Lecture 4 - DNA Repair
1/52
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
53 Terms
DNA errors can be _____ or _____ and can be introduced by several factors.
induced; spontaneous
can be induced by:
DNA replication
free radicals
ionizing radiation
induced mutations occur due to a physical or chemical external agent and spontaneous mutations occur because of natural processes in cells
Mutations are not always bad
some have consequences like being deleterious or advantageous
major source of genetic variation which drives evolutionary change to allow organisms to respond to changing environments
hotspots in chromosomes have more mutations than other areas
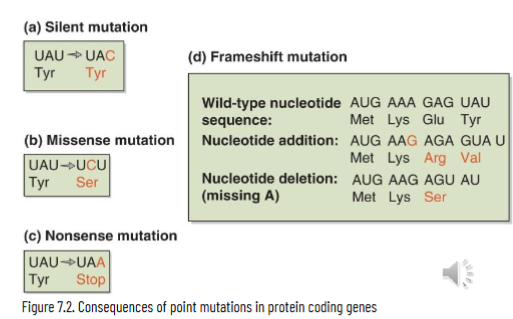
Point Mutations
single nucleotide change
transition mutation: pyrimidine replaced by pyrimidine or purine for purine
transversion mutation: purine is substituted for pyrimidine (A/G ←→ T/C)
protein coding genes with point mutations can be classified as:
silent/synonymous: nucleotide substitution that doesn’t change the amino acid encoded
missense/non-synonymous: nucleotide substitution that results in change of amino acid encoded
frameshift: insertion or deletion of nucleotide(s) that alters reading frame
nonsense: nucleotide substitutions that results in creation of stop codon and makes a truncated protein
Phenotypic Consequences of Point Mutations
consequences are based on where the mutation happens in the cell
non-coding RNA: may alter structure and function
non-coding gene: affect gene regulation and splicing
protein coding gene: alters DNA sequence but specific effect varies
Trinucleotide Repeats
type of microsatellite
increases genetic instability in some areas of the genome since they can adopt a variety of secondary structures
repeats can expand or contract from slippage of the newly made strand on the original strand during DNA synthesis so that misaligned repeats pair up
considered a dynamic mutation because the number of triplets in the disease gene continues to increase as the disease gene is inherited
Consequences of Trinucleotide Expansions
inverted repeats → cruciform structure
repeat with mirror symmetry → triplex DNA
G-rich sequences → G-quadruplex
3 Classes of DNA Damage
single base changes
structural distortion
DNA backbone damage
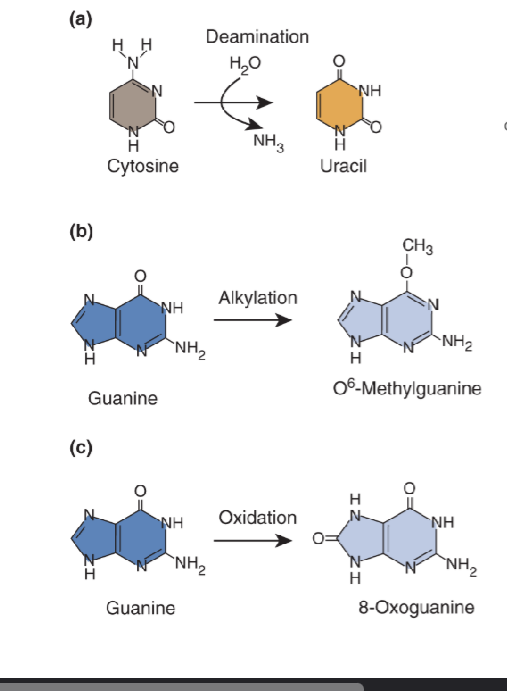
Single Base Changes
conversion: mutagen changes the base on the nucleotide in a DNA sequence
generally causes minor structural distortion so might not block replication or transcription
change base-pairing can result in transition or transversion after replication
Intrinsic and Extrinsic Agents
extrinsic agents: X-rays (oxidiser)
intrinsic agents: free radicals from metabolic processes (oxidisers), replication mistakes
DNA Backbone Damage
structural changes in helix impede replication or transcription
may happen as a consequence of pyrimidine dimers generated by UV light where adjacent pyrimidines on the same strand are covalently linked to each other preventing their base pairing with purines in the complementary strand
intercalating agents change the base stacking interactions
base analogs → different base pairing interactions that affect the shape of DNA
structural changes may also be caused by DNA adducts when DNA is chemically bonded to a cancer-causing substance
other bulk may be introduced by covalently attaching proteins to nitrogenous bases of DNA (disrupts helical structure)
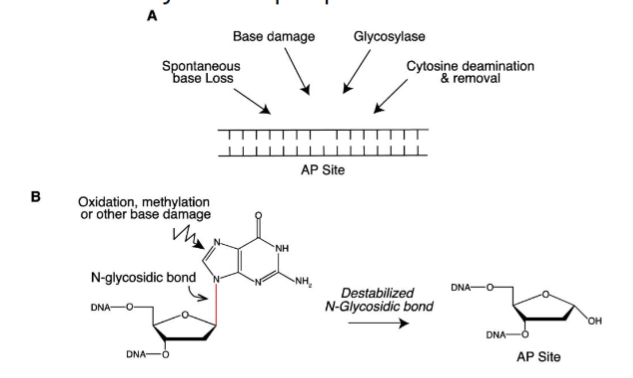
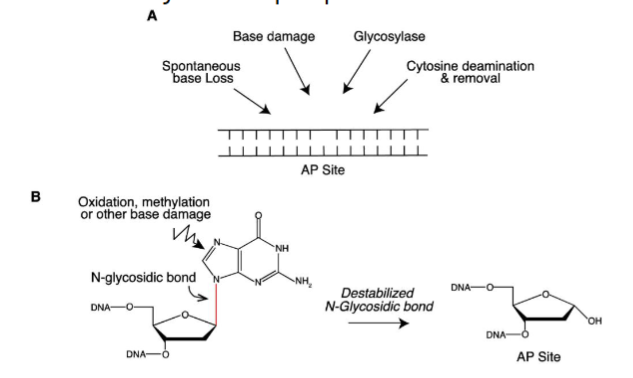
Abasic Sites
most common DNA lesions from cleavage of the N-glycosidic bond between the nitrogenous base and deoxyribose sugar, leaving an intact phosphodiester bond but no base
apurinic or apyrimidinic
Important DNA Polymerases
there are 17 in mammalian cells - 4 replicative and 13 repair
pol alpha: primes DNA synthesis during replication and repair
pol delta: replication of lagging strand during replication and repair
pol epsilon: replication of LEADING strand during replication and repair
pol gamma: mitochondrial DNA replication and repair
repair polymerases respond to damage by:
bypassing damage
reversing damage
removing damage
Damage Bypass
replication forks can encounter structural problems that impair the ability of the fork to progress:
DNA lesions (T-T dimers)
non-B DNA (triplex, slipped DNA, cruciform, G-quad)
DNA polymerases replicate through lesion by inserting an incorrect base or skipping the base → translesion synthesis
Translesion Synthesis
DNA replication factories (clusters of many replication forks in discrete subnuclear compartments) let polymerase read through the error
gap filling: replication machinery leaves ssDNA opposite the lesion to be filled by a translesion polymerase later
polymerase switching: replicative polymerase is displaced at the lesion and PCNA is loaded with a translesion polymerase with the aid of Rev1
how the lesion is filled is dependent on which polymerase is recruited - in prokaryotes pol IV and V are only part of SOS response as response to environmental stress or chemical mutagens
high fidelity in eukaryotes is achieved by pol eta
Damage Reversal
done by a distinct set of repair enzymes
in proks and some euks, Pyrimidine-pyrimidine dimers caused by UV light are directly repaired by photoreactivation
in proks and euks, reversal of base methylation (O6-methylguanine) is done by methyltransferases
In multicellular eukaryotes, DNA-protein crosslinks are reversed by _____.
SPTRN
Photoreactivation
damage and agent: UV light damage results in the formation of pyrimidine-pyrimidine dimers
repair:
DNA photolyases use visible light (near-UV to blue or 300-500nm) to break the covalent bonds holding the adjacent pyrimidines together
takes 2 co-factors: FADH for electron transfer and a chromophore for pigment
this mechanism is NOT present in placental mammals
Which mechanism for damage reversal is not present in placental mammals?
photoreactivation
Demethylation of O6-methylguanine
damage and agent: O6-methylation of guanine by alkylating agents
repair:
o6-methylguanine-DNA-methyltransferse MGMT binds DNA in minor groove, widens it, and bends the DNA to flip out the damaged bases in the major groove
active site of methyltransferase contains a cysteine residue that accepts the methyl group from the guanine (removes group from guanine)
terminal reaction (kills enzyme after protein accept methyl)
Removal of DNA Protein Crosslinks
damage and agent: induced when proteins become and remain covalently attached to the nitrogenous bases, sugar, or broke phosphodiester bond on the DNA backbone
repair:
SPRTN associates with replication machinery to degrade proteins associated with DNA and associated conformational change are essential for SPRTN activation
DNA-dependent metalloprotease that chews up any protein associated with DNA so must be regulated
regulated by ubiquitin
unubiquitinated = on
Damage Removal
simple overview:
DNA repair machinery gains access to the DNA (chromatin remodelling, histone ejection)
DNA repair machinery removes the damage
gap refilled
histones redeposited to reform nucleosome
Types of Single Base Pair Changes
mismatch repair
base excision repair
Base Excision Repair
initiated by DNA glysolyases, leaves an abasic site when damaged base is excised → no base is available for pairing even though phosphodiester bond still exists
shortpatch repair for single nucleotide
long patch repair for 2-10 nucleotides
Shortpatch Repair
glycosylase-associated beta-lyase and apurinic/apyrimidinic endonuclease (APE1) make nicks 3’ and 5’ to the abasic site, respectively
pol-beta replaces missing nucleotide and ligase 3-XRCC1 complex seals the gaps in the sugar-phosphate backbone
Long Patch Repair
apurinic/apyrimidinic endonuclease (APE1) makes a 5’ nick to the abasic site in DNA → pol delta or epsilon (based on leading or lagging strand) and PCNA displace the strand 3’ to the nick to make a flap of 2-10 nucleotides
flap is cut at the junction of the single-to-double strand transition by flap endonuclease 1 (FEN-1)
synthesized fill in is ligated by DNA ligase I
Mismatch Repair
requires strand identification to see which strand is correct - unclear in eukaryotes but in E. coli can differentiate by presence of methylated GATC, which is the strand that gets nicked
once mismatch is recognized, more ATP-bound-MSH sliding clamps are leaded (slows down replication at mismatch and recruits proteins for repair)
DNA is then excised either 5’ or 3’ from the mismatch
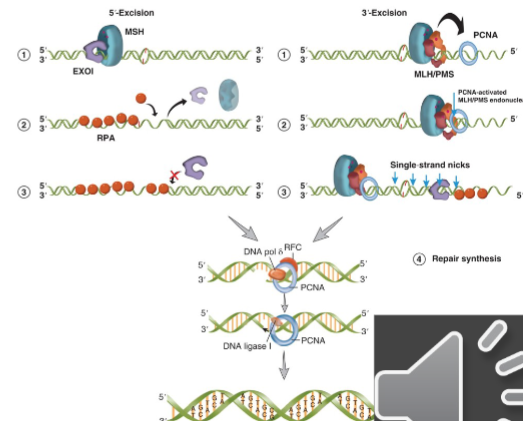
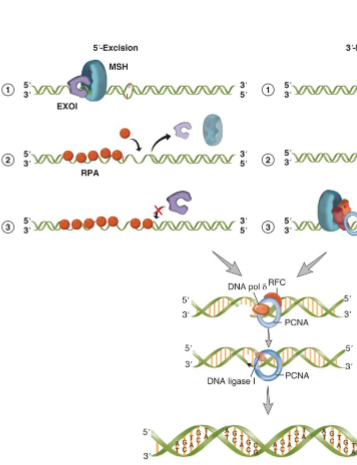
5’ Mismatch Repair
in 5’ excision ATP-bound MSH sliding clamp holds EXO1 on the DNA at the 5’ strand nick and helps it move from 5’-3’
EXO1 can then digest the strand of the DNA that contains the damage
continues for up to several thousand nucleotides
as ssDNA is generated it is coated by RPA
EXO1 can associate with different ATP-bound MSH on the DNA to digest the mismatched strand
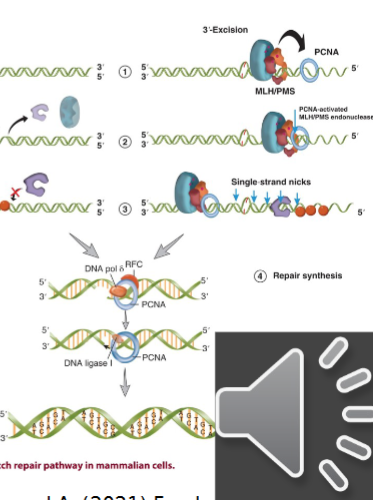
3’ Mismatch Repair
PCNA is bound to 3’ nick
ATP bound MSH sliding clamp associates with MLH/PMS and this complex diffuses along the DNA toward PCNA
interaction between MSH-MLH/PMS and PCNA activates the endonuclease activity of MLH/PMS
active MLH/PMS endonuclease introduces multiple single strand breaks in the 5’ direction from 3’ end
these nicks act as substrates for the EXO1 5’ exonuclease → repeated until the mismatch is release and no more MSH sliding clamps are loaded
once mismatched strand is excised the new DNA can be made by pol-delta and its associated factors
ligase I seals the nick by forming the last phosphodiester bond
Repair Proteins in Damage Bypass
DNA polymerases IV and V in e. coli
DNA polymerase eta in humans
Repaid Proteins in Damage Reversal
DNA photolyase for photoreactivation due to pyrimidine dimers
methyltransferase for removal of methyl groups due to O6-methylguanine
SPRTN protease for removal of DNA-protein crosslinks due to DNA-protein crosslinks
Repair Proteins in Damage Removal
DNA glysosylases for base excision repair due to damaged base
MutSa, MutLa and EXO1 for nucleotide excision repair due to replication errors
XPA, XPB, XPC, XPD, ERCC1/XPF, XPG for nucleotide excision repair due to pyrimidine dimer bulky adduct on base
MRN complex, Rad51, Rad 52, BRCA1, BRCA2, XRCC3 in humans for homologous recombination
Ku proteins, Artemis/DNA-PKCS, XRCC4 in humans for nonhomologous end joining for double strand break repair due to double stranded breaks
DNA Repair Summary
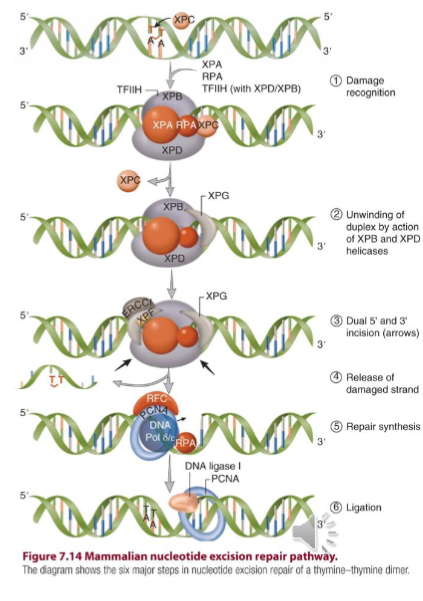
Nucleotide Excision Repair
pathway that DNA causing structural distortion (like pyrimidine dimers) are recognized by cooperative binding of RPA, XPA, XPC, TFIIH complex to facilitate excision of damaged base and filling in of the gap and ligation by pol and ligase
deficiency in NER → xeroderma pigmentosum, trichothiodystrophy, Cockayne syndrome
Responses to Pyrimidine-Pyrimidine Dimers
bypass: replicated/synthesized through by pols, generally error prone unless it’s pol eta
reversal: photoreactivation by DNA photolyase but not in placental mammals
removal: NER (highly conserved) - XP type of proteins
Xeroderma Pigmentosum (XP)
unusually sensitive to UV light and not able to efficiently repair UV damage - rare autosomal recessive disorder
7 of the genes associated with Xeroderma pigmentosum are involved in NER (XPA, XPB, XPC, XPD, XPE, XPF, XPG)
8th gene encodes for DNA pol eta
Nucleotide Excision Repair Pathway
mutations in this - XP
key players are: RPA, XPA, XPC, TFIIH, XPG, XPF/ERCC
NER is coupled to transcription and thus NER pathway repairs lesions by:
global genome repair: NER pathway responsible for recognizing DNA damage in the whole genome
transcription coupled repair: NER pathway responsible for recognizing DNA damage in the transcribed strand of active genes
XPG nicks 3’ of damage; XPF nicks 5’ of the damage
repair is completed by pol epsilon or pol-delta RFC, RPA, and PCNA
RPA
replication protein A
TFIIH
complex of proteins including XPB and XPD to unwind DNA double helix and allow for damaged strand to be released
XPF/ERCC
excision repair cross complementing rodent repair deficiency also important for homologous recombination
Global Genome Repair (GG-NER)
NER pathway responsible for recognizing DNA damage in the whole genome accomplished by RPA, XPA, XPC, TFIIH complex
XPC rapidly scans DNA for structural distortions and opens up DNA after recognizing the target thymine-thymine dimer
indirect damage sensor
pauses when it finds mismatched base to verify a relevant lesion exists before making erroneous incisions
flips out damaged bases
Transcription Coupled Repair
NER pathway responsible for recognizing DNA damage in the transcribed strand of active genes
bulky lesion halts progress of polymerase and another pair of proteins appear to contribute to the recognition
XPG
an endonuclease
nicks DNA 3’ of the damage
XPF
an endonuclease
nicks the DNA 5’ of the damage
Double Strand Breaks and Repair
breaks are induced by things like reactive oxygen species, ionizing radiation such as X-rays, and chemicals that generate reactive oxygen species
repaired by:
homologous recombination - high fidelity, requires template, typically restricted to late S-G2
major role in prokaryotes and lower eukaryotes
non-homologous end joining - error prone, template-free, functions throughout the cell cycle
somatic cells of higher eukaryotes including mammals
both mechanisms start with detection of the break and subsequent DNA processing
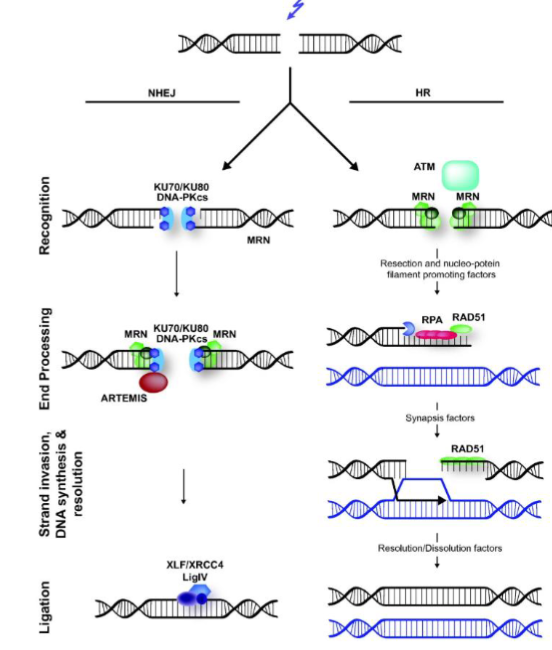
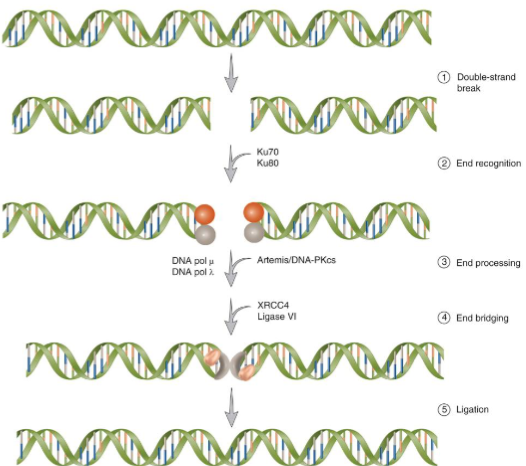
Non-Homologous End Joining
requires heterodimer KU and the catalytic subunit of the DNA-dependent protein kinase, DNA-PKcs
DNA ligase 4-XRCC4 complex ligates the DNA
Homologous Recombination
repairs double strand breaks by getting genetic info from an undamaged homologue (sister chromatic or homologous chromosome) and this requires DNA sequence homology
ataxia telangiectasia mutated (ATM) is the key signal transducer
most insight about this comes from E. coli (prokaryote) and S. cerervisiae (eukaryote) - not the only homology-directed repair, other includes single-strand annealing (SSA)
Steps of Homologous Recombination
initiation
resection
strand invasion
DNA synthesis
branch migration
holiday junction resolution
Initiation of HR
MRN complex = MRE11, RAD50, NBS1 → binds to broken ends of the DNA
recruits ataxia telangiectasia mutated, a serine threonine kinase that phosphorylates proteins involved in the pathway
Resection of HR
starts by MRN complex (MRE11, RAD50, NBS1)
single-stranded 3’ tails are rapidly bound by RPA
Strand Invasion in HR
nucleoprotein filaments subsequently form after replacement of RPA by RAD51
nucleoprotein filaments promote invasion of the undamaged homolog, strand displacement, and D-loop formation
displacement of RPA is facilitated by BRCA1-BARD1-PALB2 and BRCA2-DSS1 complex
DNA Synthesis in HR
displaced strand pairs with the broken and abandoned strand. making a heteroduplex molecule and a Holiday Junction
synthesis using the undamaged strand as a template occurs, involving PCNA, DNA pol-epsilon, pol-delta
at the end of synthesis, a double Holiday junction is formed
Holiday Junction Resolution in HR
the resolvasome (RuvABC)
non-crossover (default pathway in S. cerevisiae) requires BTR complex (BLM, TOPIIIa, RMI1-RMI2)
The Holiday Junction
distinct intermediate in which the two recombining duplexes are joined covalently by single-strand crossovers
junction is resolved into two duplexes by an enzyme complex called the resolvasome
mammals and yeast: holiday junction may be dissolved of this intermediate in 3 ways
non crossover only
unbiased (leads to crossover and non crossover)
cross over specific - only in meiotic cells