
Looks like no one added any tags here yet for you.
What is protein concentration a balance between?
Synthesis and degradation
What are the 2 types of protein degradation?
Lysosomal
Proteases in an acidic organelle
Cytosolic
In proteasomes, multienzyme complexes in the cytoplasm after ubiquitination
Or by proteolytic enzymes (trypsin, chymotrypsin, pepsin…)
What are lysosomes?
Major digestive organelle for both cytosolic and extracellular molecules
Single membrane
How do lysosomes work?
Acidification needed for proper activity. Low pH environment
Vacuolar H+ pump to keep pH at ~4.5 (helps unfold target proteins)
Most proteins will be unfolded – acidic
Primarily through hydrolytic activity – add water to amide bond to break it
How do targets for degradation enter lysosomes?
Either heterophagy or autophagy
What do lysosomes contain?
Contains many hydrolytic enzymes (hydrolases)
o Proteases, lipases, glycosidases, nucleases, amylase
o Can digest lipids, sugars, mRNA ect
Work optimally at low pH
What targets proteins into lysosomes from ER?
Mannose-6-phosphate tag
How are proteins to be degraded delivered to lysosomes?
By endocytosis
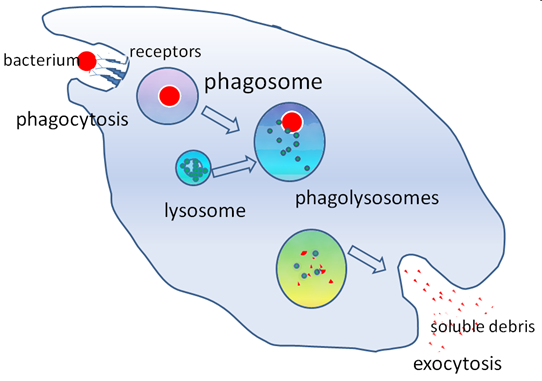
What is heterophagy?
Digestion within a cell of a substance taken in by phagocytosis from the cell's environment. e.g. bacteria
How does heterophagy work?
Receptor recognizes bacteria
Create invagination
Phagosome forms
Lysosome combines with phagosome to form phagolysosomes to degrade
What is autophagy?
• Degradation of cell's own components using lysosomes
• Lysosomes fuse with vesicles and degrade contents
• Stimulated by starvation
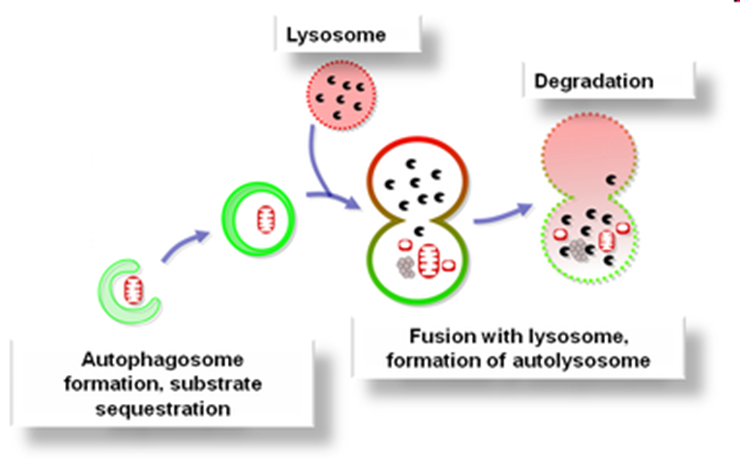
How does autophagy work?
Autophagosome forms
Substrate sequestered
Fuses with lysosome to for autolysosome
What is autolysis?
• Cell destroys itself by releases lysosome contents into cytoplasm
• Occurs in injured cells or dying tissue
• Webbed fingers or feet occur due to incomplete autolysis in a fetus
• Sometimes when proteins don’t go into autolysis they become cancerous
What is the proteasome?
Machine for degrading proteins located in the nucleus and cytosol of eukaryotic cells
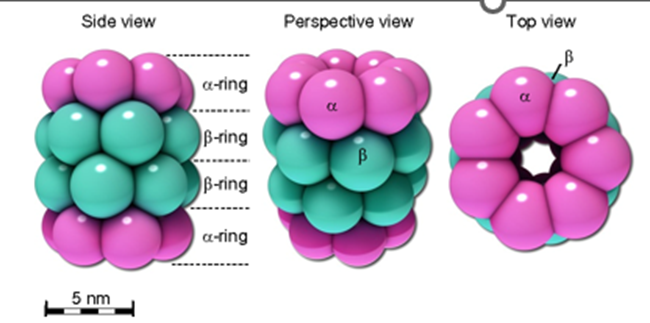
What is the structure of the core particle (20S proteasome)
4 stacked rings, forming a cylinder
Each ring made of 7 polypeptides
2 outer rings made of α-subunits, and the inner two rings consist of β-subunits, which are involved in the proteolytic activity.
The inner cavity of the proteasome contains the proteolytic sites, where protein degradation occurs.
What are the beta subunits of the proteasome?
Proteasome hydrolases - threonine proteases
Conserved N-terminal threonine is involved in catalysis at each active site
Why are the threonine proteases not active all the time?
have a sequence at the end that caps the OH of the N terminus
When removed, threonine proteases are active
How are the beta subunits synthesised and activated?
Synthesised as inactive pre-proteins
Activated when N-terminal removed, making threonine the N terminal residue
Catalytic threonines are then exposed at the inner surface
What do the threonine proteases do and then what happens?
Threonine proteases chops the protein substrate into 7-9 residue peptides
Another set of enzymes makes them 3 aa long
Another set chops them into single aa
Step process to make sure we don’t degrade the wrong protein or at the wrong time
What happens to a ubiquitin tagged protein in a proteasome?
An ubiquitin-tagged protein is unfolded and de-ubiquitinated in the cap, then threaded through the core, where it is digested into peptides
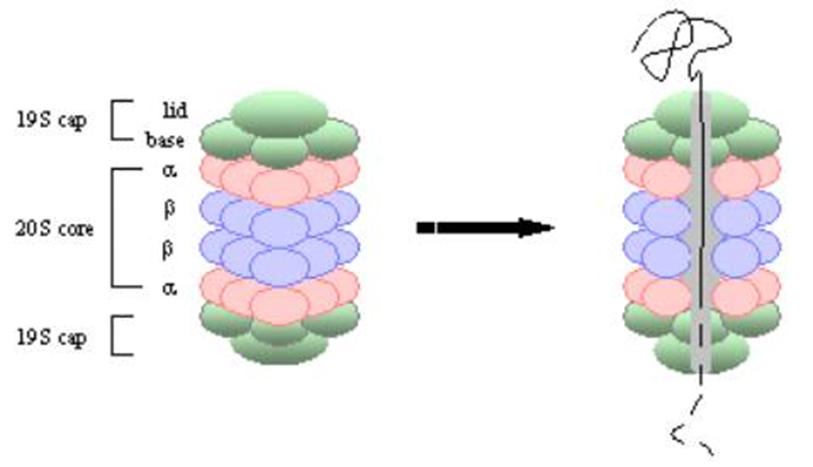
What is the regulatory particle (19S proteasome)
The regulatory particle is composed of additional subunits that control the entry of substrates into the proteasome.
It contains a lid that helps recognize polyubiquitinated proteins (those tagged for degradation) and a base that unfolds and translocates the protein substrate into the core for degradation.
Opens the 20S so a substrate can enter
What is protease activity done by?
3 of the beta subunits - each with different specificities
What are the specificities of the beta subunits?
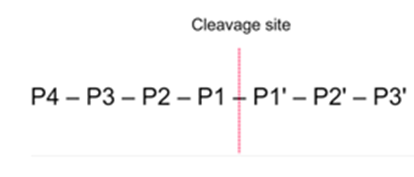
• One catalytic b subunit has a chymotrypsin-like activity with preference for tyrosine or phenylalanine at the P1 (peptide cabonyl) position.
• One has a trypsin-like activity with preference for arginine or lysine at the P1 position.
• One has a post-glutamyl activity with preference for glutamate or other acidic residue at the P1 position.
How is progression through the cell cycle controlled?
Regulated degradation of cyclins
How can proteasome inhibitors cause cell cycle arrest?
Stop cyclins being degraded
Cause cell cycle arrest and induction of apoptosis when added to rapidly dividing cells
Potential anti-cancer drug
How is proteasome controlled by glucose?
When extracellular glucose is high, several subunits of the proteasome are glycosylated with GlcNAc (N-acetylglucosamine. Intracellular proteolysis then decreases.
Conversely: low extracellular glucose leads to removal of GlcNAc which increased proteolysis
What is rhabdomyolysis?
o When we starve for a long time, so body starts eating at protein from muscles
o Releases toxins into the blood
o Can be reversed when start eating protein again
What are proteinases?
Enzymes that break proteins into shorter fragments (peptides) and eventually into their component amino acids
How are proteinases classified?
Classified by how they work:
o Serine proteases
o Threonine proteases
o Cysteine proteases
o Aspartate proteases
o Metalloproteases
o Glutamic acid proteases
where Serine etc. is the side chain in the active site that does the chemistry
What is the preferred side chain (r group) of chymotrypsin?
Aromatic - Phe and tyr
What is the preferred side chain (r group) of trypsin?
Positive charge - lys and arg
cleaves peptide bonds at the C-terminal side of lysine or arginine residues
Serine protease
What is the preferred side chain (r group) of elastase?
Small hydrophobic eg ala
How do proteinases work?
• Different enzymes are specific for different side chains (R groups)
• Can work in combination with metal
• Add water and break a peptide bond
• Usually not active on own, need triad
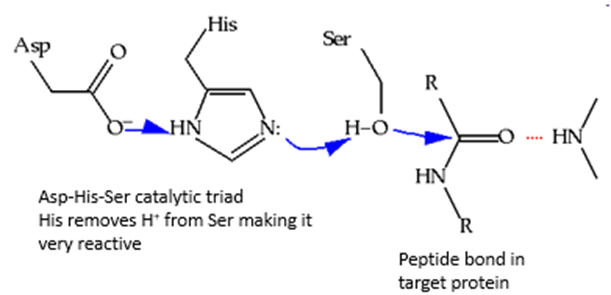
How do serine proteases work?
Active site contains catalytic triad - serine, histidine and aspartate
To activate serine, histidine removes hydrogen so have a negative oxygen
Aspartate takes away hydrogen from histidine which becomes positive and takes the hydrogen away from serine
Attach the peptide bond and break it
Ultimately, proteins are chopped up into their amino acids, ready to be used again to make new proteins
What does each member of the catalytic triad do?
Serine (Ser): The nucleophilic residue that attacks the peptide bond.
Histidine (His): Acts as a base, accepting a proton from serine, making serine a better nucleophile.
Aspartate (Asp): Stabilizes the positive charge on histidine, aiding in its role as a base.
More detailed serine protease digestion
Substrate Binding:
The substrate protein (the peptide or protein to be cleaved) binds to the enzyme's active site.
The scissile bond (the peptide bond to be cleaved) is positioned near the active site, and specific binding interactions help orient the substrate for catalysis.
Nucleophilic Attack:
The serine hydroxyl group (OH), activated by the histidine residue, performs a nucleophilic attack on the carbonyl carbon of the scissile peptide bond.
This attack forms a tetrahedral intermediate, which is a temporary structure with four bonds around the carbonyl carbon.
Formation of Acyl-Enzyme Intermediate:
The tetrahedral intermediate quickly collapses, leading to the breakage of the peptide bond and the formation of an acyl-enzyme complex. The leaving group is typically the amino portion of the substrate.
The carboxyl group of the cleaved peptide (the part after the scissile bond) is released as a free product.
Water Attack:
A water molecule is then activated by the histidine residue, and the water molecule attacks the acyl-enzyme complex.
This step regenerates the active serine hydroxyl group and releases the remaining cleaved peptide as a product.
Regeneration of Active Site:
After the hydrolysis of the acyl-enzyme intermediate, the active site is regenerated, ready to cleave another peptide bond.