Actin Binding Proteins & Microtubule Associated Proteins
0.0(0)
Card Sorting
1/27
Earn XP
Description and Tags
Last updated 12:28 AM on 3/25/23
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
28 Terms
1
New cards
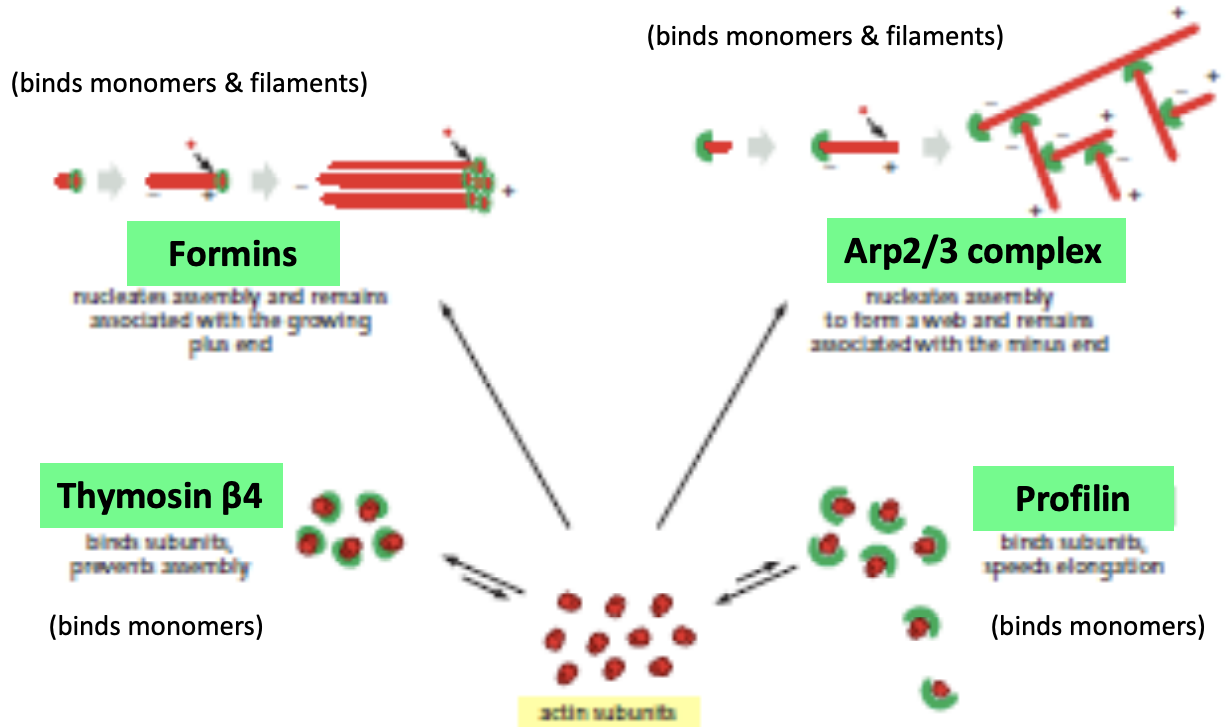
Actin binding proteins (ABPs)
\
* control the assembly, disassembly, organization, and movement of actin filaments
* Formins, Arp2/3, thymosin, profilin
* \
* Actin binding proteins perform a variety of functions
* Control of polymerization and depolymerization: monomer sequestering proteins, nucleating factors, capping proteins, stabilizing proteins, depolymerizing proteins, severing proteins
* Organization of filaments: network forming crosslinking proteins, bundle forming crosslinking proteins, membrane linker proteins
* Movement of filaments: motor proteins
* \
* Actin binding proteins cap, stabilize, sever, and depolymerize filaments
* Capping proteins stabilize filaments by binding to their ends. Filaments are stabilized because the capping proteins prevent addition or loss of subunits from the filament ends. Examples include CapZ, which caps the (+) end of filaments, and tropomodulin, which caps the – ends of actin filaments. (Can you think of another group of proteins that caps the (–) ends of filaments?)
* Stabilizing proteins bind along the length of filaments. Tropomyosin is a long snake-like molecule that binds several subunits along the filament and stabilizes the polymerized state.
* Severing/Depolymerizing proteins disassemble actin filaments and higher order structures. The assembly of actin filaments in the cell must be balanced by filament disassembly, which maintains the monomer pool and thus replenishes the substrates for polymerization. A protein called cofilin enhances the rate of actin depolymerization by severing actin filaments and creating more (–) ends. A protein called gelsolin also severs actin filaments and breaks them into short fragments. After severing, gelsolin remains bound to the + end of the filament. The severing activity of gelsolin is activated by increases in the intracellular concentration of Ca+2.
* control the assembly, disassembly, organization, and movement of actin filaments
* Formins, Arp2/3, thymosin, profilin
* \
* Actin binding proteins perform a variety of functions
* Control of polymerization and depolymerization: monomer sequestering proteins, nucleating factors, capping proteins, stabilizing proteins, depolymerizing proteins, severing proteins
* Organization of filaments: network forming crosslinking proteins, bundle forming crosslinking proteins, membrane linker proteins
* Movement of filaments: motor proteins
* \
* Actin binding proteins cap, stabilize, sever, and depolymerize filaments
* Capping proteins stabilize filaments by binding to their ends. Filaments are stabilized because the capping proteins prevent addition or loss of subunits from the filament ends. Examples include CapZ, which caps the (+) end of filaments, and tropomodulin, which caps the – ends of actin filaments. (Can you think of another group of proteins that caps the (–) ends of filaments?)
* Stabilizing proteins bind along the length of filaments. Tropomyosin is a long snake-like molecule that binds several subunits along the filament and stabilizes the polymerized state.
* Severing/Depolymerizing proteins disassemble actin filaments and higher order structures. The assembly of actin filaments in the cell must be balanced by filament disassembly, which maintains the monomer pool and thus replenishes the substrates for polymerization. A protein called cofilin enhances the rate of actin depolymerization by severing actin filaments and creating more (–) ends. A protein called gelsolin also severs actin filaments and breaks them into short fragments. After severing, gelsolin remains bound to the + end of the filament. The severing activity of gelsolin is activated by increases in the intracellular concentration of Ca+2.
2
New cards
Cofilin
severs end of actin filament
3
New cards
Gelsolin
severs and caps + end, can’t grow, rapid disassembly
4
New cards
CapZ
caps + end, inhibit growth
5
New cards
Tropomyosin
bind to side of existing filaments, stabilizes
6
New cards
Fimbrin
cross links multiple actin filaments, short sequence, tightly packed
\
\
* Short spacer cross linking proteins
* is short with 2 binding domains that creates tight packed actin fibers, parallel
* seen in microvilli, filopodia, focal adhesions (links outside to inside)
* Cross-linking proteins with shorter, more rigid spacers tend to organize filaments into aligned bundles. Bundling proteins with very short spacers (fimbrin, villin, and fascin) often lead to tight bundles. α-actinin can crosslink filaments into bundles that are arranged into parallel and anti-parallel arrays involved with contraction.
\
\
* Short spacer cross linking proteins
* is short with 2 binding domains that creates tight packed actin fibers, parallel
* seen in microvilli, filopodia, focal adhesions (links outside to inside)
* Cross-linking proteins with shorter, more rigid spacers tend to organize filaments into aligned bundles. Bundling proteins with very short spacers (fimbrin, villin, and fascin) often lead to tight bundles. α-actinin can crosslink filaments into bundles that are arranged into parallel and anti-parallel arrays involved with contraction.
7
New cards
A-actinin
cross links multiple actin filaments, long sequence, longer distance
\
* Long spacer cross linking proteins
* Dimer in head to tail fashion, 2 binding domains, longer so spacing is wider, proteins can go through
* Seen in stress fibers (contract-motor proteins get through), filopodia, muscle Z line
\
* Long spacer cross linking proteins
* Dimer in head to tail fashion, 2 binding domains, longer so spacing is wider, proteins can go through
* Seen in stress fibers (contract-motor proteins get through), filopodia, muscle Z line
8
New cards
Filamin
long flexible that interacts with actin fibers to make flexible mesh
\
* Long, flexible spacer cross linking proteins
* Long and flexible
* Seen in leading edge, stress fibers, filopodia
\
Cross-linking proteins with longer, more flexible spacers usually function to arrange filaments into gel-like networks. These include filamin and spectrin. Some actin-binding proteins have multiple functions. For example, the Arp2/3 complex can nucleate and cross-link actin filaments.
\
* Long, flexible spacer cross linking proteins
* Long and flexible
* Seen in leading edge, stress fibers, filopodia
\
Cross-linking proteins with longer, more flexible spacers usually function to arrange filaments into gel-like networks. These include filamin and spectrin. Some actin-binding proteins have multiple functions. For example, the Arp2/3 complex can nucleate and cross-link actin filaments.
9
New cards
Spectrin and ERM proteins
link to plasma membrane
spectrin:
\
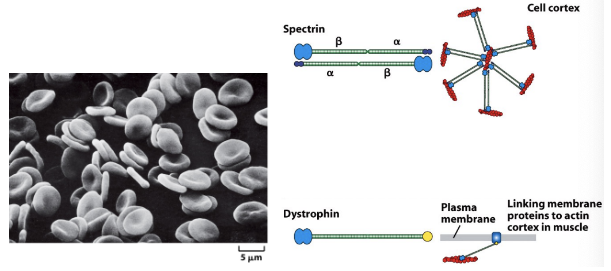
* Membrane linking proteins
* Dimer in head to tail fashion, interact with cytoskeleton
* Seen in Cell cortex providing rigidity
\
\
* Membrane-actin linker proteins attach the actin cytoskeleton to the plasma membrane. The simplest connections entail indirect interactions between integral membrane proteins and actin filaments. These interactions are often mediated by peripheral membrane proteins such as the ERM proteins (e.g., ezrin), which act as a linker between F-actin and the integral membrane proteins. Spectrin also connects actin to membrane proteins. Dystrophin links cortical actin to muscle cell membranes via the Dystroglycan complex.
spectrin:
\
* Membrane linking proteins
* Dimer in head to tail fashion, interact with cytoskeleton
* Seen in Cell cortex providing rigidity
\
\
* Membrane-actin linker proteins attach the actin cytoskeleton to the plasma membrane. The simplest connections entail indirect interactions between integral membrane proteins and actin filaments. These interactions are often mediated by peripheral membrane proteins such as the ERM proteins (e.g., ezrin), which act as a linker between F-actin and the integral membrane proteins. Spectrin also connects actin to membrane proteins. Dystrophin links cortical actin to muscle cell membranes via the Dystroglycan complex.
10
New cards
Dystrophin
\
* Membrane linking proteins
* Linking membrane proteins to actin cortex in muscle
* Membrane linking proteins
* Linking membrane proteins to actin cortex in muscle
11
New cards
ABPs link actin filaments to membranes
\
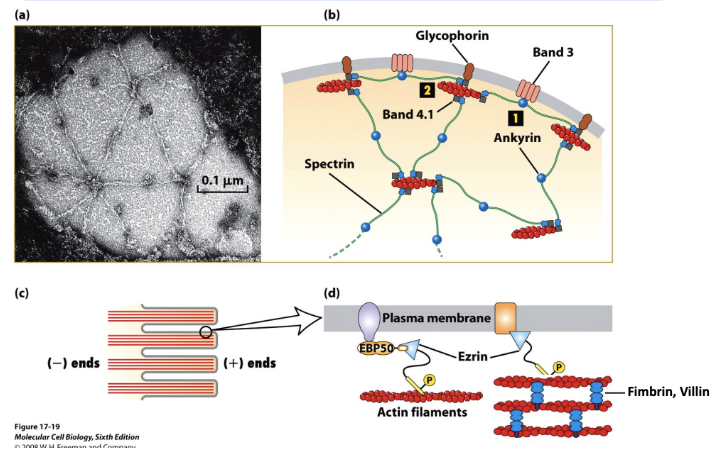
* Many actin structures are found at the cell periphery in the actin-rich layer just beneath the plasma membrane called the cell cortex. This layer gives an animal cell mechanical strength and enables it to form surface structures and perform surface movements (phagocytosis, cytokinesis, and locomotion). The Spectrin-based membrane skeleton lies immediately adjacent to the plasma membrane. Ezrin links bundled actin to the membrane in microvilli.
* Spectrin binding protein called ankyrin interact with membrane protein called band 3, links to cell membrane
* Spectrin and ankyrin also interact with band 4.1 which interacts with another membrane protein called glycophorin
* Leads to spectrin based membrane skeleton
* Microvilli with actin and how that interacts with plasma membrane
* Filaments held together using fimbrin
* Membrane linker protein called ezrin to stabilize
* Many actin structures are found at the cell periphery in the actin-rich layer just beneath the plasma membrane called the cell cortex. This layer gives an animal cell mechanical strength and enables it to form surface structures and perform surface movements (phagocytosis, cytokinesis, and locomotion). The Spectrin-based membrane skeleton lies immediately adjacent to the plasma membrane. Ezrin links bundled actin to the membrane in microvilli.
* Spectrin binding protein called ankyrin interact with membrane protein called band 3, links to cell membrane
* Spectrin and ankyrin also interact with band 4.1 which interacts with another membrane protein called glycophorin
* Leads to spectrin based membrane skeleton
* Microvilli with actin and how that interacts with plasma membrane
* Filaments held together using fimbrin
* Membrane linker protein called ezrin to stabilize
12
New cards
ABPs and disease
\
* Mutations in the genes encoding proteins of the red blood cell membrane skeleton (spectrin, ankyrin, band 4.1) can result in cells that rupture easily, giving rise to diseases known as hereditary spherocytic anemias. - cell shape is important
* Muscular dystrophies are genetic diseases often characterized by the progressive weakening of skeletal muscle. Duchenne muscular dystrophy (DMD) affects the protein dystrophin. Without functional dystrophin, the plasma membrane of muscle cells weakens and eventually ruptures. - membrane linkage is important
* Mutations in the genes encoding proteins of the red blood cell membrane skeleton (spectrin, ankyrin, band 4.1) can result in cells that rupture easily, giving rise to diseases known as hereditary spherocytic anemias. - cell shape is important
* Muscular dystrophies are genetic diseases often characterized by the progressive weakening of skeletal muscle. Duchenne muscular dystrophy (DMD) affects the protein dystrophin. Without functional dystrophin, the plasma membrane of muscle cells weakens and eventually ruptures. - membrane linkage is important
13
New cards
Microtubule Proteins
\
* Microtubule associated proteins (MAPs) control the assembly, disassembly, organization, and movement of microtubules
* y-TURC, +TIPS, Stathmin, Kinesin-13, Katanin, MAPs, Tau and MAP2, Plectin
* Microtubule associated proteins (MAPs) control the assembly, disassembly, organization, and movement of microtubules
* y-TURC, +TIPS, Stathmin, Kinesin-13, Katanin, MAPs, Tau and MAP2, Plectin
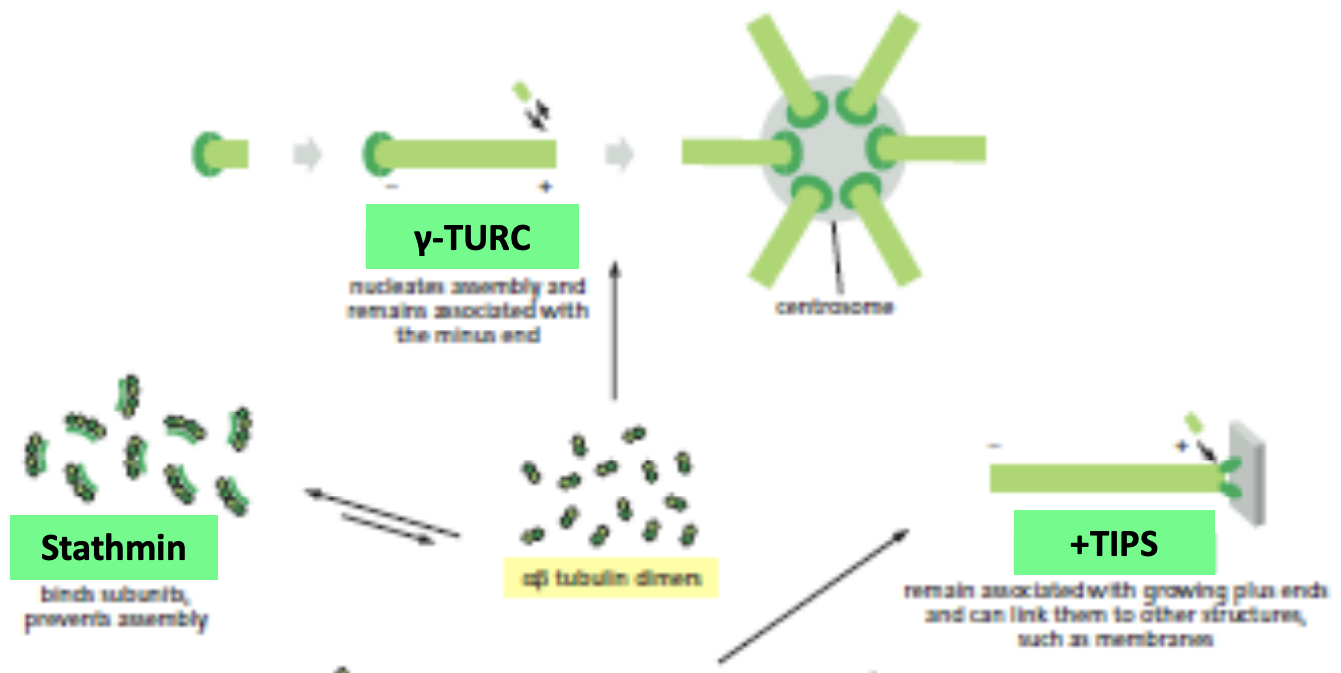
14
New cards
y-TURC
\
* Nucleating protein
* helps in nucleation from centrosome
* Nucleating protein
* helps in nucleation from centrosome
15
New cards
\+TIPS
\
* Interact with + end if has a GTP cap
* Can link to membrane
* Interact with + end if has a GTP cap
* Can link to membrane
16
New cards
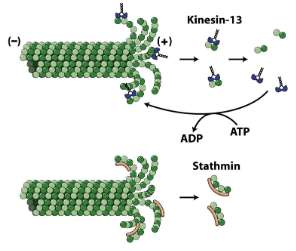
Stathmin
\
* Bind to fragments of microtubules
* Difficult to grow microtubules and promotes catastrophe
* Bind to fragments of microtubules
* Difficult to grow microtubules and promotes catastrophe
17
New cards
Kinesin-13
Causes depolymerization
18
New cards
Katanin
Causes depolymerization by cutting microtubules in the middle and those pieces undergo catastrophe
\
* Fluorescent and see how it works
* Creates Gaps and then shrinking of fragments
\
* Fluorescent and see how it works
* Creates Gaps and then shrinking of fragments
19
New cards
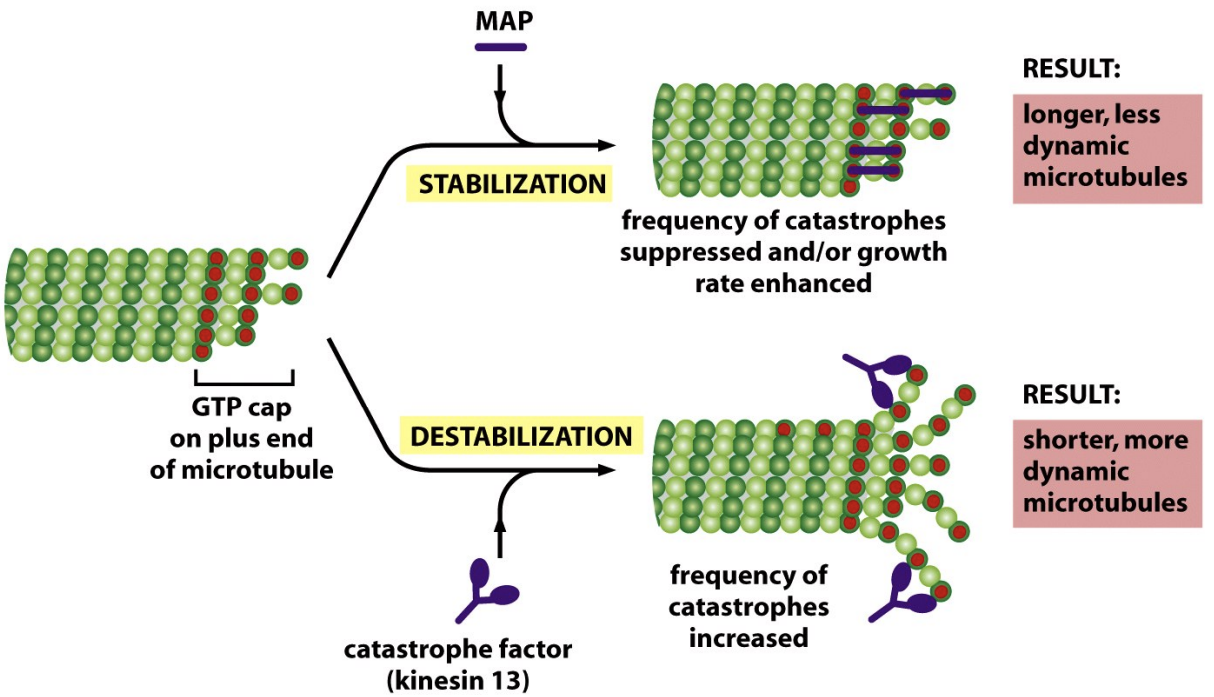
MAPs
\
* Stabilize microtubules to prevent dissasembly
* \
* enhance growth
* Bind to multiple subunits at + end to make catastrophe harder
* Stabilize microtubules to prevent dissasembly
* \
* enhance growth
* Bind to multiple subunits at + end to make catastrophe harder
20
New cards
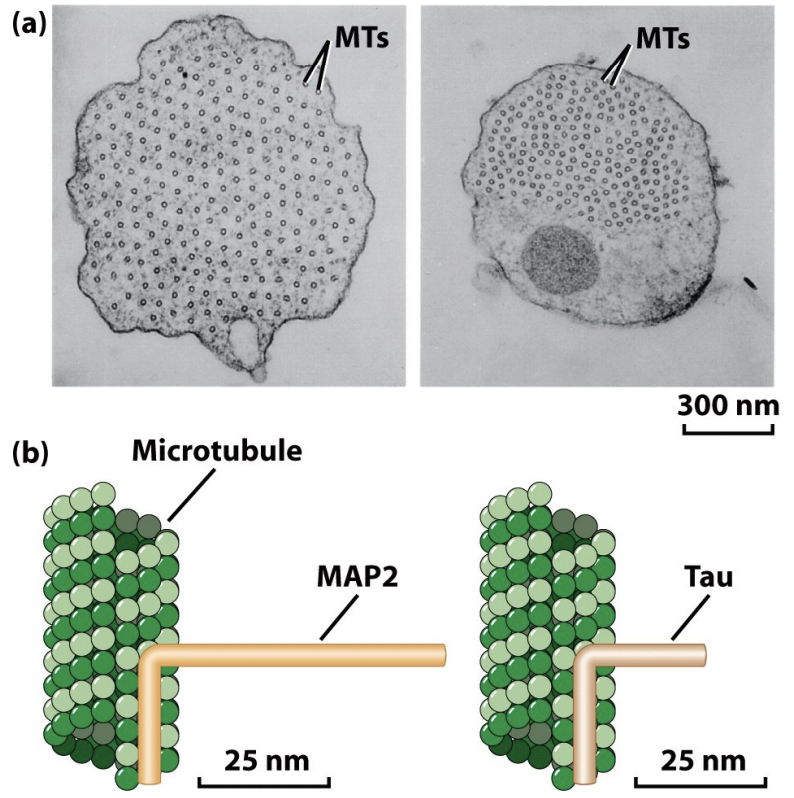
Tau and MAP2
\
* Stabilize microtubules by binding to sides
* Create spacing - Tau is tight, MAP2 has more space
\
\
* MAPs like Tau stabilize and control the spacing of microtubules
* Neurons have these
* MAP2 and Tau bind to microtubule (microtubule binding domain) and project away (projection domain)
* MAP2 has longer projection domain, a lot of MAP2 spaces aout microtubules, Tau has closer spacing
* Overexpression makes spacing change
* Stabilize microtubules by binding to sides
* Create spacing - Tau is tight, MAP2 has more space
\
\
* MAPs like Tau stabilize and control the spacing of microtubules
* Neurons have these
* MAP2 and Tau bind to microtubule (microtubule binding domain) and project away (projection domain)
* MAP2 has longer projection domain, a lot of MAP2 spaces aout microtubules, Tau has closer spacing
* Overexpression makes spacing change
21
New cards
Plectin
\
* Microtubule crosslinked with filaments
* Microtubule crosslinked with filaments
22
New cards
Microtubule associated proteins perform a variety of functions
\
* Control of polymerization and depolymerization: oligomer sequestering proteins, nucleating factors, end-binding (+TIP) proteins, stabilizing proteins depolymerizing proteins severing proteins
* Organization of microtubules: cross-linking proteins bundling proteins membrane linker proteins?
* Movement of microtubules: motor proteins
* (As you can see, MAPs (with a few exceptions) fall into the same broad categories as ABPs.)
* Control of polymerization and depolymerization: oligomer sequestering proteins, nucleating factors, end-binding (+TIP) proteins, stabilizing proteins depolymerizing proteins severing proteins
* Organization of microtubules: cross-linking proteins bundling proteins membrane linker proteins?
* Movement of microtubules: motor proteins
* (As you can see, MAPs (with a few exceptions) fall into the same broad categories as ABPs.)
23
New cards
Microtubule associated proteins cap, stabilize, sever, and depolymerize filaments
\
* Tubulin oligomer binding proteins: Stathmin can bind and sequester tubulin heterodimers or oligomers and thus promote catastrophe and depolymerization.
* Nucleating proteins: The γ-TURC nucleates MT assembly at the centrosome.
* End binding proteins: +TIPs bind to the (+) ends of MTs. EB1 is a +TIP that can regulate (+) end dynamics and can link MTs to organelles. CLIP-170 is thought to mediate the interaction of chromosomes and membranes with the (+) end.
* Severing proteins: Katanin severs MTs and may promote depolymerization at (-) ends.
* Depolymerizing proteins: A subset of kinesins (e.g., Kin13) promote MT depolymerization at the (+) end by binding to and inducing protofilament curling. This results in destabilization of the MT end and subsequent depolymerization. Kin13 can promote the depolymerization of GTP-tubulin, so the mechanism does not involve hydrolysis of the GTP cap.
* Tubulin oligomer binding proteins: Stathmin can bind and sequester tubulin heterodimers or oligomers and thus promote catastrophe and depolymerization.
* Nucleating proteins: The γ-TURC nucleates MT assembly at the centrosome.
* End binding proteins: +TIPs bind to the (+) ends of MTs. EB1 is a +TIP that can regulate (+) end dynamics and can link MTs to organelles. CLIP-170 is thought to mediate the interaction of chromosomes and membranes with the (+) end.
* Severing proteins: Katanin severs MTs and may promote depolymerization at (-) ends.
* Depolymerizing proteins: A subset of kinesins (e.g., Kin13) promote MT depolymerization at the (+) end by binding to and inducing protofilament curling. This results in destabilization of the MT end and subsequent depolymerization. Kin13 can promote the depolymerization of GTP-tubulin, so the mechanism does not involve hydrolysis of the GTP cap.
24
New cards
\+TIPs like EB1
\
* influence the dynamics of the plus ends of microtubules
* EB1 is at ends because that is where GTP caps are (red)
* influence the dynamics of the plus ends of microtubules
* EB1 is at ends because that is where GTP caps are (red)
25
New cards
Dynamic instability can be visualized in cells
Importantly, due to dynamic instability, proteins that interact with the ends of microtubules are able to influence microtubule stability and the organization of microtubule arrays involved in diverse functions like chromosome segregation and neuronal axon extension. (We will learn about these microtubule-associated proteins, and these cellular functions, in later lectures.)
26
New cards
Some microtubule associated proteins can promote depolymerization
\
* Kinesin 13 promotes catastrophe
* Stathmin interacts with fragments and prevents them from being available to rescue growth
* Kinesin 13 promotes catastrophe
* Stathmin interacts with fragments and prevents them from being available to rescue growth
27
New cards
Microtubule associated proteins organize microtubules into bundles and promote interactions with other filaments
\
* Polymer-binding MAPs can have different activities. They can stabilize MTs by binding to their sides and inhibit disassembly, enhance assembly by stabilizing nuclei and thus facilitate nucleation, organize MTs into bundles in various cellular structures, and mediate MT interactions with other proteins in the cell, including intermediate filaments and actin.
* In general, polymer-binding MAPs have two major domains. The microtubule binding domain binds several tubulin dimers at once and thus helps to stabilize the polymer. The projection domain can interact with MTs or other structures such as intermediate filaments.
* MAP2 and Tau are responsible for organizing microtubules in neuronal axons and dendrites. When MAP2 and Tau are expressed in cells that do not normally form axons, long axon like structures are induced. The organization of MTs in the MAP2- and Tau-induced structures are similar to one another with one exception: the projection domain of MAP2 is longer, so the spacing between microtubules is greater in the MAP2 expressing cells.
* Polymer-binding MAPs can have different activities. They can stabilize MTs by binding to their sides and inhibit disassembly, enhance assembly by stabilizing nuclei and thus facilitate nucleation, organize MTs into bundles in various cellular structures, and mediate MT interactions with other proteins in the cell, including intermediate filaments and actin.
* In general, polymer-binding MAPs have two major domains. The microtubule binding domain binds several tubulin dimers at once and thus helps to stabilize the polymer. The projection domain can interact with MTs or other structures such as intermediate filaments.
* MAP2 and Tau are responsible for organizing microtubules in neuronal axons and dendrites. When MAP2 and Tau are expressed in cells that do not normally form axons, long axon like structures are induced. The organization of MTs in the MAP2- and Tau-induced structures are similar to one another with one exception: the projection domain of MAP2 is longer, so the spacing between microtubules is greater in the MAP2 expressing cells.
28
New cards
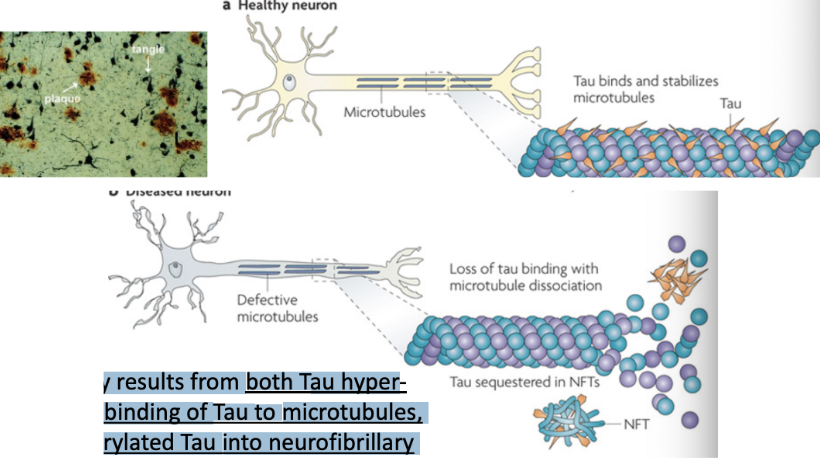
Alzheimer’s disease pathogenesis
\
* Tau aggregation in an insoluble form may be involved in Alzheimer’s disease pathogenesis
* Tau is particularly abundant in neurons and facilitates microtubule stabilization and is normally phosphorylated. Tau function is compromised in Alzheimer's disease and other Tauopathies. This probably results from both Tau hyperphosphorylation, which reduces the binding of Tau to microtubules, and sequestration of hyper-phosphorylated Tau into neurofibrillary tangles (NFTs), which reduces the amount of Tau available to bind microtubules. Loss of Tau function leads to microtubule instability and reduced axonal transport, which could contribute to neuropathology.
* Tangles are aggregates of Tau when they fall off of microtubules
* Tau aggregation in an insoluble form may be involved in Alzheimer’s disease pathogenesis
* Tau is particularly abundant in neurons and facilitates microtubule stabilization and is normally phosphorylated. Tau function is compromised in Alzheimer's disease and other Tauopathies. This probably results from both Tau hyperphosphorylation, which reduces the binding of Tau to microtubules, and sequestration of hyper-phosphorylated Tau into neurofibrillary tangles (NFTs), which reduces the amount of Tau available to bind microtubules. Loss of Tau function leads to microtubule instability and reduced axonal transport, which could contribute to neuropathology.
* Tangles are aggregates of Tau when they fall off of microtubules