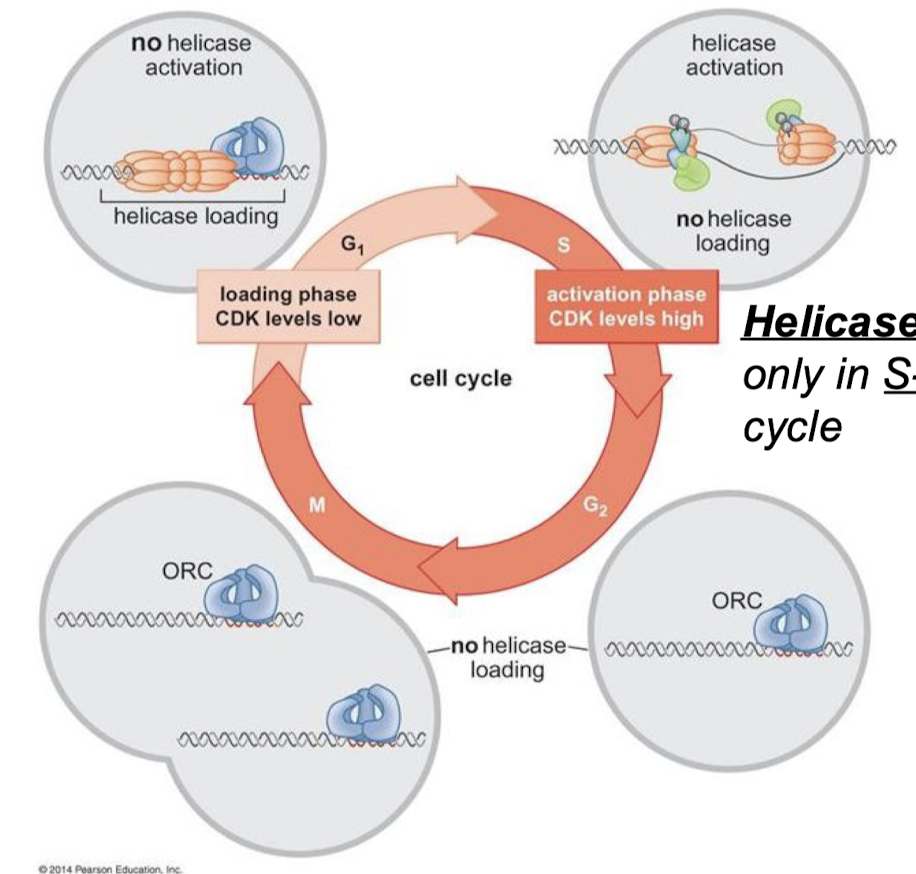
Does high or low CDK levels correspond with the loading phase? With the activation phase?
Loading phase = CDK levels low
Activation phase = CDK levels high
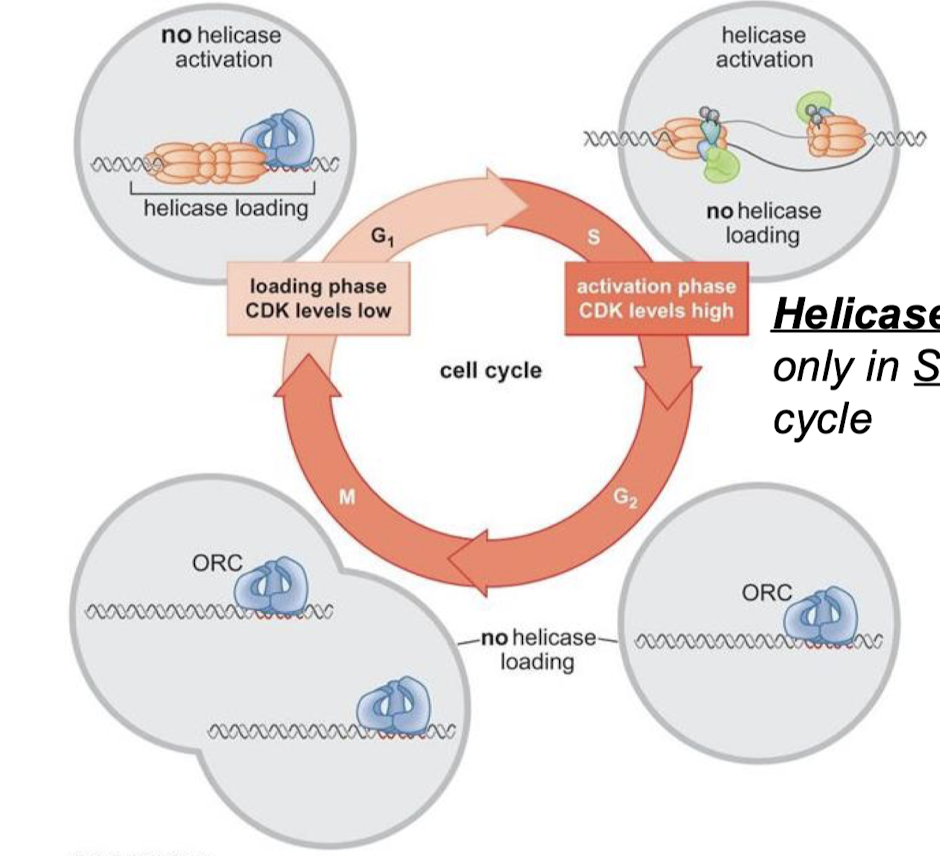
For each cell cycle phase (G1, S, G2, M), describe if there is or is not helicase loading and/or activation.
G1: Helicase loading, no helicase activation (CDK levels low)
S: No helicase loading, yes helicase activation (CDK levels high).
G2: No helicase loading or activation.
M: No helicase loading or activation.
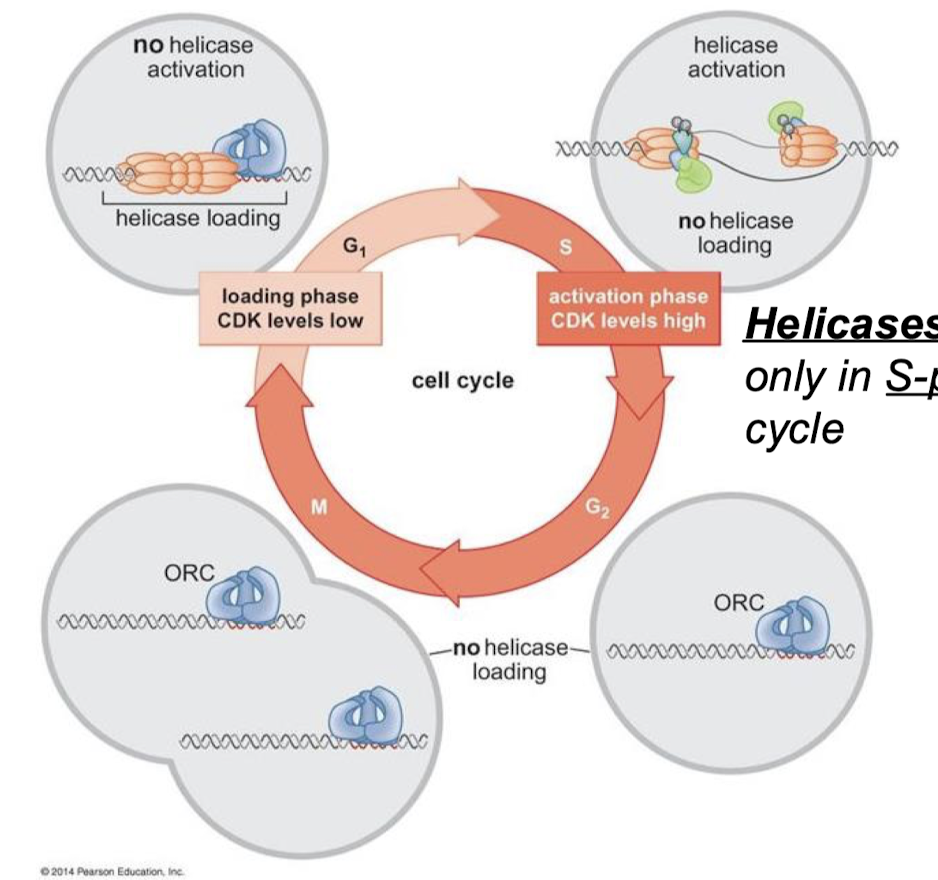
Eukaryotic DNA replication: How many origins of replication? Are initiations random or specific? What complexes bind DNA at the origin?
Around 30,000 origins of replication in humans. Initiations are not random and have certain specificity. The origin of replication complex (ORC) is a multi-subunit DNA binding complex.
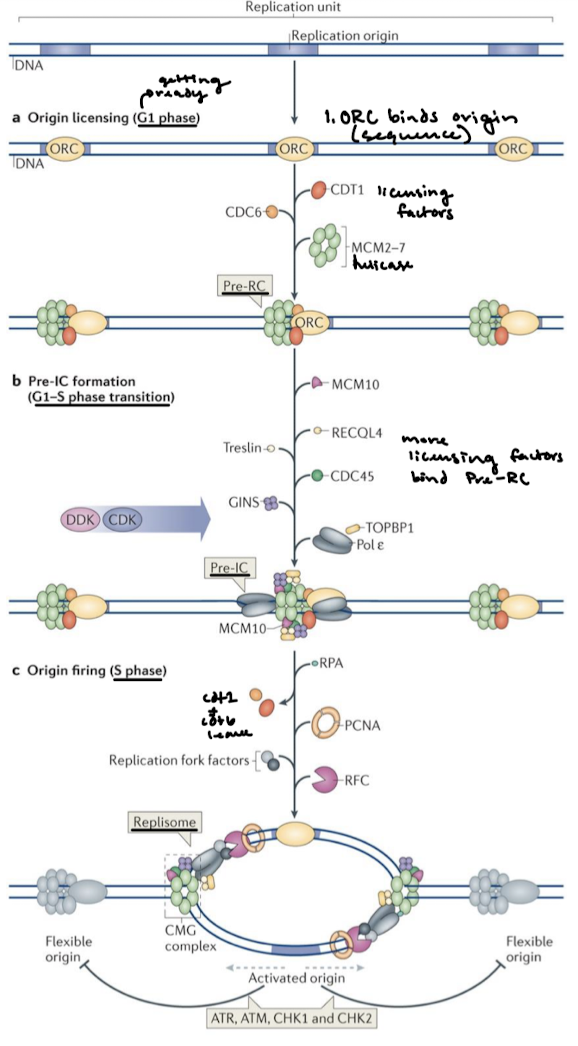
Eukaryotic DNA replication: What complex contains licensing factors Cdc6, Cdt1, and Mcm2-7 (helicase)? What do licensing factors do? How is the helicase complex activated in eukaryotes?
The pre-replicative (initiation) complex (pre-RC) contains licensing factors (Cdc6, Cdt1, Mcm 2-7 (helicase)).
Licensing factors permit the formation of pre-RC and ensure a single round of replication.
Origin firing in the S phase is how the helicase complex is activated. Helicases separate the strands and replication protein A (SSB) binds ssDNA.
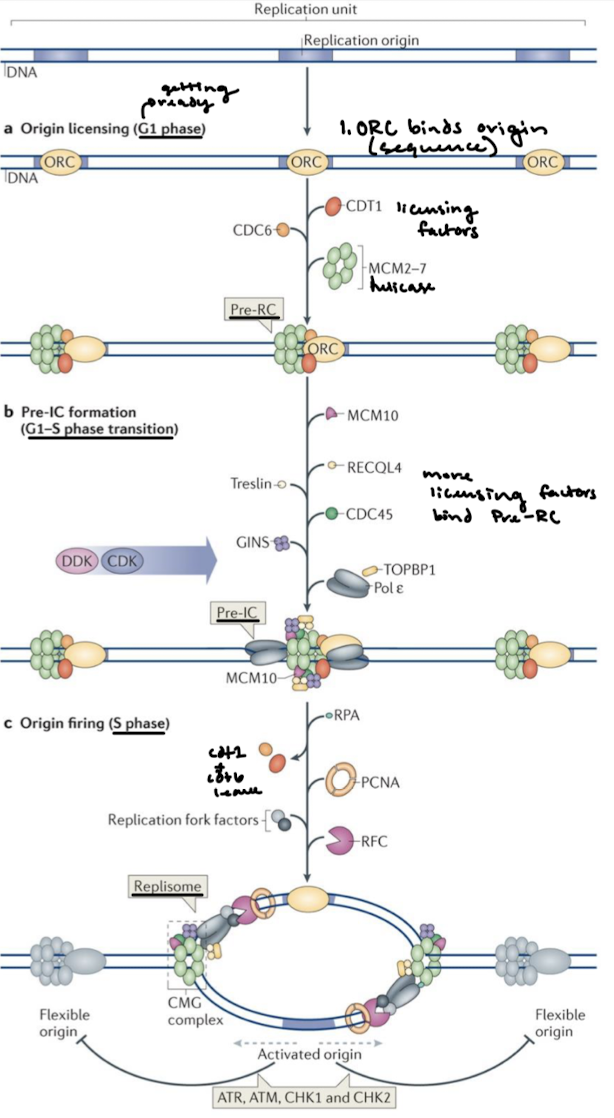
Describe origin licensing, pre-IC formation, and origin firing.
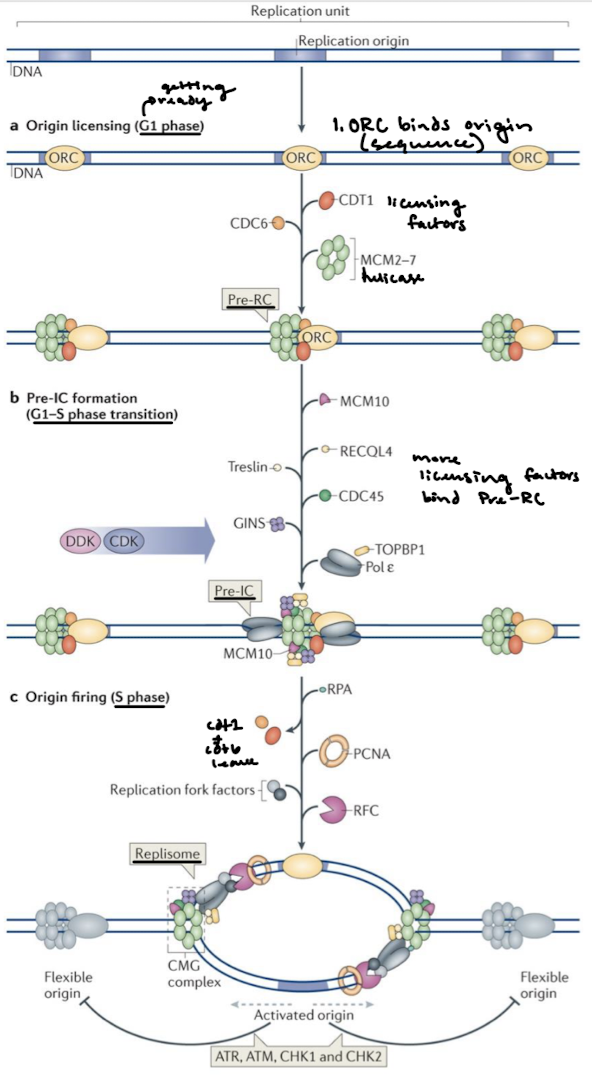
What complex functions like bacterial DnaA? What does it do?
The Origin Recognition Complex (ORC) loads helicase onto the DNA.
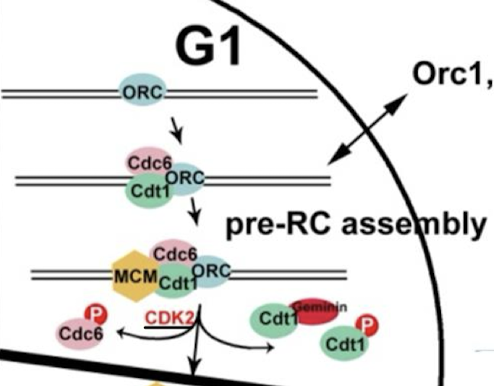
Helicase is a hexamer of ______ proteins, which functions like bacterial ______ helicase.
MCM2-7 (mini-chromosome maintenance proteins); DnaB
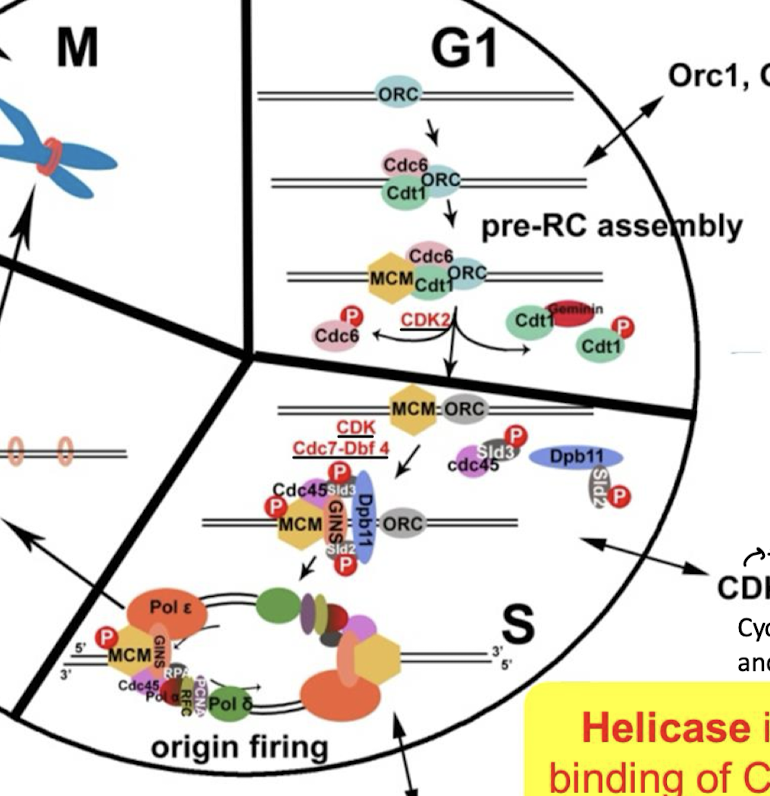
Helicase is activated by the binding of what two proteins?
Cdc45 and GINS
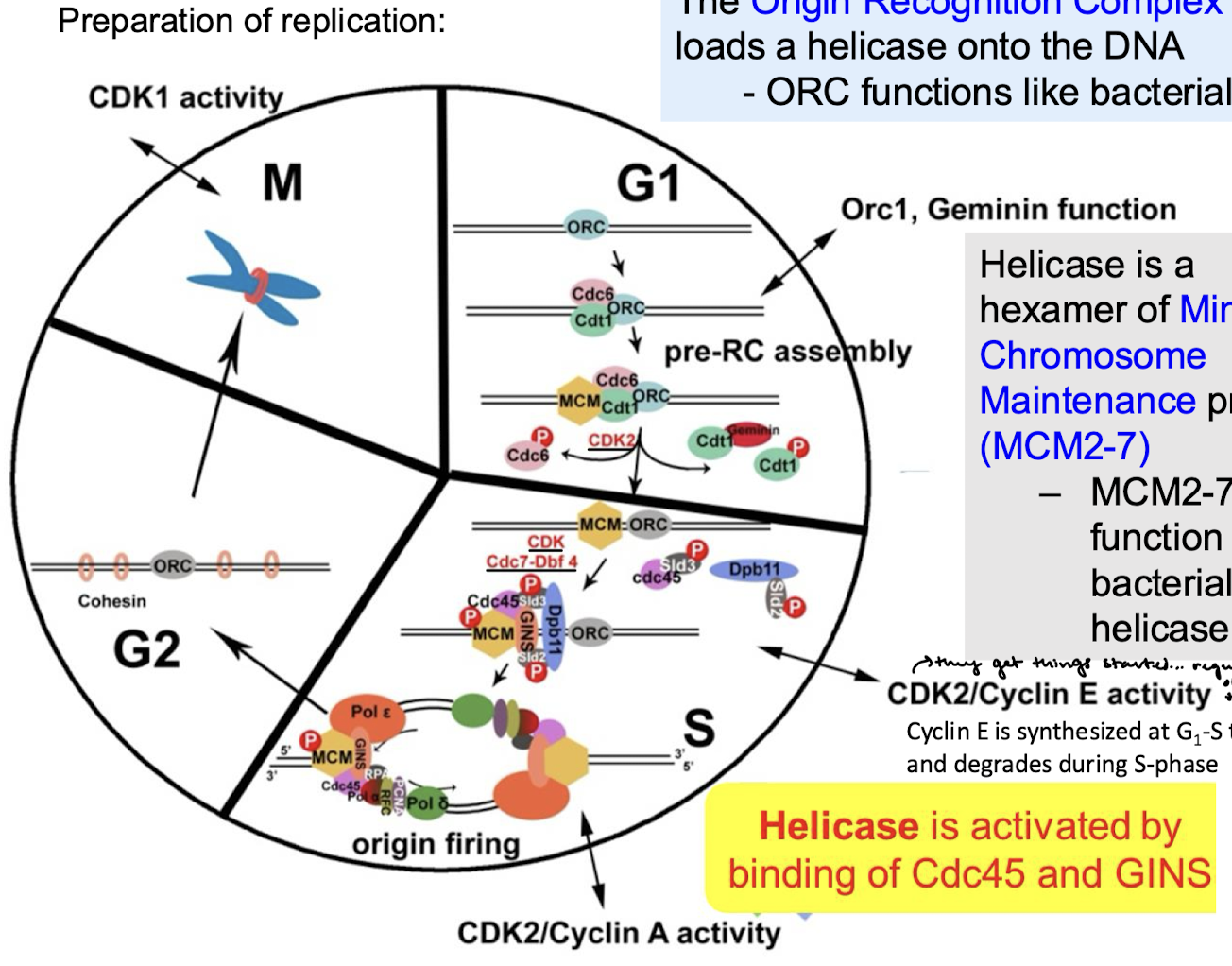
Describe what happens for the preparation of replication in the G1, S, G2, and M phases.
What is DNA polymerase switching in eukaryotic DNA replication? Which polymerases are involved?
Pol alpha is switched out for either Pol delta (lagging) or Pol epsilon (leading) for greater processivity and exonuclease activities (fidelity).
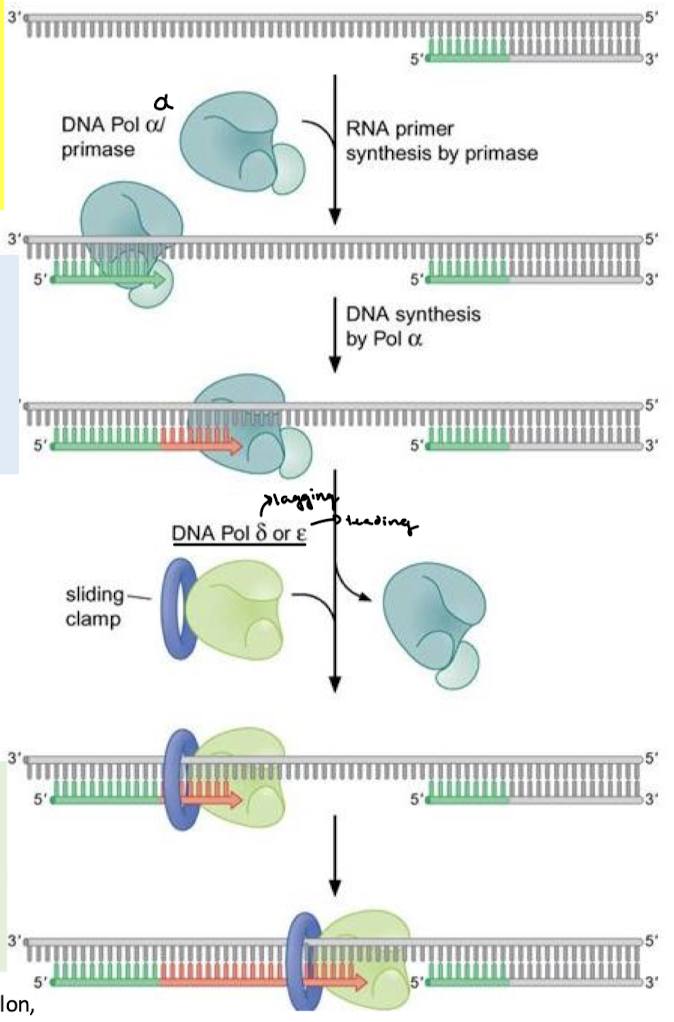
Describe the functions of DNA Pol alpha.
Primase, initiation, Okazaki fragment priming. It lacks a 3’—>5’ exonuclease activity and adds only about 20 dNTPs at a time.
What is the name of the clamp loader and the sliding clamp? What is the clamp loader’s function in DNA polymerase switching?
Protein replication factor C (RFC) is the clamp loader. It displaces Pol alpha and binds proliferating cell nuclear antigen (PCNA), or the sliding clamp.
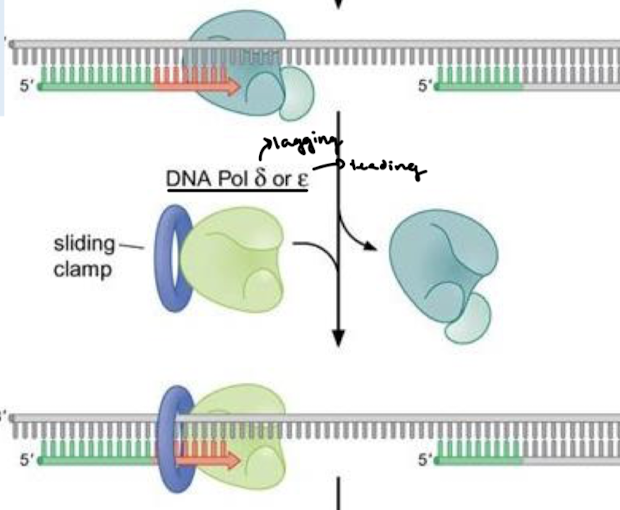
Describe the functions of DNA Pol delta and epsilon.
Delta = lagging strand and epsilon = leading strand. They are highly processive and have 3’ to 5’ exonuclease activity.
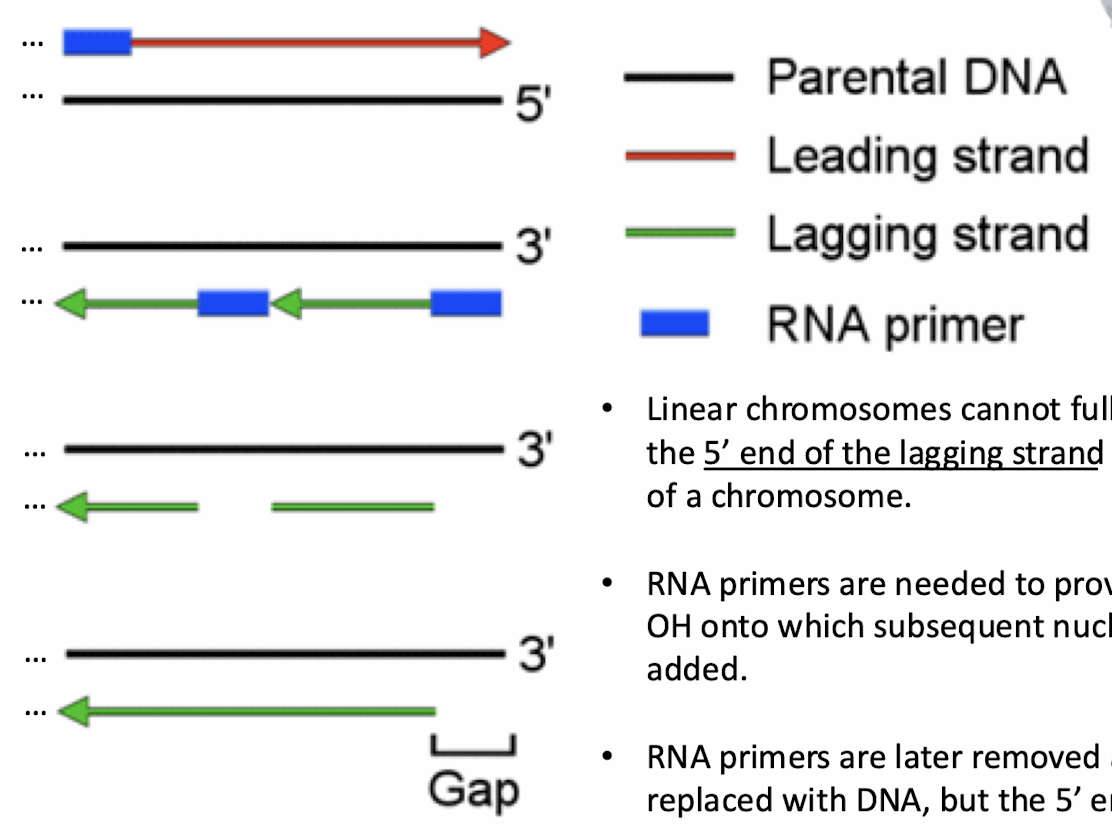
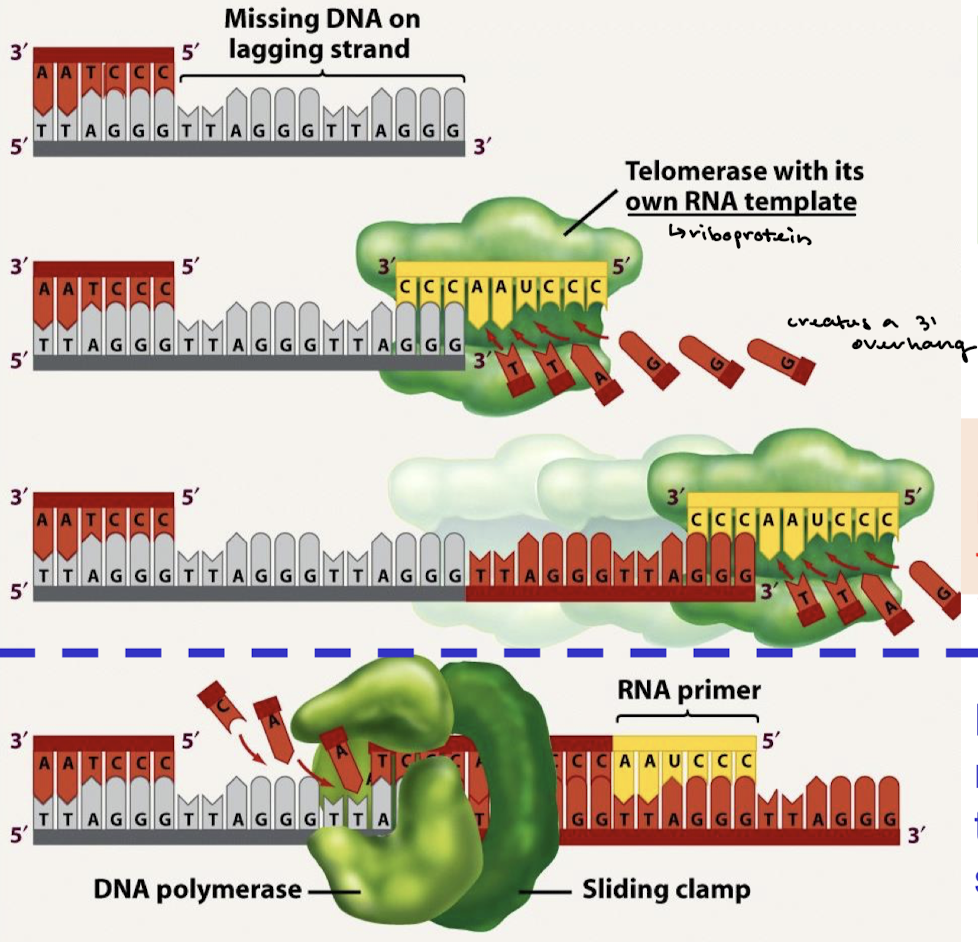
What is the end replication problem?
Linear chromosomes cannot fully replicate the 5’ end of the lagging strand at each end of a chromosome, leaving a 3’ overhang. RNA primers are needed to provide a 3’ OH onto which subsequent nucleotides are added. RNA primers are later removed and replaced with DNA, but the 5’ end cannot be replaced with DNA due to lack of a 3’ OH.
What are telomeres? What is a good analogy for telomeres?
The protective caps consist of (TTAGGG) repeats at the ends of chromosomes in cells. “They are like the tips of shoelaces. If you lose the tips, the ends start fraying” - Elizabeth Blackburn.
What is the function of telomeric DNA? (2)
Protects the organism’s genes from being eroded through successive rounds of DNA replication and shortening.
Aids in preventing separate chromosomes from fusing.
Genome instability is seen in cancers. Why might this be?
Telomeres can become dysfunctional in cancer cells and lead to chromosome instability.
The number of telomeric repeats in human somatic cells appears to decrease with… (2)
Cell divisions
Chronological Age
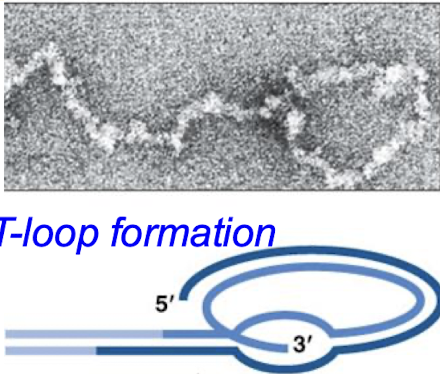
What is telomerase? What kind of enzyme is telomerase? What are its two components?
The enzyme that restores the length of telomeres. Telomerase is an RNA-dependent DNA polymerase (a specialized reverse transcriptase). The two components are the RNA template and the protein (RT) component.
At what end does telomerase bind and increase the length of the end? What enzyme is recruited to synthesize the complementary strand?
Telomerase binds and increases the length at the 3’ end. DNA Pol alpha-primase is recruited to synthesize the complementary strand.
When and where is telomerase active? Which cells contain telomerase activity?
Telomerase is on during fetal development and remains active in various proliferative cells (stem cells, activated lymphocytes, hair follicles, etc.). Germ-line cells (cells involved in the production of sperm and egg) do possess active telomerase activity.
Is telomerase active in adult cells? Is there telomerase activity in cancers?
Telomerase is down-regulated BUT STILL ACTIVE in many adult cells. Telomerase is hyperactive in 80-90% of invasive cancers (which can make the cell line “immortal”).

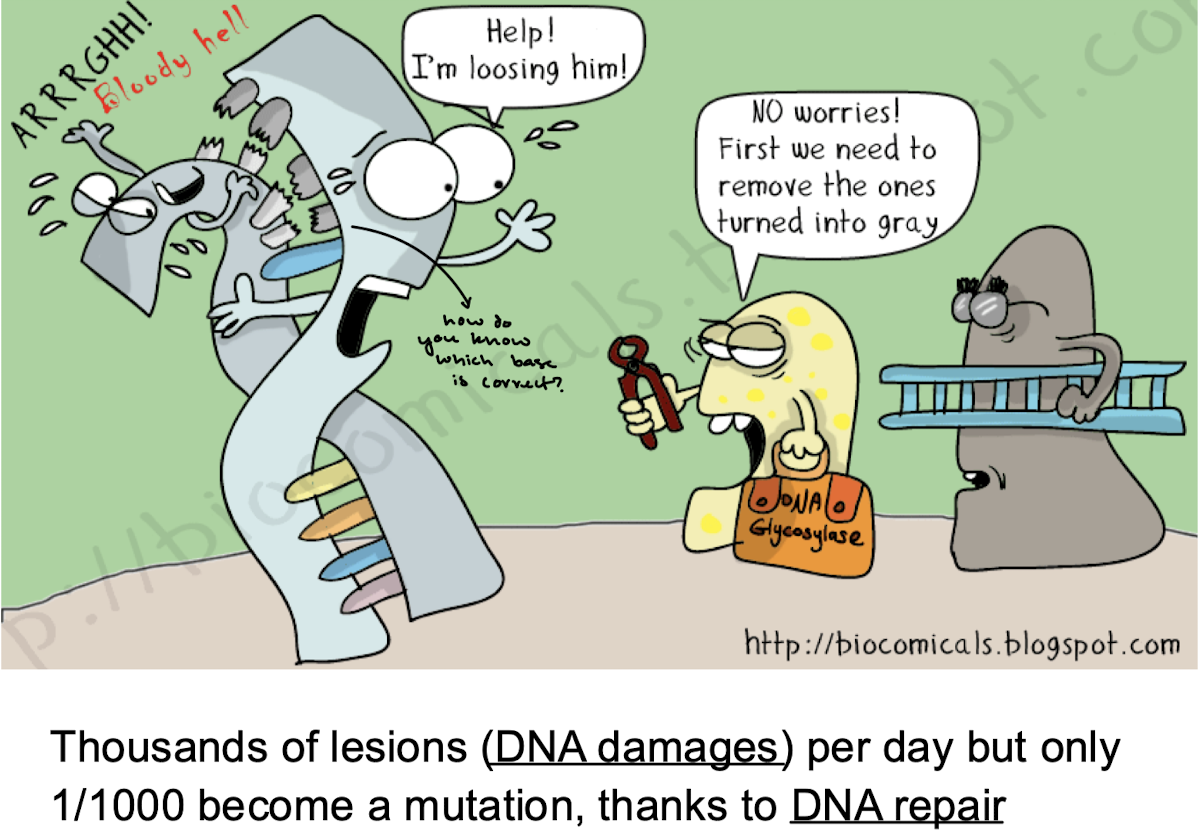
What are the two major problems with mutations? How many damages are there a day and how many become mutations?
A mistake in the germ-line cells will affect all of the following generations.
Normal non-dividing DNA mutations means changes to enzymes and proteins.
Thousands of lesions per day but only 1/1000 become a mutation, thanks to DNA repair.
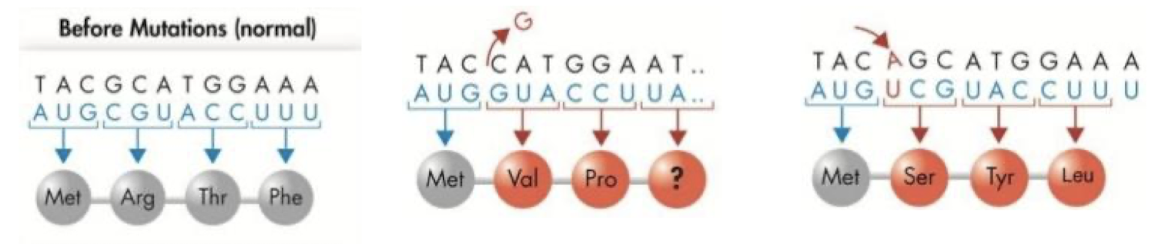
What are the three types of mutations?
Substitutions (point mutations), base deletion, base insertion.
What are the two kinds of substitutions/point mutations?
Transition: Purine to purine or pyrimidine to pyrimidine.
Transversion: Purine to pyrimidine or vice versa.
What is a frame shift and what mutations can result in a frame shift mutation?
Frame shifts change the trinucleotide reading frame for all codons that follow the mutation. Base deletions and insertions can cause frame-shift mutations.
What are the two major sources of DNA damage?
Spontaneous hydrolysis of DNA under normal conditions and in response to mutagens (chemical or radiation).
What are the two kinds of spontaneous hydrolysis of DNA?
Depurination and deamination
What are the three mutagens (chemical or radiation) that result in DNA damage?
Mutagenic chemicals, ultraviolet radiation, and ionizing radiation.
What are three types of mutagenic chemicals?
Base analogues (5-bromouracil; 2-aminopurine)
Base-modifying agents (oxidation, alkylation)
Intercalating agents (ethidium bromide)
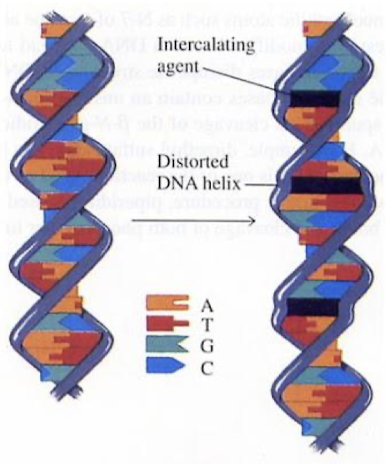
How does Ultraviolet (UV) radiation cause DNA damage?
Sunlight can cause pyrimidine dimer formation.
How does ionizing radiation result in DNA damage?
Such as X-rays, ionizing radiation knocks electrons off of biomolecules to generate highly reactive intermediates.
What mutagenic base analogue is used experimentally to induce mutations?
5-bromouracil
Describe whether the keto or enol form of 5-bromouracil will base pair with adenine or guanine and what makes this possible.
Tautomeric shift from the normal keto form to the enol form changes the 5-bromouracil from an H-bond acceptor to an H-bond donor. The keto form will base pair with adenine and the enol form with guanine.
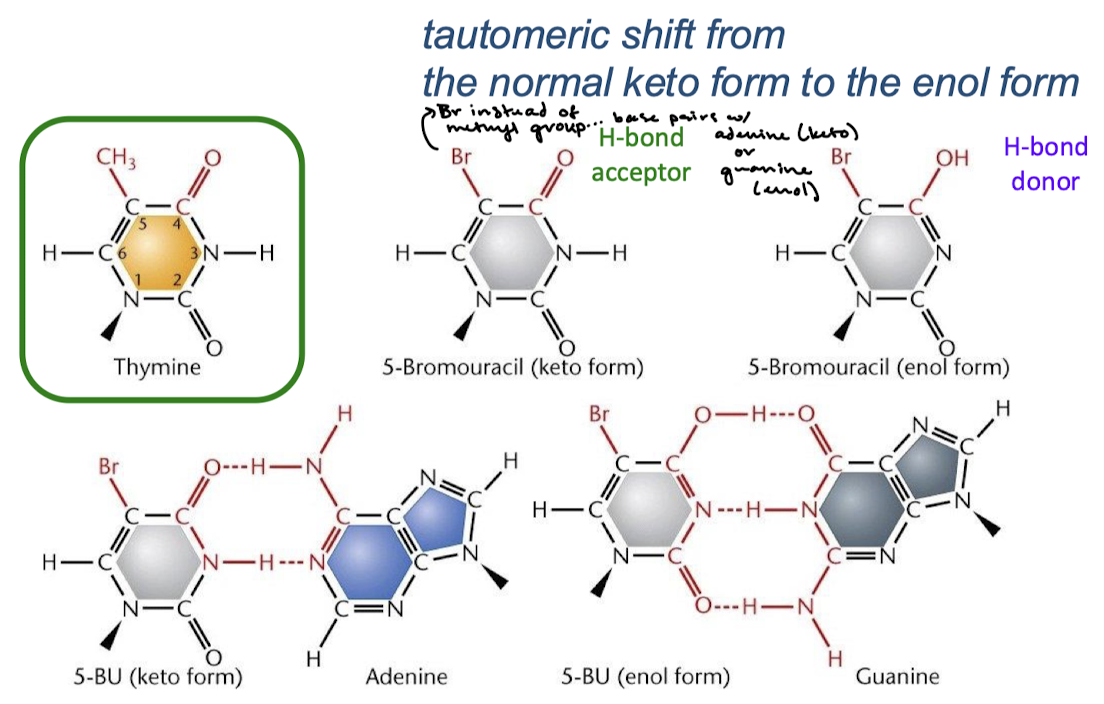
What mutation results from 5-bromouracil.
After one round of replication, an A-T to G-C transition mutation results.
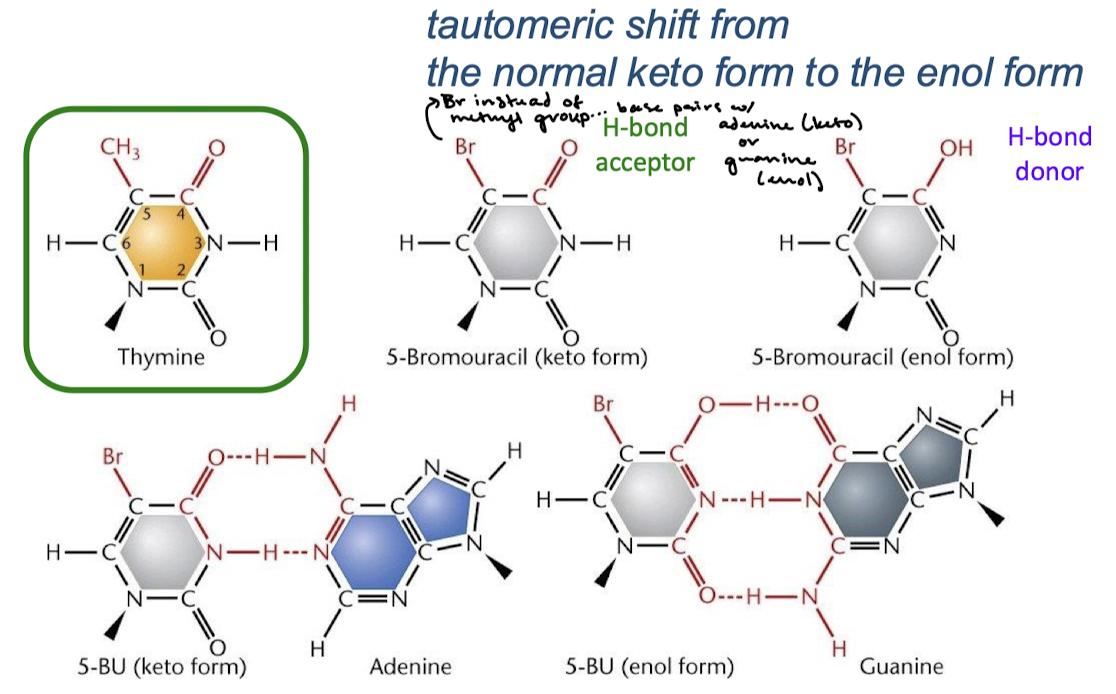
Base modifying agents: Reactive oxygen species (ROS), hydrogen peroxide and hydroxyl radicals (free radicals) chemically modify _______, and is the most common DNA damage from ROS.
Guanine, the oxidation of guanine to 8-oxoguanine.
With what base does the modified guanine pair with? What mutation does this cause?
The modified guanine pairs with adenine. This causes C-G —> A-T transversion.
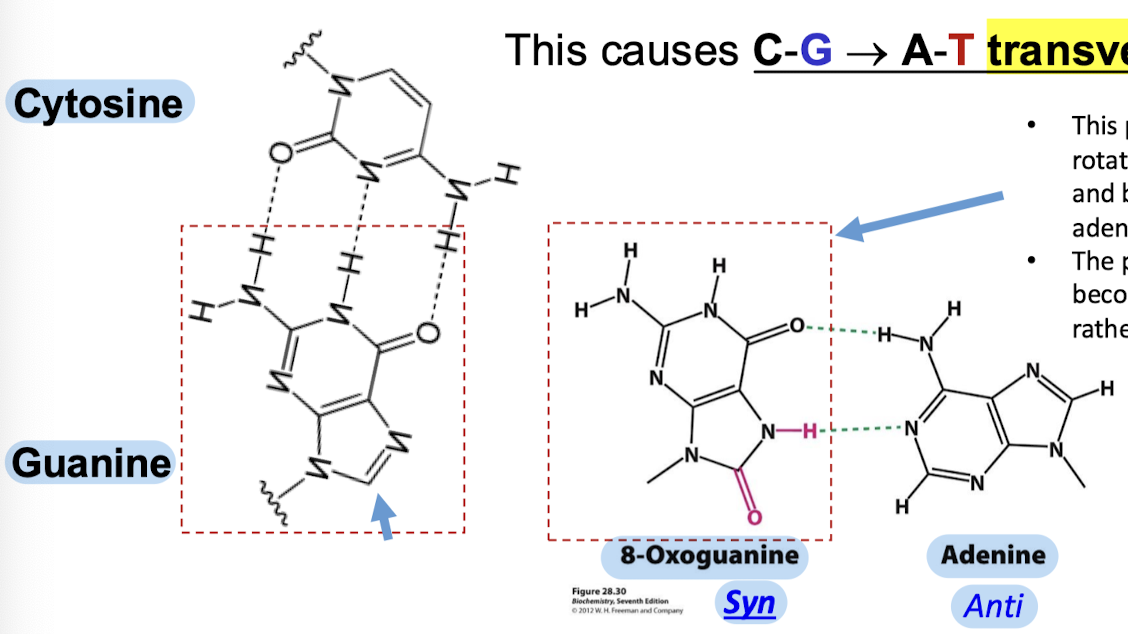
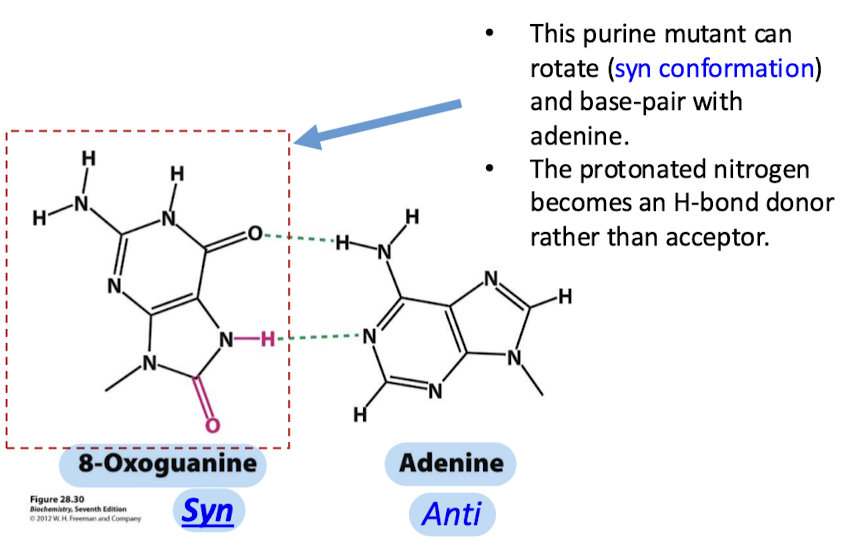
How is 8-oxoguanine able to base-pair with adenine?
The purine mutant can rotate (syn conformation) and base-pair with adenine (anti conformation). The protonate nitrogen becomes an H-bond donor rather than an acceptor.
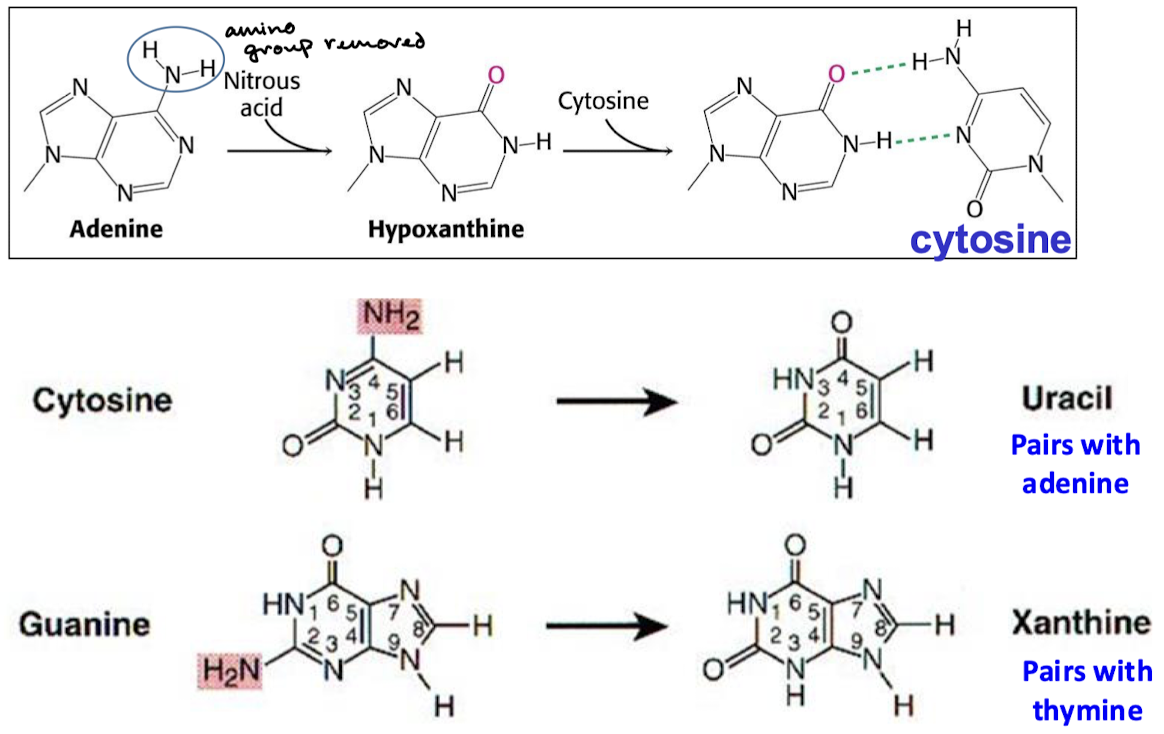
What is the effect of nitrous acid (HNO2)? What are the different results whether this happens with adenine, cytosine, or guanine?
Results in oxidative deamination of the amino group. Adenine becomes hypoxanthine and base pairs with cytosine. Cytosine becomes uracil and base pairs with adenine. Guanine becomes xanthine and base pairs with thymine.
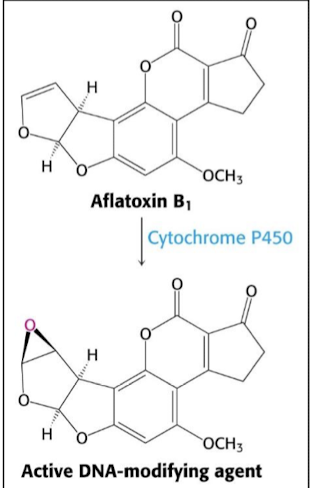
What kind of agent is aflatoxin B1? What does it promote? Where does it bind (what residues)?
Aflatoxin B1 is a powerful depurinating agent that promotes carcinogenesis via apurinic mutations. It binds directly to DNA at guanine residues (at the N7 position), breaking them off and creating an apurinic site.
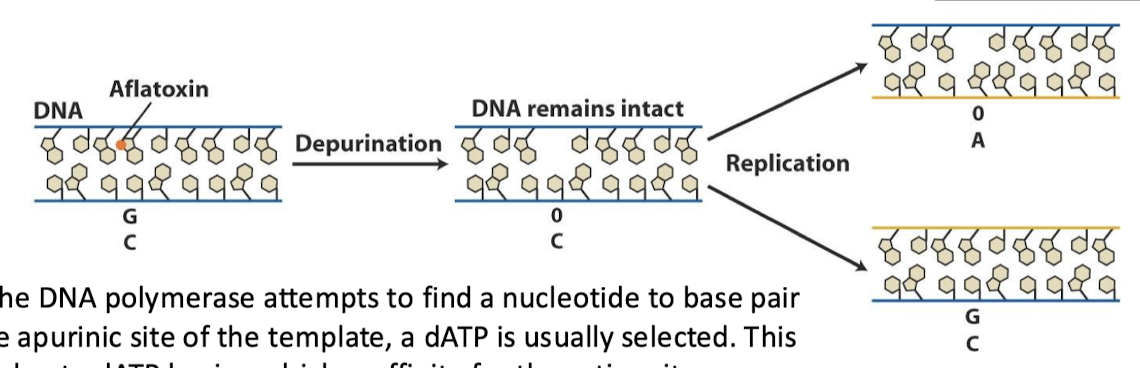
What is the result of an apurinic site induced by aflatoxin B1? What is the mutation that occurs?
When the DNA polymerase attempts to find a nucleotide to base pair with the apurinic site of the template, a dATP is usually selected. This may be due to dATP having a higher affinity for the active site. So, after the second replication, duplex with 0-A will give T-A base pairing. The net effect is a G-C —> T-A transversion.
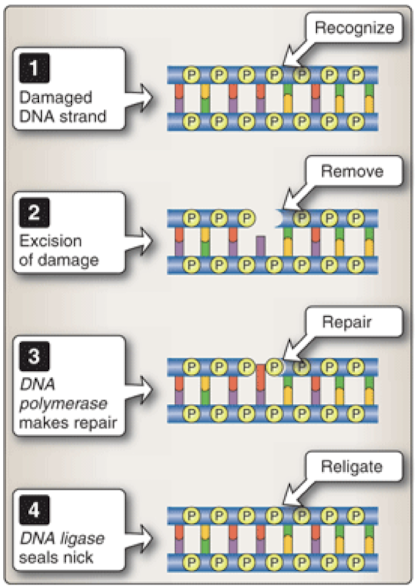
What is the mechanistic outline of DNA damage repair? When are errors repaired?
RECOGNIZE —> REMOVE —> REPAIR
The errors are repaired after replication (post-replication repair).
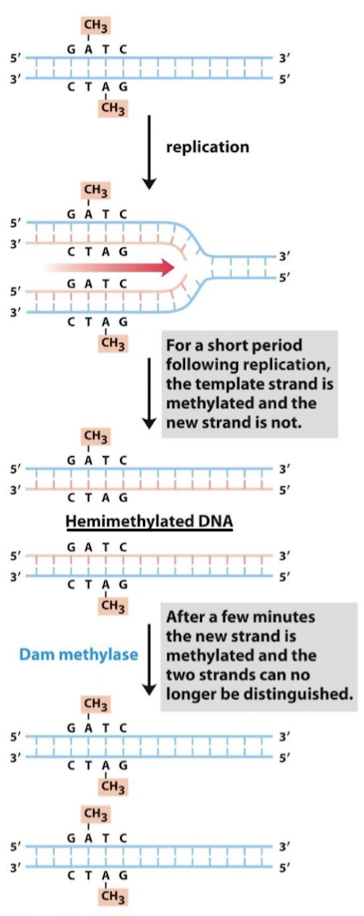
How do repair enzymes “know” which strand is the correct one? What enzyme induces this and where?
In E. coli, the parent strand is methylated. Dam methylase inserts CH3 at adenines in GATC sequence. The newly synthesized strand is unmethylated for a short period after synthesis. Any replication errors must reside in the unmethylated strand.
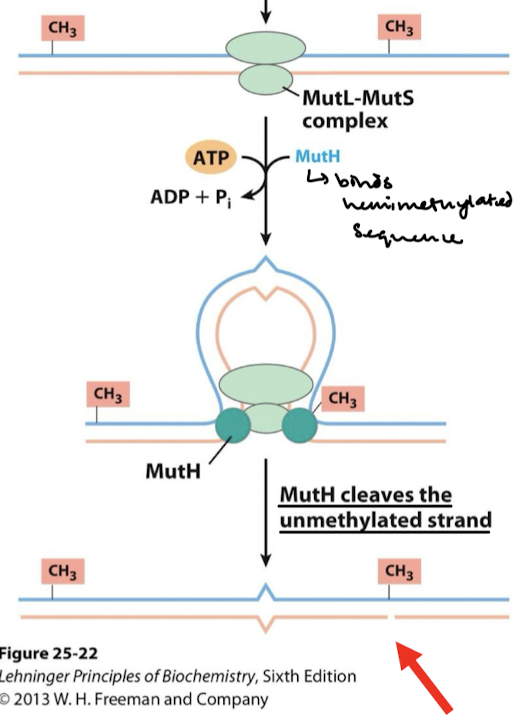
What cleaves the unmethylated strand in the initial part of the repair process?
Methylation-directed mismatch repair system
What are the first two steps in methylation-directed mismatch repair in E. coli?
MutS binds to mismatched base pairs.
MutL is recruited to the complex and activates MutH which binds to hemimethylated GATC sequence.
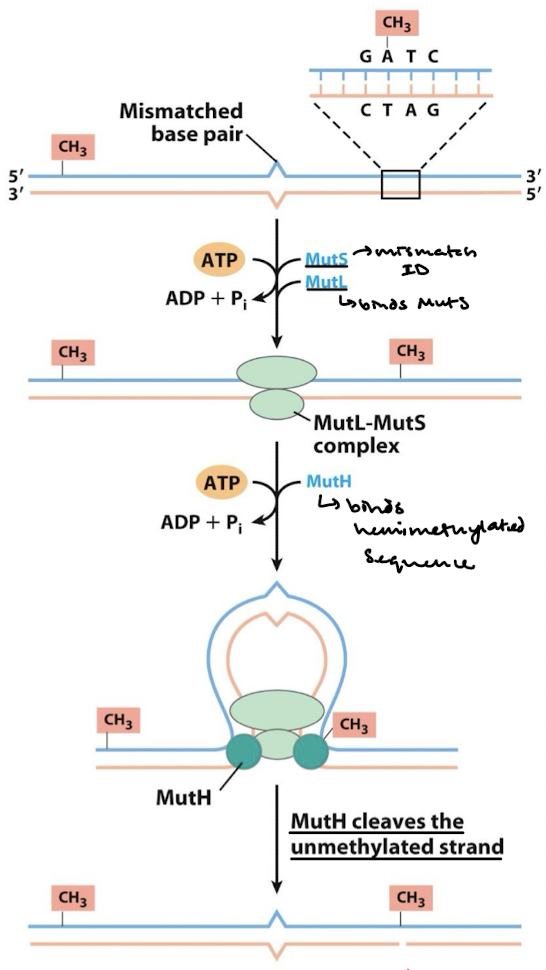
What are the third and fourth steps in methylation-directed mismatch repair in E. coli?
DNA strands thread through the complex until methylated GATC is encountered. The mismatch could be thousands of base pairs away from GATC.
Activation of MutH cleaves the unmethylated strand at the GATC site.
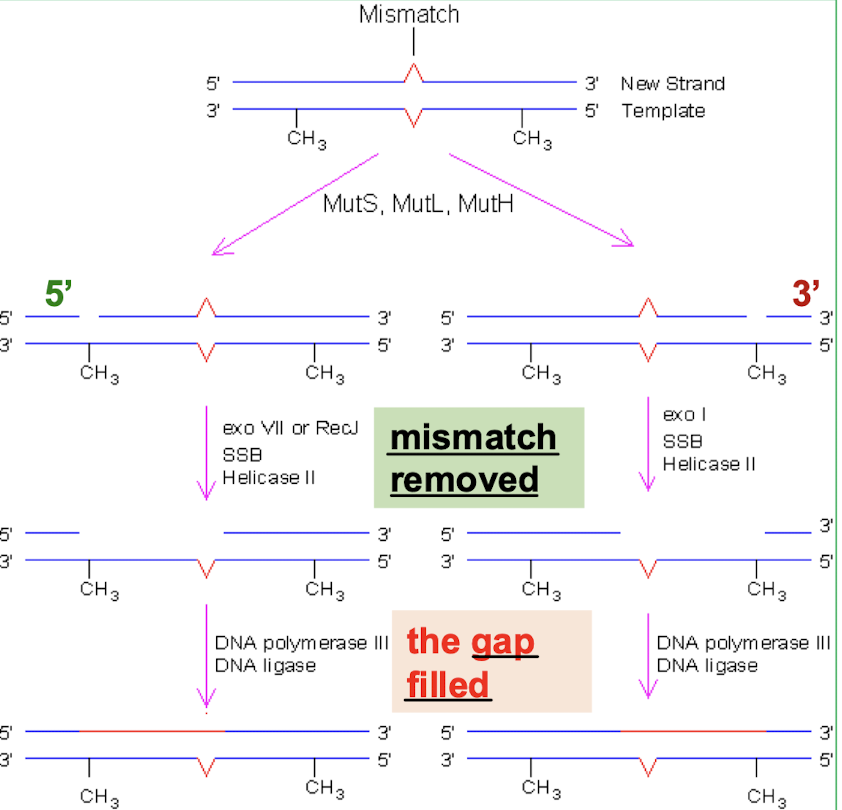
What are the last/later steps in methylation-directed DNA repair in E. coli?
The segment from the cleavage site to the mismatch is removed by exonuclease (and helicase II and SSB proteins) and the gap is filled by DNA Pol II and DNA ligase. There is a difference if the cleavage occurs on the 3’ or the 5’ side.
E. coli and salmonella methylate at _____. Eukaryotes and most other prokaryotes do not. Instead, they have homologs. Are these homologs incolved in mismatch repair?
GATC. These homologs are involved in mismatch repair but are not methylation-directed.
MSH1 to MSH5 are homologs in eukaryotes to ____. MLH1, PMS1, and PMS2 are eukaryotic homologs to ____.
MutS; MutL
Mutations of MSH2, PMS1, and PMS2 are related to…
colon cancer.
Is base-excision repair (BER) in prokaryotes or eukaryotes?
Both (but we are describing using prokaryotic system).
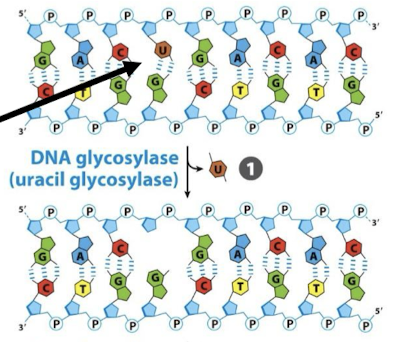
Are the DNA glycosylases in BER generic or specific? What does DNA glycosylase do and where? This is the first step in BER.
Specific! Each type of DNA glycosylase recognizes a specific kind of damaged base and cleaves between the base and deoxyribose in the backbone.
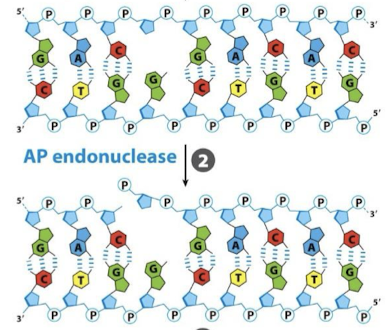
What is the second step in BER?
An apurinic/apyrimidinic (AP) endonuclease cuts the phosphodiester backbone.
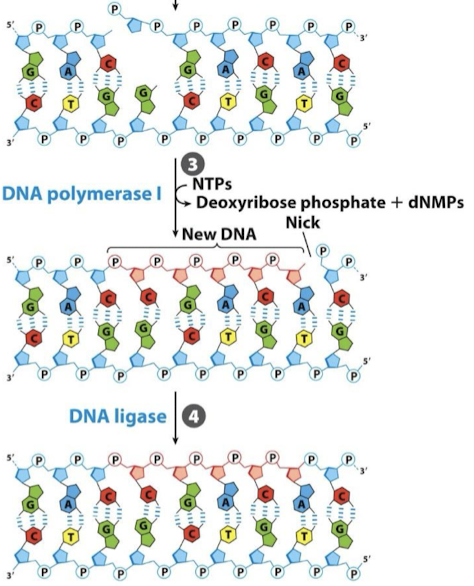
What is the third and fourth step in BER?
DNA Pol I fills the gap (has 5’ to 3’ exonuclease activity). DNA ligase seals the backbone with a phosphodiester bond.
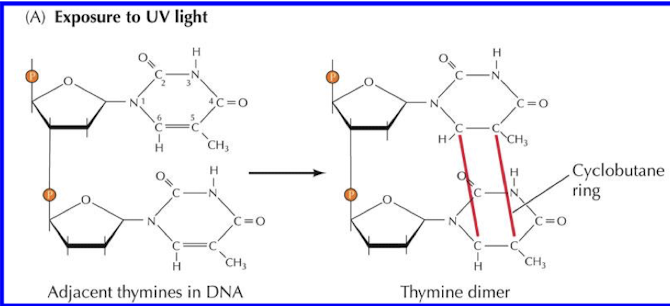
Why do thymine-thymine (pyrimidine) dimers need to be repaired?
Pyrimidine dimers stop replication and transcription.
What are the two ways of repairing a pyrimidine dimer?
Direct/photoreactivation repair (light reaction): Reverses the mutation with light energy.
Nucleotide-excision repair (NER): Excision of dimer (uvr gene products), filling the gap with DNA Pol I, and sealing the gap with ligase.
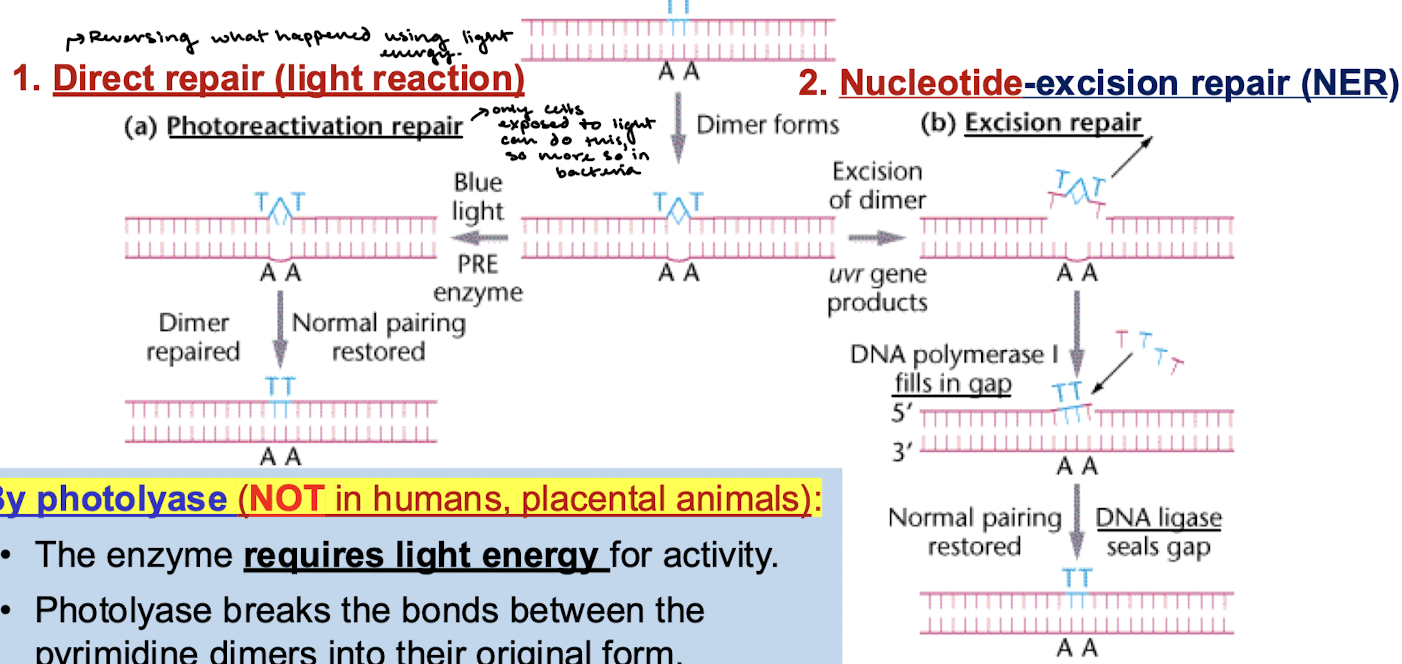
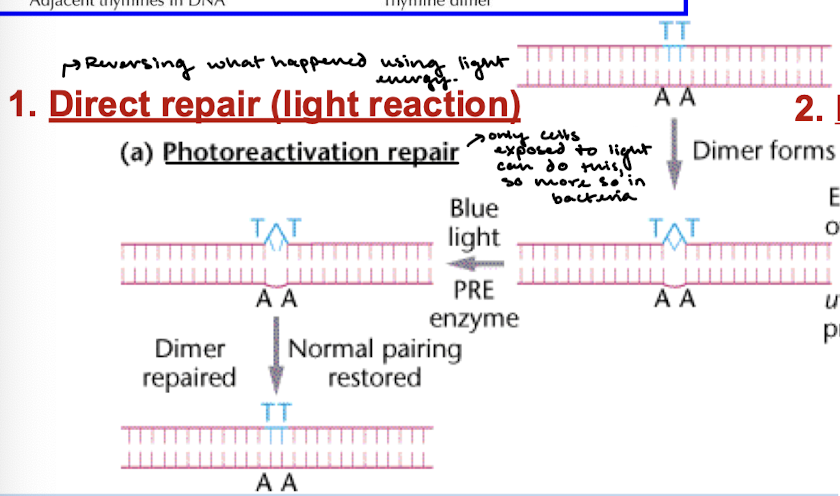
In what organisms does photoreactivation repair occur? By what enzyme? What does this enzyme require?
Repair is catalyzed by photolyase and does not occur in humans and other placental animals. The enzyme requires light energy for activity. Photolyase breaks the bonds between the pyrimidine dimers into their original form.
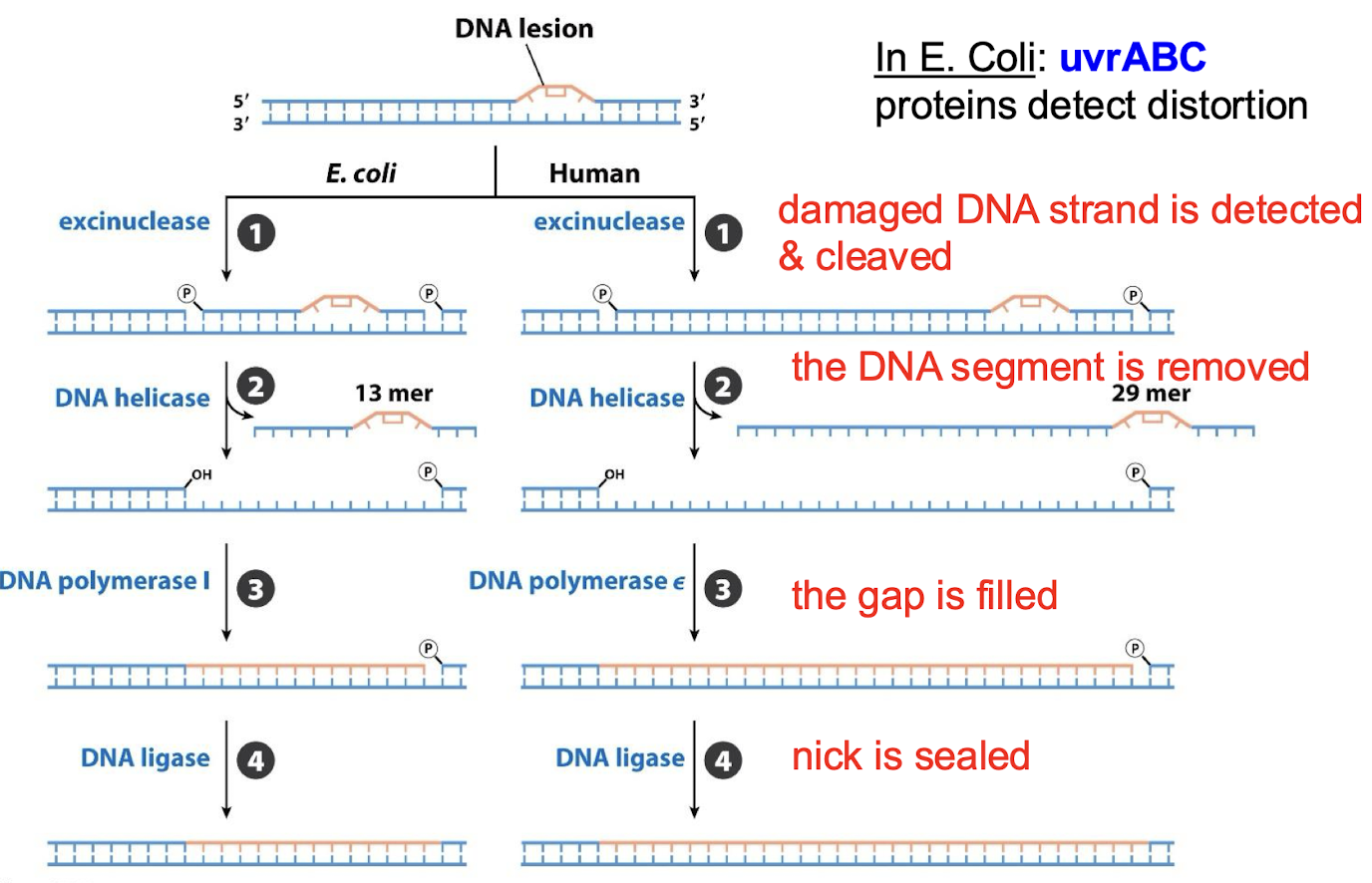
What are the four steps in nucleotide-excision repair (NER)?
The damaged DNA strand is detected and cleaved by an exconuclease.
The DNA segment is removed by DNA helicase.
The gap is filled by DNA polymerase I
The nick is sealed by DNA ligase.
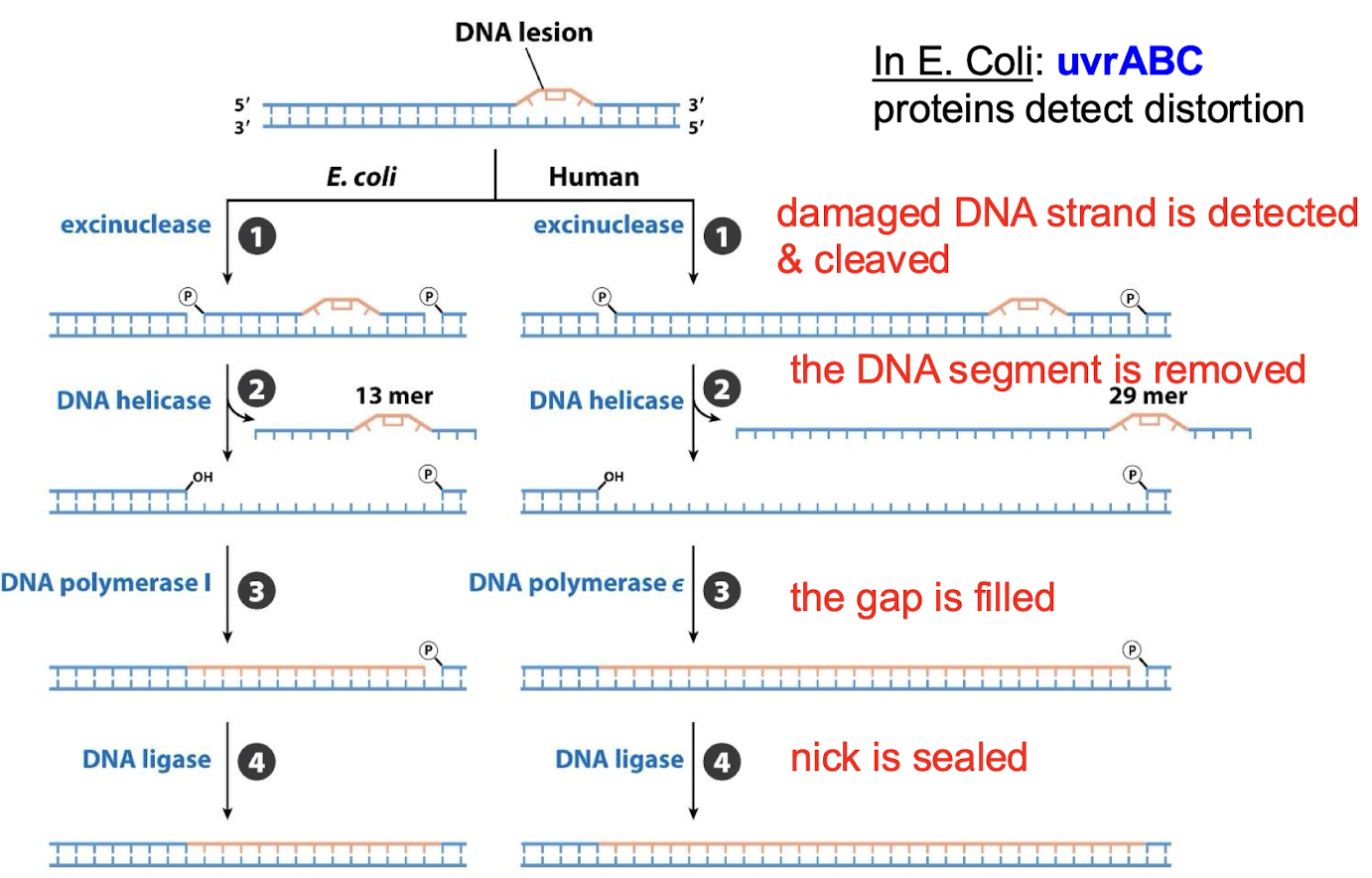
Is NER pathway similar across organisms? What are the proteins that detect distortion in E. coli NER?
The general pathway is similar in all organisms. In E. coli: uvrABC proteins detect distortion.
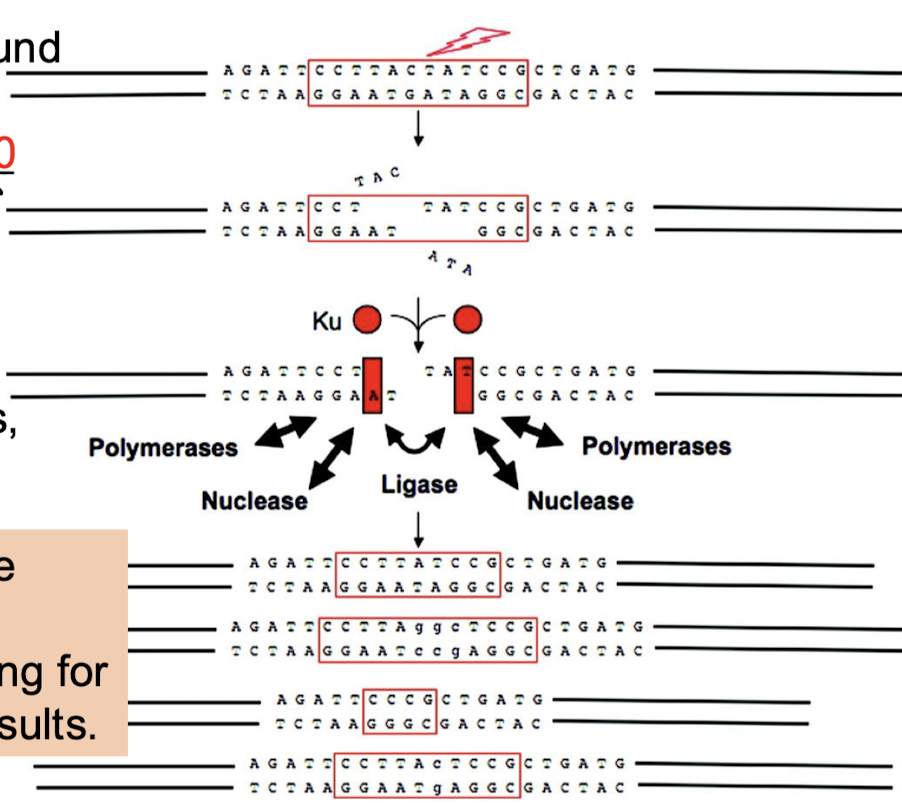
What is the function of non-homologous end joining (NHEJ)?
Repairs double-stranded breaks in DNA. NHEJ does not return the local DNA to its original sequence and is highly error-prone, thus accounting for the wide range of end results.
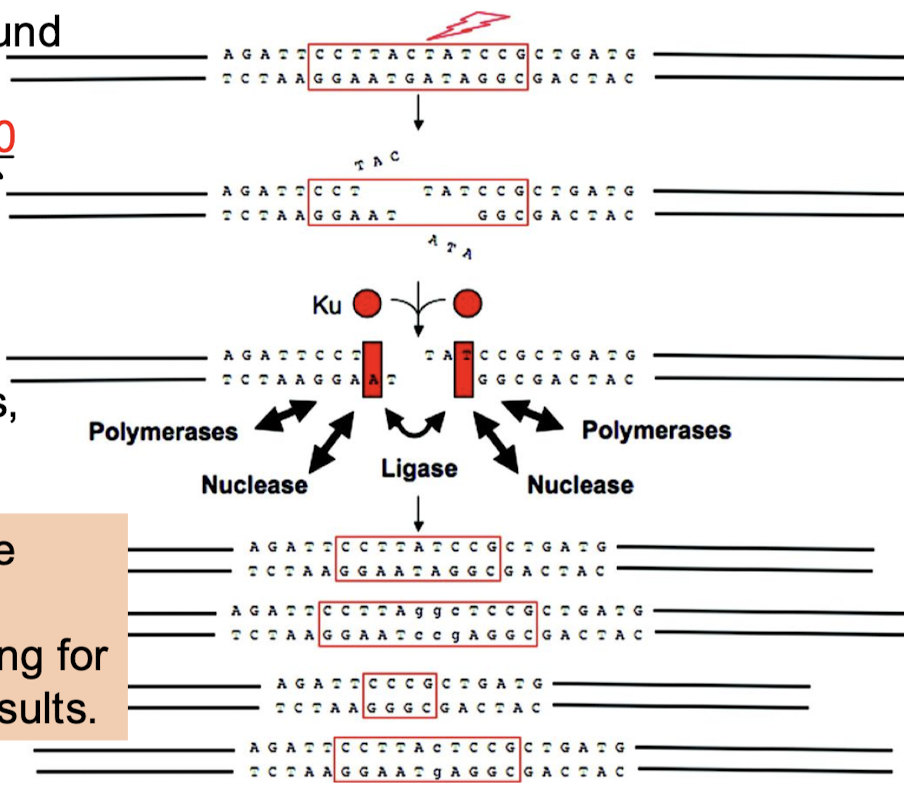
What is the function of Ku70 and Ku80 in NHEJ?
The free ends are bound by heterodimers of Ku70 and Ku80, which keep the DNA close together to stabilize the ends and improve the binding equilibrium of the nuclease, polymerases, and ligase.
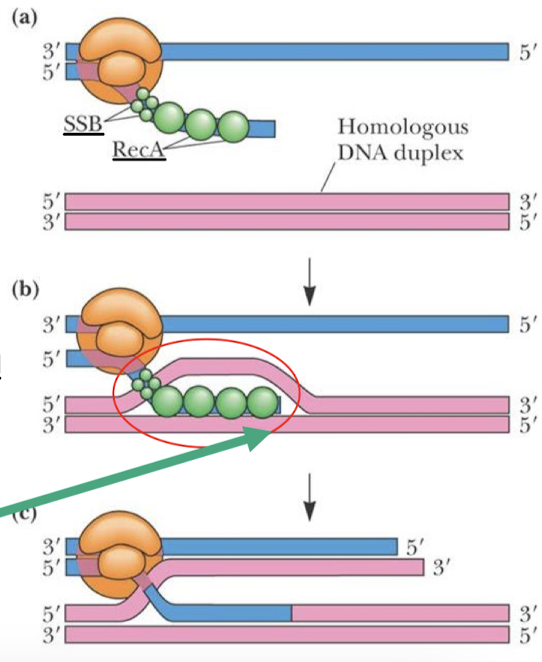
What is the purpose of homologous recombination?
To repair double-stranded breaks. Homologous recombination is an accurate and efficient system for repair because it uses homologous chromosomes as a template.
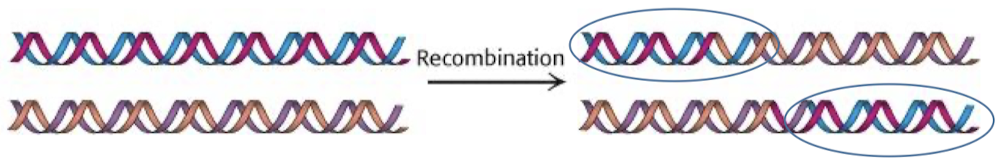
What strands are involved in homologous recombination?
DNA molecules with a free end (after ds break) can recombine with a DNA molecule that does not have free ends. A free 3’ is base-paired to contiguous strand, which can now act as a primer for DNA synthesis.
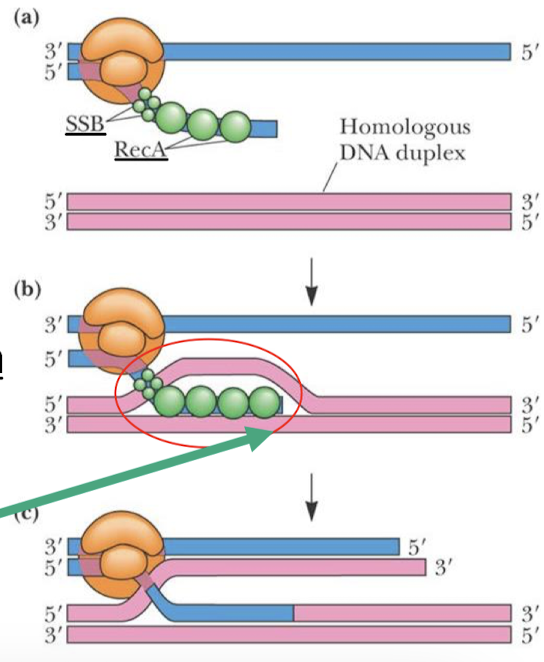
What enzyme is involved in homologous recombination in prokaryotes and in humans? What does it promote?
RecA (RAD51 in humans) can initiate recombination by promoting strand invasion —> D (displacement) and loop formation.