Biol 214 - Regulation of Gene Expression
1/54
Earn XP
Description and Tags
Exam 3 material. The process of controlling which genes are turned on or off at any given time to regulate the cell's function, cell differentiation, and response to stimuli.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
55 Terms
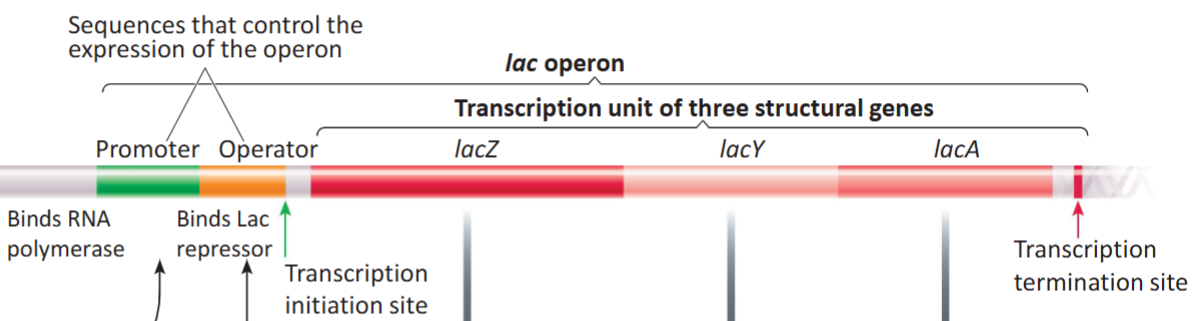
Operon
A prokaryotic gene organization system (DNA) containing a cluster of genes under the control of a single promoter, meaning the genes are transcribed together as a single mRNA strand.
Eukaryotes do not have operons in most cases.
Structural Gene
A gene that encodes the proteins needed to perform a certain function. Transcribed together as a single mRNA molecule.
Regulatory Gene
Typically located outside the operon. Encodes a regulatory protein, such as a repressor or activator, that controls the transcription of structural genes. Transcribed as an mRNA molecule. Never turned “off.”
Promoter
Binds RNA polymerase onto an operon. A DNA sequence, along with the operator, that controls the expression of the operon.
Operator
Binds a repressor onto an operon. A DNA sequence, along with the promoter, that controls the expression of the operon.
Inducible Operon
A set of genes that is normally “off” but can be switched “on” by an inducer molecule.
Ex: lactose operon
Allosteric Protein
A protein that can change their shape and function in response to the binding of certain molecules.
Repressor Molecule
An allosteric protein that inhibits gene expression by binding to the operon region on an operator when active. Blocks transcription.
An active repressor occasionally falls off the operator, allowing a very low rate of transcription (which results in a few molecules of each protein being made).
Repressors and their mRNAs are always made, since they originate from regulatory genes.
Constitutively Expressed
Always expressed.
Repressor Molecule Activity in an Inducible Operon
Whether the repressor is normally active or normally inactive depends on the operon type (inducible vs. repressible).
Normally active (constitutively expressed), operon off.
Repressor Molecule Activity in an Repressible Operon
Whether the repressor is normally active or normally inactive depends on the operon type (inducible vs. repressible).
Normally inactive, operon on.
Permease/Transport Protein
A membrane-bound protein that allows specific molecules to move across a cell membrane. Transports certain molecules into the cell to be made into inducer molecules.
Inducer Molecule
A substance (usually a sugar or hormone) that is converted into an inducer and binds to a repressor protein, causing a change in its shape that prevents it from binding to the DNA.
Allows polymerase to read the full operon and increases mRNA transcription.
Inducer Molecule Activity in an Inducible Operon
Whether the inducer is normally active or normally inactive depends on the operon type (inducible vs. repressible).
Usually absent, repressor is attached to operator, operon is off.
Inducer Molecule Activity in an Repressible Operon
Whether the inducer is normally active or normally inactive depends on the operon type (inducible vs. repressible).
Usually present, repressor is not attached to operator, operon is on.
Activator Protein
A protein that must bind to a DNA sequence promoter proximal region to increase the transcription of target genes. Works when activated by getting DNA polymerase to bind more efficiently to the promoter, but gene expression is not possible until all specific activators are bound to their specific enhancer regulatory sequences (a form of coordinate regulation).
Often used to amplify the creation of needed protein blueprints (mRNA), making transcription more efficient for the specific situation a prokaryote may be in and need.
Ex. CAP protein activated with cAMP
Positive Gene Regulation System
A system where transcription is increased.
A signal molecule (ex. cAMP) indicates an environmental condition and binds to an activator protein (ex. CAP). The activated activator binds to a DNA site near the promoter, which allows enhances RNA polymerase binding. Transcription rate increases.
Repressible Operon
A set of genes that is normally “on” but can be switched “off” by a repressor and corepressor molecule.
Ex: tryptophan (trp) operon
Corepressor
A molecule that inhibits gene expression by binding to a repressor protein, activating it to turn off expression of the operon. Some can bind to the repressor without processing, but some need to.
Ex. tryptophan
Negative Gene Regulation System
A system where transcription is decreased.
A repressor protein binds DNA to block transcription after being activated, either with a modified molecule or a corepressor. The system is turned off when the repressor is active, and on when the repressor is inactive.
Regulation of Gene Expression in Eukaryotes
• Transcriptional regulation
• Posttranscriptional regulation
• Translational regulation
• Posttranslational regulation
Transcriptional Regulation
Determines breakdown which genes are transcribed.
• Chromatin remodeling to make genes accessible for transcription
• Regulatory events at a gene’s promoter and regulatory sequences
Posttranscriptional Regulation
Determines types and availability of mRNAs to ribosomes.
• Variations in pre-mRNA processing
• Variations in rate of mRNA breakdown
Translational Regulation
Determines the rate at which proteins are made.
• Variations in rate of initiation of
protein synthesis
• RNA interference
Posttranslational Regulation
Determines availability and activity of functional proteins.
• Chemical modification of proteins
• Processing of proteins to activate them
• Variations in rate of protein degradation
Nucleosome in Inactive Chromatin
Consists of negatively-charged DNA looped around a positively-charged histone octamer, and positively-charged histone tails wrapped around the DNA.
Nucleosome in Active Chromatin
Consists of negatively-charged DNA looped around a a positively-charged histone octamer, and positively-charged histone tails bound to negatively-charged acetyl groups.
Histone Acetyltransferase
A type of enzyme that aids in acetylation, a form of gene regulation controlling chromatin structure. Frees the histone tails from the DNA backbone by attaching the ends to acetyl groups, changing the charge of the histone tails from positive to neutral.
Histone Deacetylase
A type of enzyme that aids in deacetylation, a form of gene regulation controlling chromatin structure. Takes away the acetyl groups and allows for the histone tails to reattach back onto the DNA backbone, changing the charge of the histone tails from neutral to positive.
Nucleosome
A strand of DNA ~147 bp long wrapped around a histone octamer (8 histones). A histone octamer is composed of two copies each of four core histone proteins: H2A, H2B, H3, and H4.
Inactive State
Nucleosomes prevent general transcription factors and RNA polymerase II from binding so transcription does not occur.
Histone tails are not acetylated: the tails form a tight association with the DNA backbone wrapped around the histone octamer of a nucleosome.
Active State
General transcription factors and RNA polymerase II bind to the promoter so transcription regulation can occur, which only happens when histone tails and the histone octamer are loosened and moved a few (to a few ten) base pairs.
Chromatin Remodeling
A process in which large multiprotein complexes/chromatin remodeling complexes use the energy from ATP hydrolysis to shift the position of nucleosome to uncover the promoter region.
Considered inducing if it facilitates gene expression, such as “sliding” to uncover promoter regions.
DNA Methylation
The process of adding a methyl group to the DNA molecule itself, which typically silences gene expression. DNA methylation modifies the DNA itself, whereas histone acetylation modifies the proteins that DNA is wrapped around.
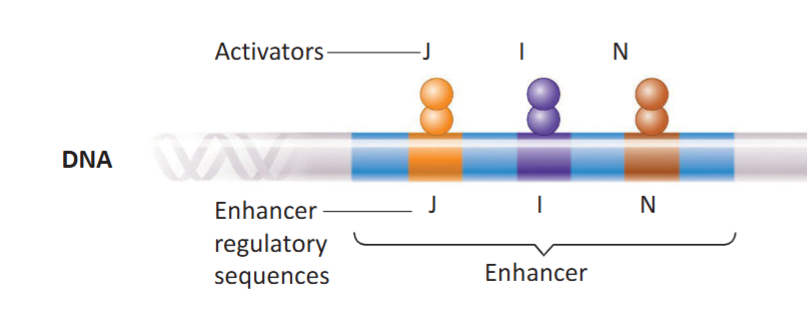
Enhancer Regulatory Sequence in Eukaryotes
A regulatory DNA sequence that increases transcription when activated. Can be far away (upstream or downstream) from the actual promoter and transcription unit of the gene.
Also the site where activator proteins attach to. Each specific regulatory sequence corresponds to a specific activator.
Ex. Human β -interferon gene in a cell infected by a virus
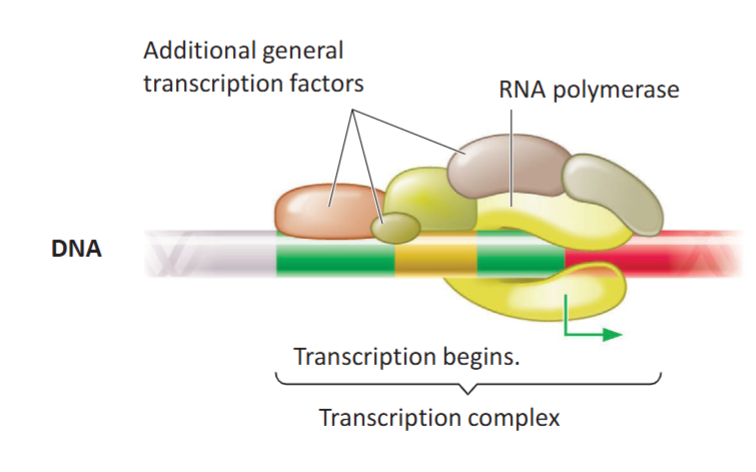
Transcription Complex
A complex of general transcription factor proteins that bind to the TATA box and themselves after the promoter region is exposed on loosened chromatin. They recruit RNA polymerase II onto the promoter region.
A particular type of transcription factor has helicase activity, which can unwind double-stranded DNA only at the promoter (not the whole gene) and initiates transcription.
DNA-Binding Motifs
The motif is a specific part of transcription factors that bind to DNA, sometimes referred to as protein domains. The DNA-binding domain shape is fixed and depends on the type of transcription factor each motif is a part of. Since alpha helices sit in major grooves of DNA (~10 bp apart), they have the ability to bind to sequence-specific areas of the DNA. There are three main shapes covered in Biol 214.
• Helix-turn-helix
• Zinc finger
• Leucine zipper
α-Helices
A common structural element in general transcription factors and are particularly well suited for fitting into sequence-specific parts of the major groove of DNA.
Represented as blue cylinders in the textbook.
Beta Sheets
A secondary structure found in general transcription factors. They look like “pleated” or folded sheets and provide rigidity and stability to protein structures.
Represented as flat arrows pointing from the N-terminus to the C-terminus.
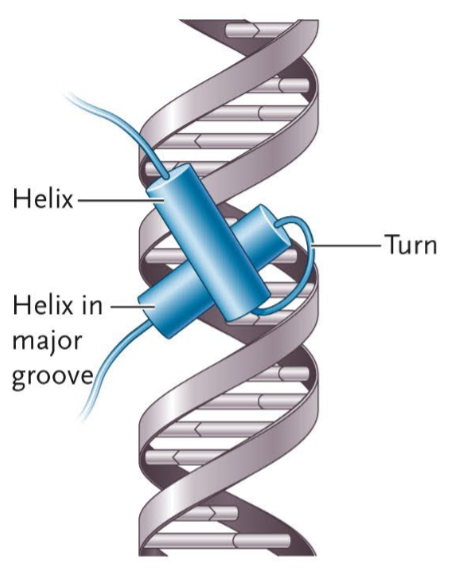
Helix-Turn-Helix Motif
A motif that has two α-helices connected by a short "turn," or a loop in the connecting region of a helix. One helix fits into the major groove of the DNA and interacts with specific base pairs, allowing the protein to recognize a DNA sequence. The other holds the first helix in place.
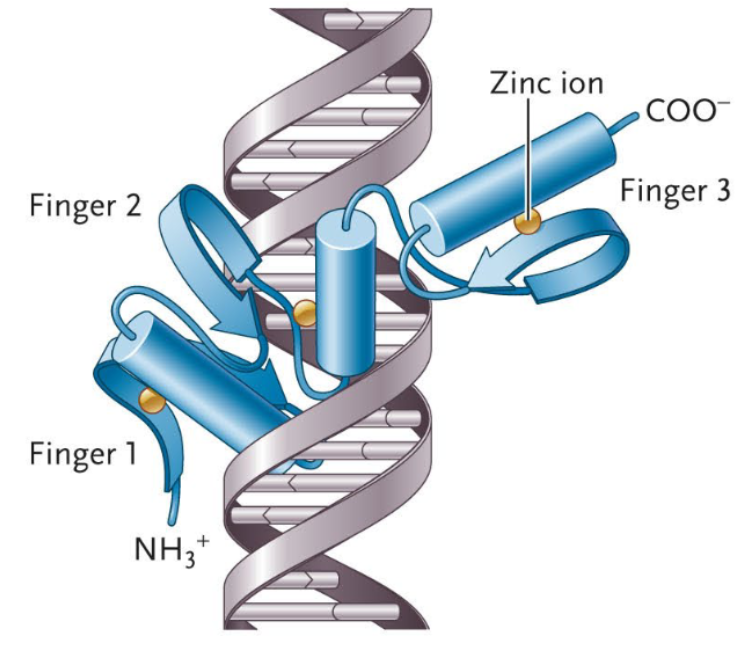
Zinc Finger Motif
A motif that has one or more α-helices and loops, often accompanied by a short β-sheet and one or more zinc ions to stabilize the structure.
“Fingers” consist of alpha helices and loops, and structures with multiple fingers can match to sequence-specific areas of DNA using each alpha helix to occupy a major groove in the DNA. The most popular example of motifs’ abilities to bind with sequence specificity.
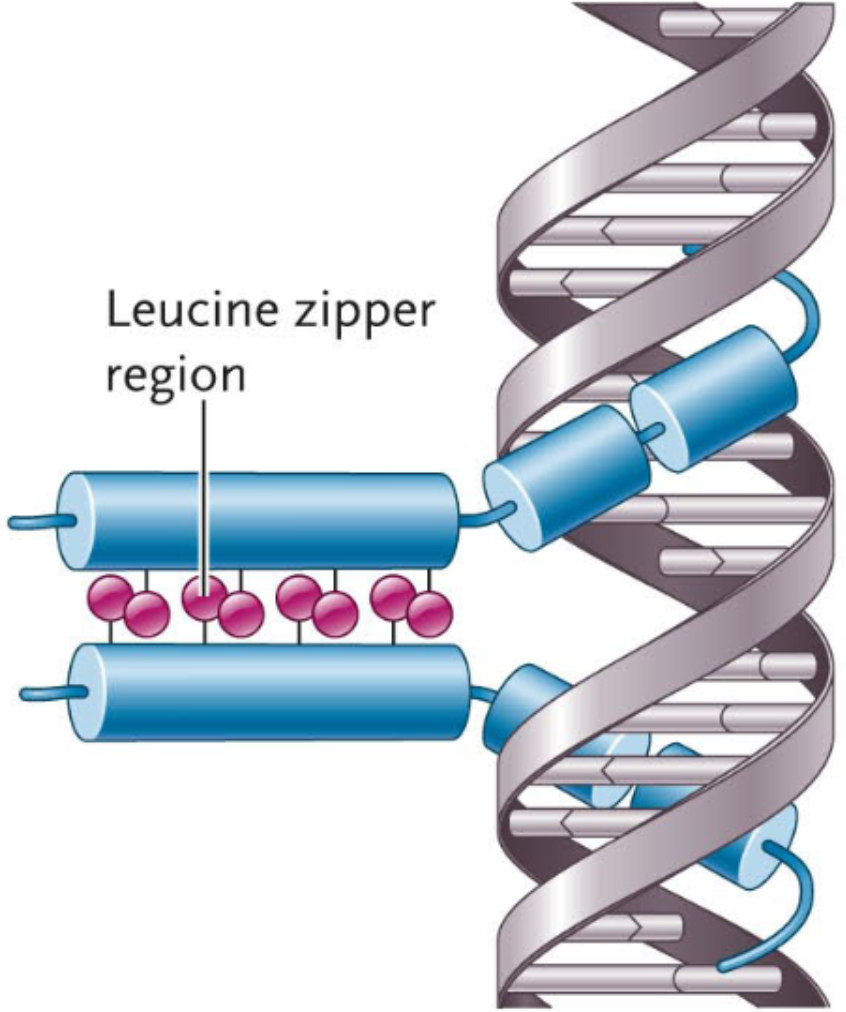
Leucine Zipper Monomer
Each monomer is one α-helix that contains a repeating pattern of leucine residues (an amino acid) every 7 amino acids along one face of the helix.
Leucine Zipper Motif
A motif that has α-helices that bind to the major grooves in DNA and a dimer: a complex formed from two α-helices (one from each monomer) that zip together via hydrophobic interactions.
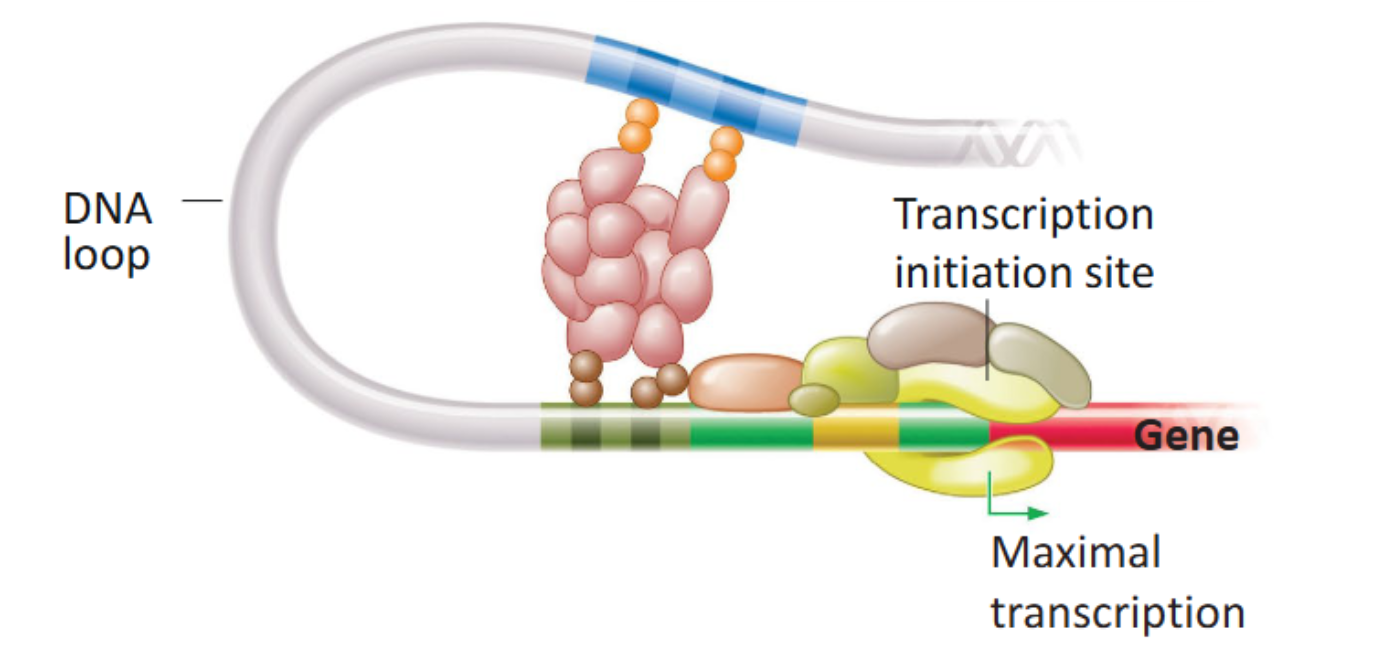
Coactivator
A large assembly of proteins that serves as a physical bridge that connects transcriptional activators bound to enhancer regions. By doing so, they form a loop in DNA, which activates transcription to the highest level, where RNA polymerase II can transcribe the gene sequence multiple times.
Hormone–Receptor Complex
A hydrophobic steroid hormone that passed through the phospholipid bilayer to activate a steroid hormone receptor in the cytoplasm. Once this structure is created, it can now pass into the cell nucleus and act as a transcriptional activator.
Steroid Hormone Response Element
The sequence of DNA that the hormone–receptor complex binds to. Once the sequence is activated, the gene controlled by the steroid hormone is transcribed.
Long Noncoding RNAs (lncRNAs)
RNA molecules transcribed from DNA (like mRNA) but do not code for proteins. Their function is regulatory: lncRNAs can interact with DNA regions or with proteins that bind them to regulate gene expression at multiple levels (chromatin, transcription, post-transcriptional).
Ex: Xist lncRNA is transcribed from the X-inactivation center on one X chromosome in females. Xist RNA coats that X chromosome, recruiting proteins that condense the chromatin and silence transcription of nearly all genes on it.
UTR/Untranslated Region
A portion of mRNA that is not translated into protein, but it plays important regulatory roles. Found in exons at the ends of a gene so they stay in mature mRNA, located before (5′ UTR) or after (3′ UTR) the coding sequence.
Only the coding region, not the UTR, gets translated into protein.
Ex: miRNAs bind to sequences in the 3′ UTR of target mRNAs (untranslated region at the 3′ end of the mRNA) to control how long the mRNA lasts or whether it gets translated into protein.
pre-miRNA
A hairpin (stem-loop) structure, transcribed from an miRNA gene it is then exported to cytoplasm and attaches to a dicer enzyme to be modified. Turned into a double-stranded RNA strand after the loop is cleaved. A protein complex binds to double-stranded RNA with the dicer still attached.
miRNA-Induced Silencing Complex (miRISC)
A form of RNA interference. The combination of a protein complex, a dicer, and a single miRNA.
The miRNA portion binds to target mRNAs that have a complementary or nearly complementary base sequence in their 3' UTRs.
Micro RNA (miRNA)
Created after an enzyme in the protein complex degrades one of the RNA strands of the double-stranded RNA hairpin structure, leaving the miRNA.
Imperfect pairing of miRNA to target mRNA causes repression of translation or shorter half-life of the target RNA. Perfect pairing causes mRNA degradation (cleavage).
Not considered long noncoding RNAs because of their short length in bp and their strictly post-translational function.
Small Interfering RNA (siRNA)
Created when double-stranded RNA (dsRNA) produced from viral RNA is cut into fragments via the dicer enzyme and reorganized.
siRNA-Induced Silencing Complex (siRISC)
One strand of the double-stranded (the dsRNA guide strand) is loaded into the RISC complex (siRISC). The passenger strand is degraded.
Single-stranded RNAs that are perfectly complementary to the siRNA in the siRISC are targeted and cleaved.
Protein Degradation
A process that starts with the consumption of ATP to add ubiquitin (a protein marker) to an unneeded protein.
Proteasome digests the protein and releases peptide fragments. These are degraded to amino acids by cytosolic enzymes. The amino acids, along with ubiquitin and the proteasome itself, are recycled for use in protein synthesis or oxidized as an energy source.
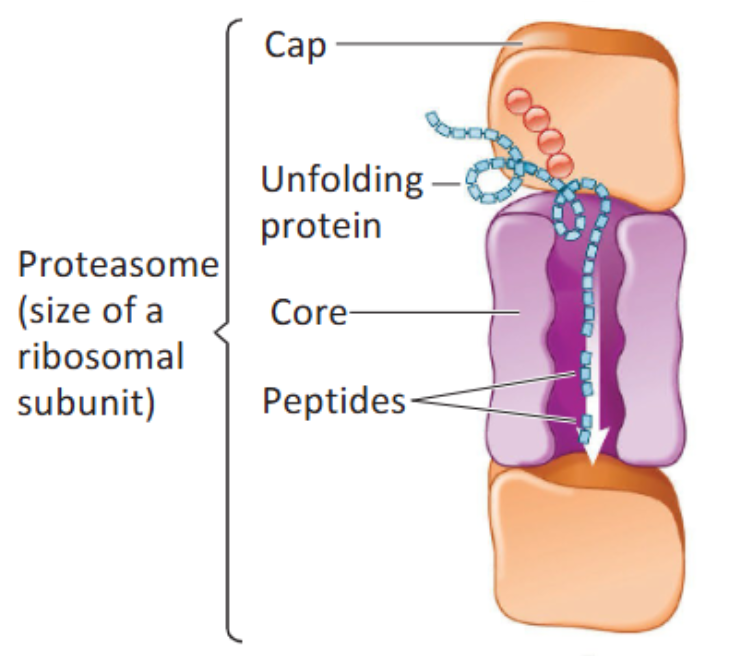
Proteasome
A large cytoplasmic complex (around the size of a ribosomal subunit) of a number of different proteins that recognizes ubiquitin-tagged protein and unfolds it. Enzymes that are part of the core digest protein to small peptides (requires ATP).