Biochem Exam 2 Stuff
1/193
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
194 Terms
Primary Structure (1°)
Amino acid sequence
Linear order of AA’s
Secondary Structure (2°)
Local spatial alignment of amino acid backbone without regard to side chains, α-helix, β-strands/sheets, random coil, and β-turns
Tertiary Structure (3°)
The 3-dimensional structure of an entire polypeptide, fold, biological function and catalytic mechanism
Quaternary Structure (4°)
The manner in which the tertiary structures of two or more polypeptide chains of a protein interact
Spatial arrangements of subunits (folded chains)
Conformation
Spatial arrangement of atoms that depend on bonds and bond rotations/torsions.
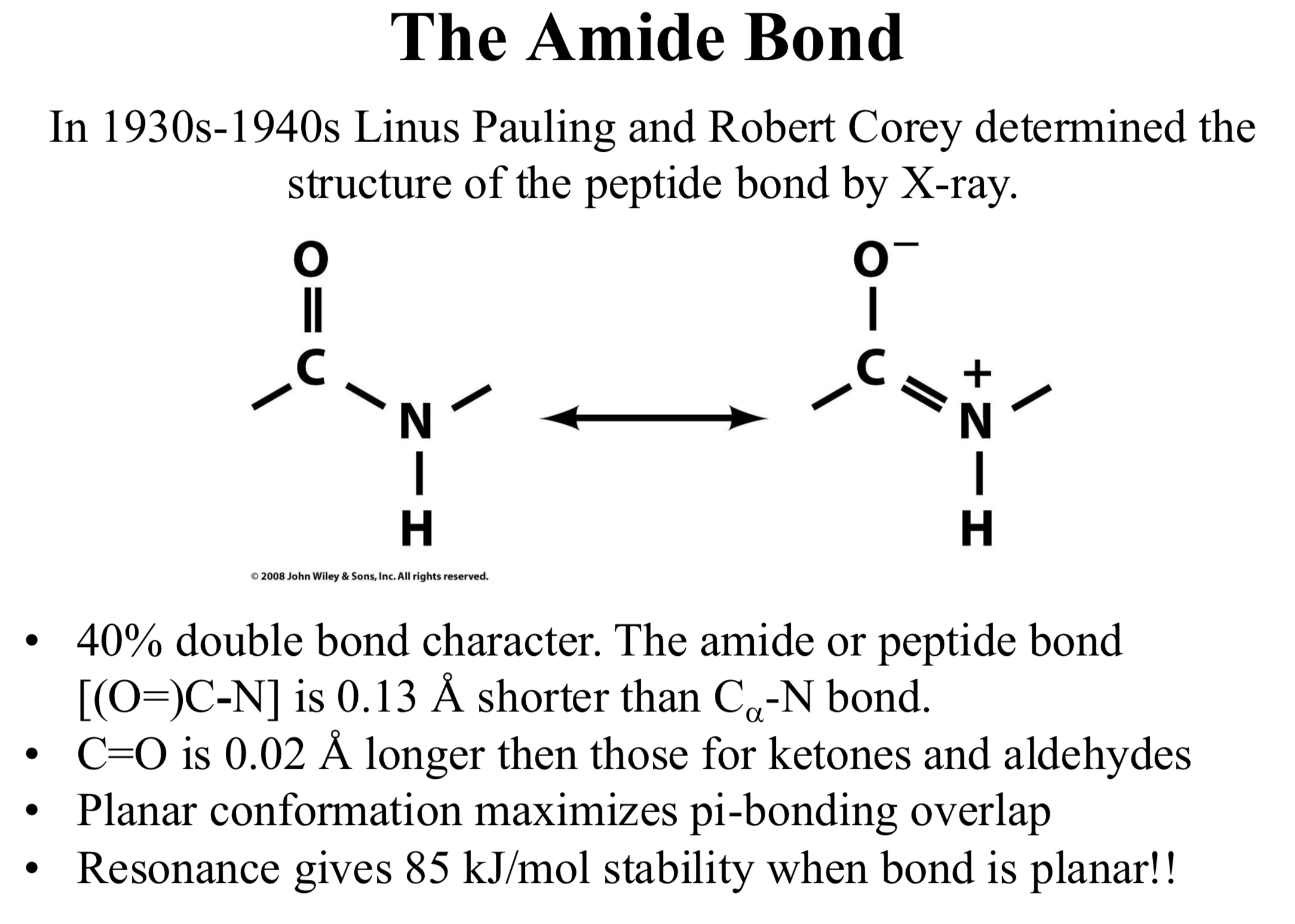
In 1930s-1940s Linus Pauling and Robert Corey determined the structure of the peptide bond by X-ray.
40% double bond character.
The amide or peptide bond [(O=)C-N] is 0.13 Å shorter than Cα-N bond
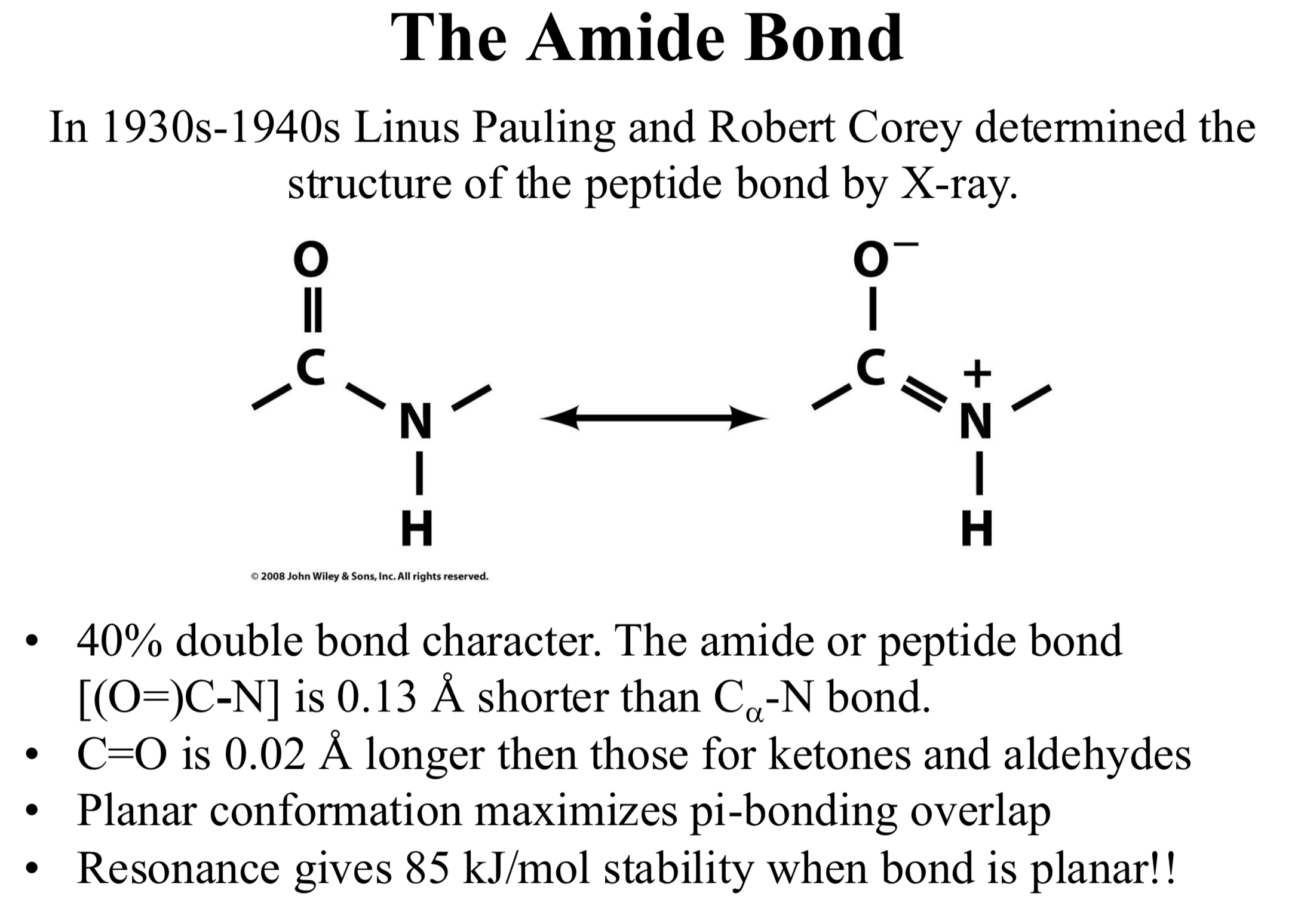
C=O is 0.02 Å longer then those for ketones and aldehydes
Planar conformation maximizes pi-bonding overlap
Resonance gives 85 kJ/mol stability when bond is planar!
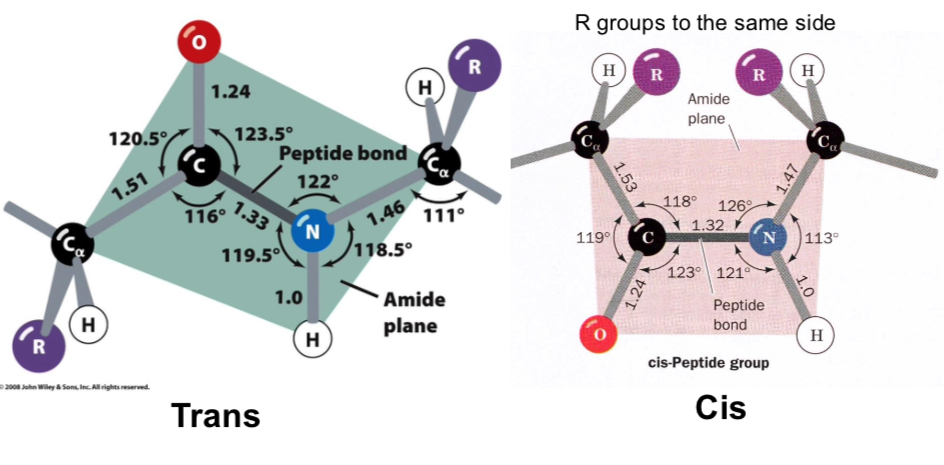
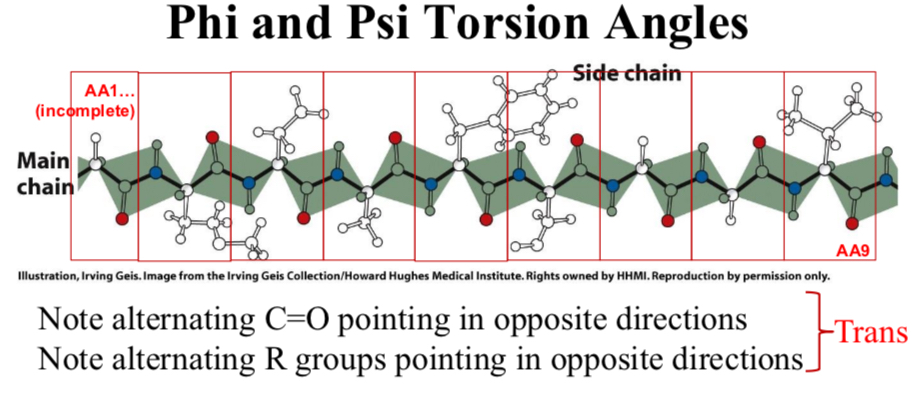
Peptide bonds are planar
Most peptide bonds are trans, 10% that follow proline may be cis
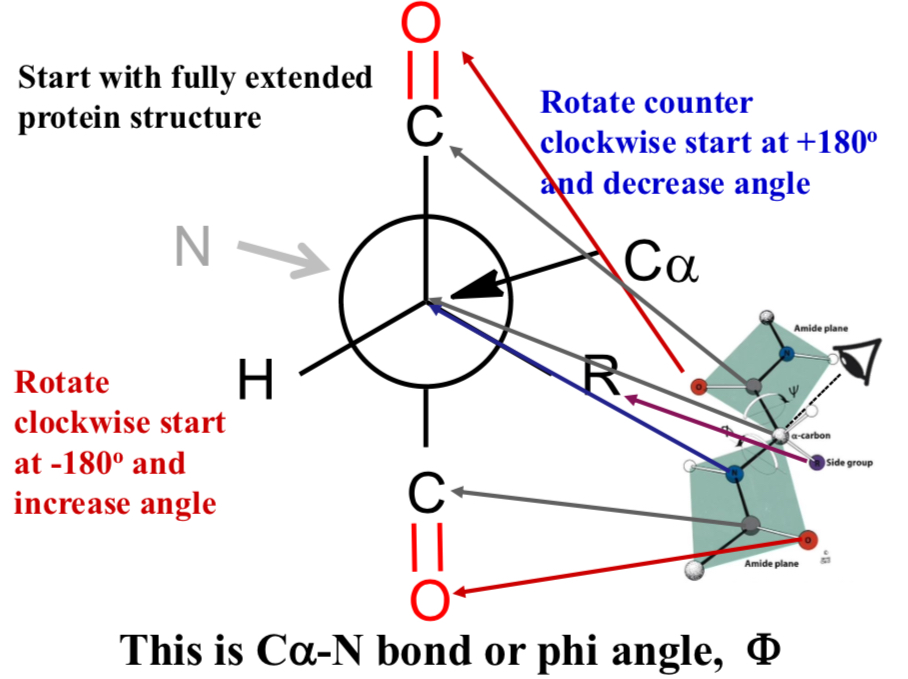
This is Cα-N bond or phi angle, Φ
Start with fully extended protein structure
Rotate counter clockwise start at +180° and decrease angle
Rotate clockwise start at -180° and increase angle
Phi and Psi Torsion Angles
For a trans peptide bond, the dihedral angle is 180° by definition (or -180°, these are the same).
In a cis peptide bond, the dihedral angle is 0° by definition
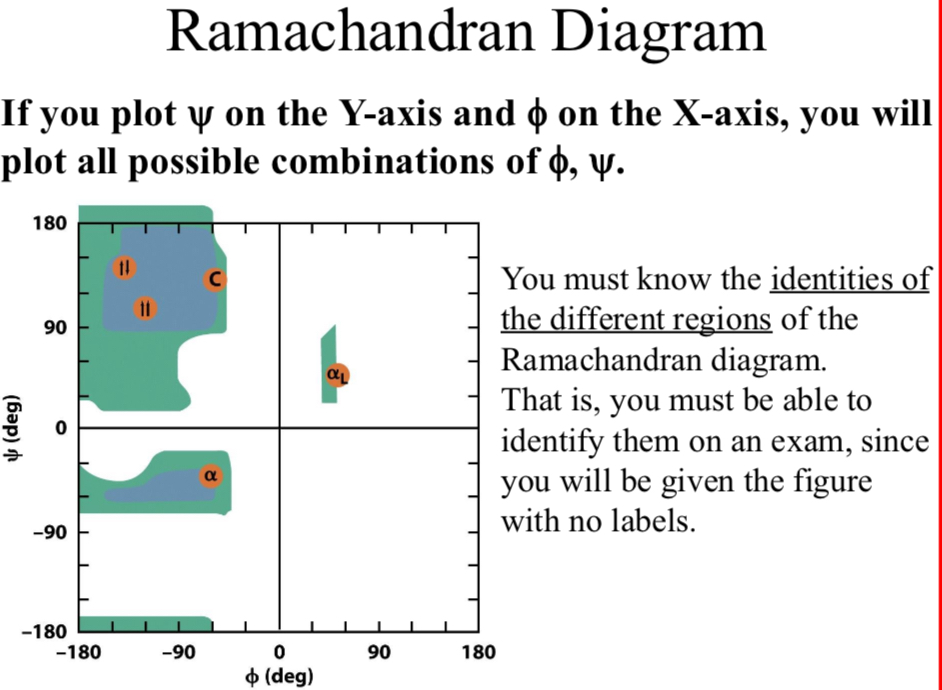
Ramachandran Diagram
If you plot Ψ on the Y-axis and Φ on the X-axis, you will plot all possible combinations of Φ, Ψ.
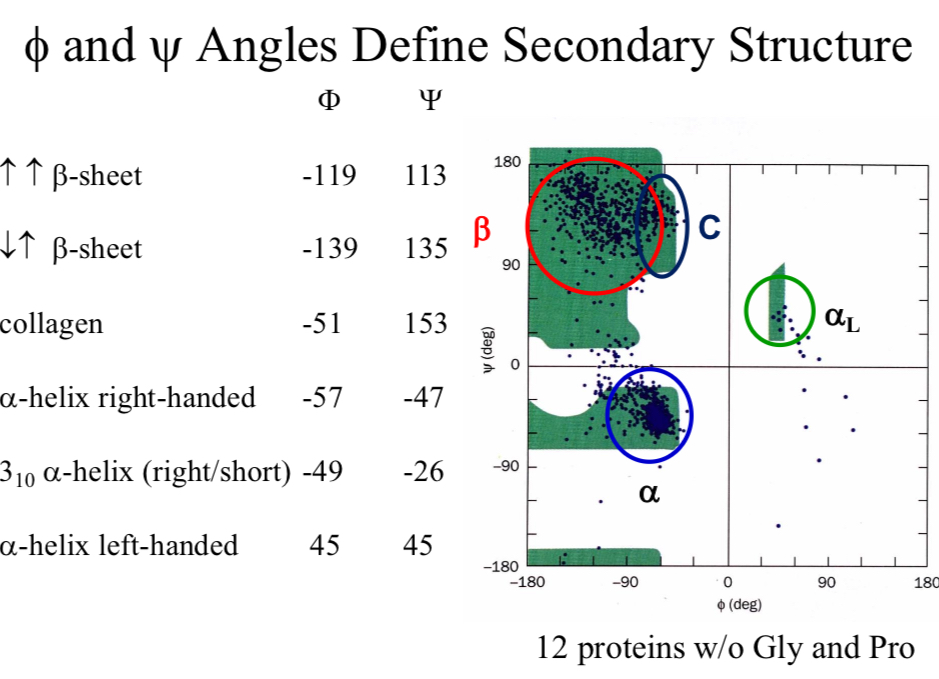
Φ and Ψ Angles Define…
Secondary Structure
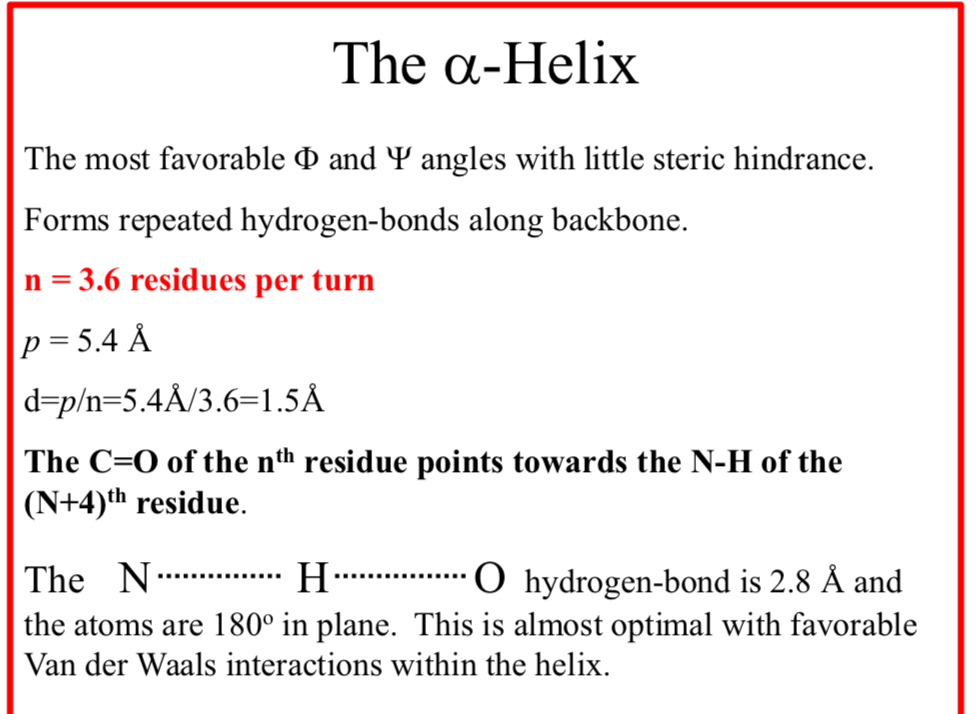
The α-Helix
The most favorable Φ and Ψ angles with little steric hindrance
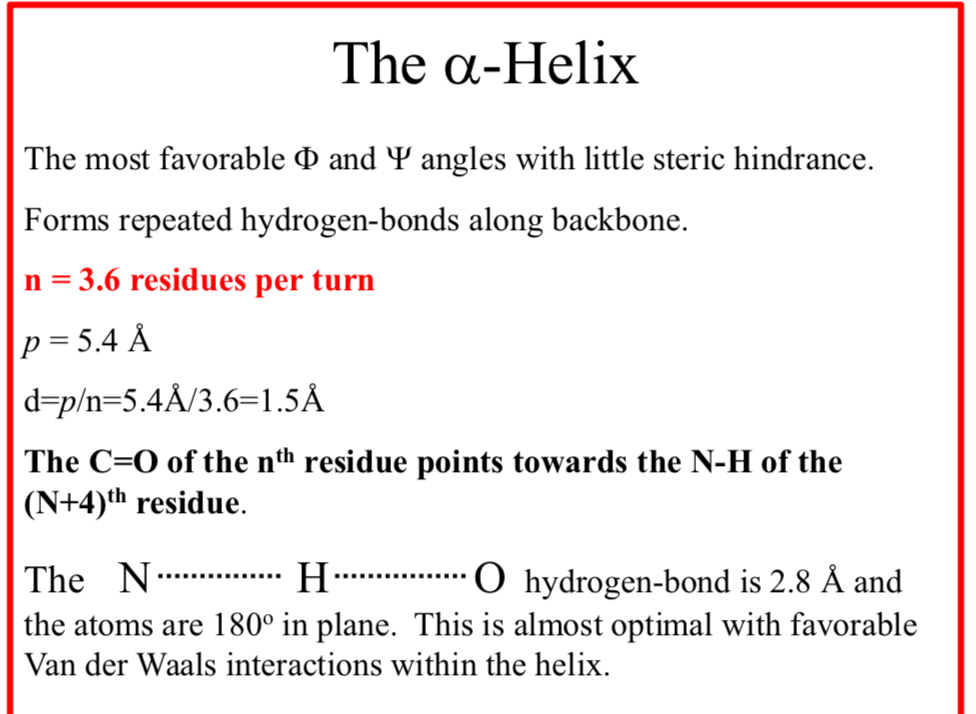
The α-Helix forms repeated hydrogen-bonds along backbone
n = 3.6 residues per turn
p = 5.4 Å
d = p/n = 5.4Å/3.6 = 1.5Å
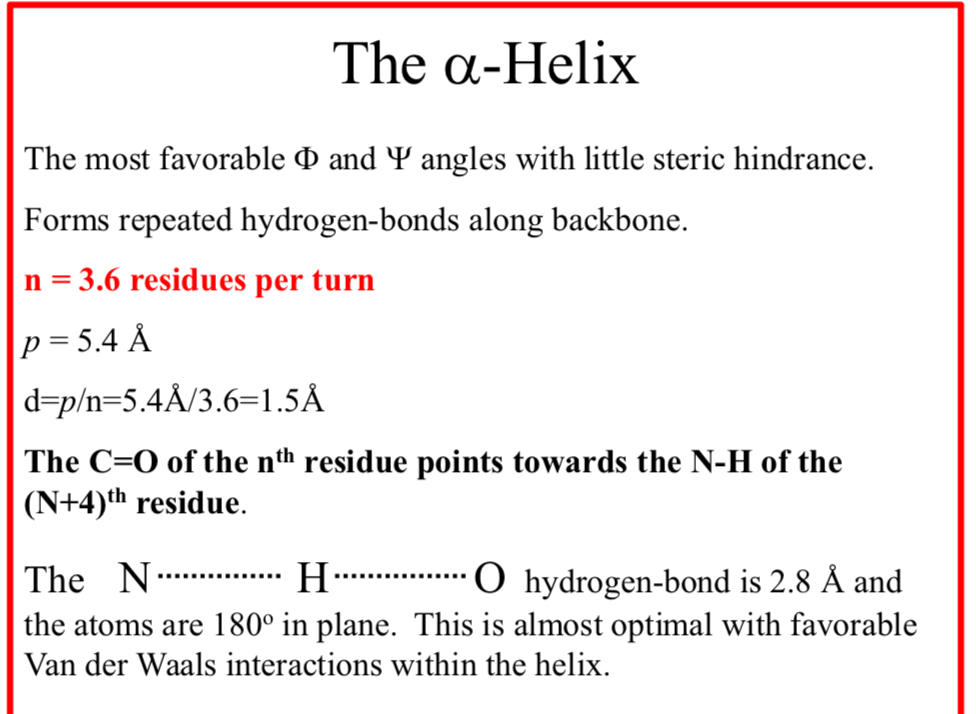
The C=O of the nth residue points towards the N-H of the (N+4)th residue
The N……..H……..O hydrogen-bond is 2.8 Å and the atoms are 180° in plane.
This is almost optimal with favorable Van der Waals interactions within the helix
The Nm Helix Nomenclature
N = the number of repeating units per turn
m = the number of atoms that complete the cyclic system that is enclosed by the hydrogen bond.
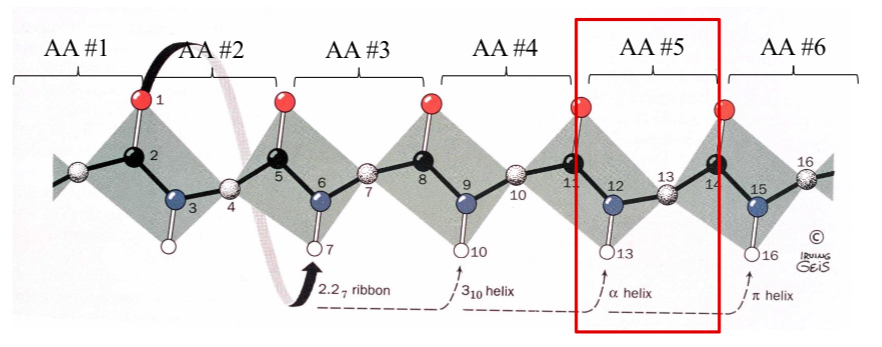
What is the designation for an α-helix?
Nm = 3.613
Helix types: The 2.27 Ribbon
Atom (1) -O- hydrogen-bonds to the 7th atom in the chain with an N = 2.2 (2.2 residues per turn)
Helix types: 310-helix
Atom (1) -O- hydrogen-bonds to the 10th residue in the chain with an N= 3.
Pitch = 6.0 Å occasionally observed but torsion angles are slightly forbidden.
Seen as a single turn at the end of an α-helix.
Helix types: π-helix 4.116
4.4 residues per turn.
Very rare!!
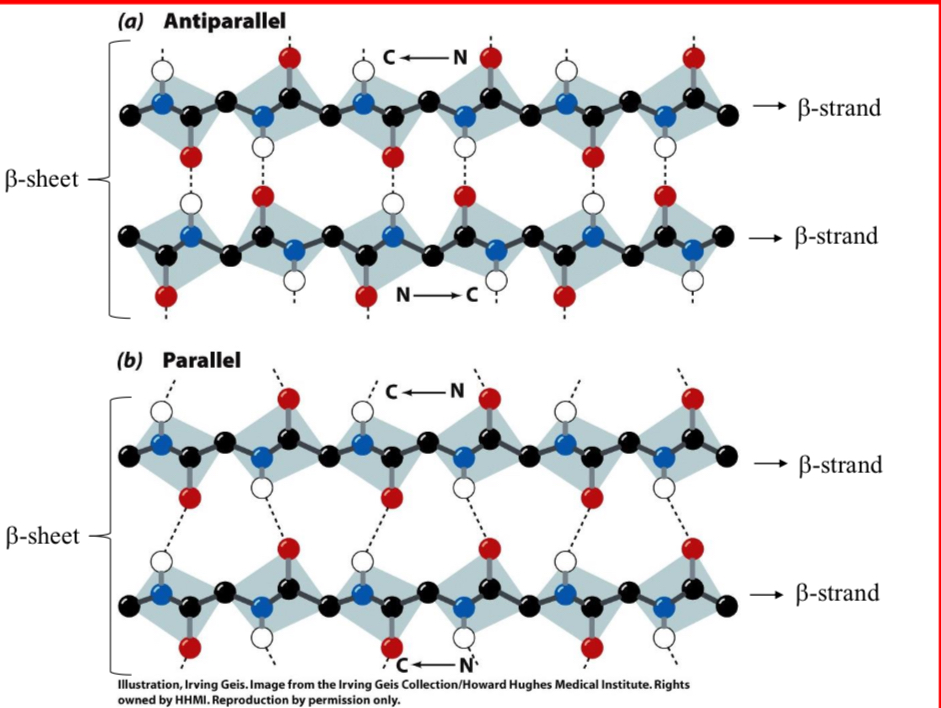
Beta Structures (β-Sheet)
Hydrogen-bonding between adjacent peptide chains.
Almost fully extended but have a buckle or a pleat.
Much like a Ruffles potato chip
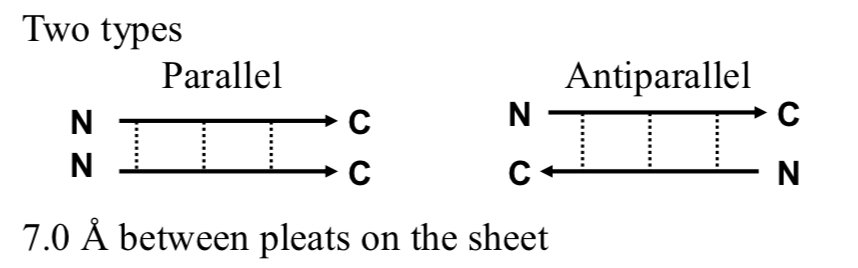
2 Types of Beta Structures
Parallel & Antiparallel
7.0 Å between pleats on the sheet
Widely found pleated sheets exhibit a right-handed twist, seen in many globular proteins
What is the repeat distance of a β-Sheet?
7.0 Å
Where is the R group at for a β-Sheet?
R group on the amino acids alternate up-down-up above and below the plane of the sheet
How long is a β-Sheet?
2 - 15 amino acids residues long
How many strands are in a β-Sheet?
2 - 22 strands per sheet
Avg. of 6 strands with a width of 25 Å
Parallel β-Sheet is less stable than antiparallel
True
Antiparallel β-Sheet needs what?
A hairpin turn
What does a tandem parallel β-Sheet need?
It needs crossover connection which is right handed sense
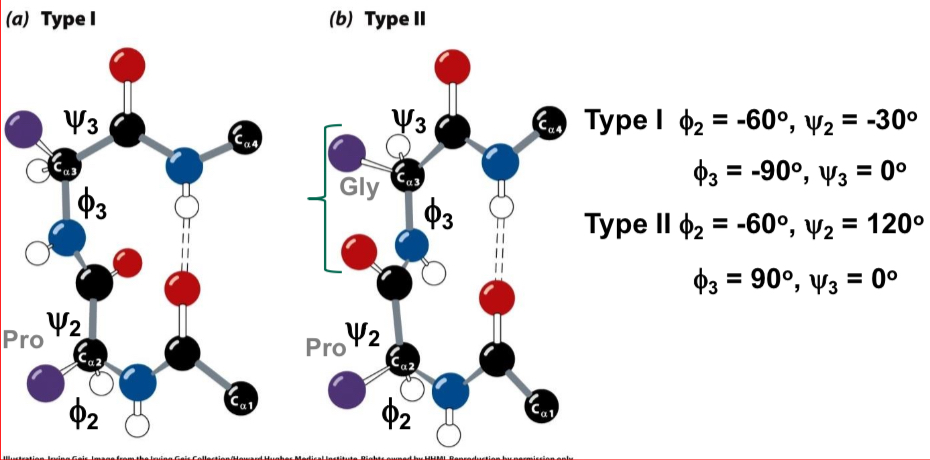
Non-Repetitive Structures
Turns - coils or loops: 50% of structure of globular proteins are not repeating structures
β-bends (b-turns) - type I and type II: hairpin turn between anti-parallel sheets
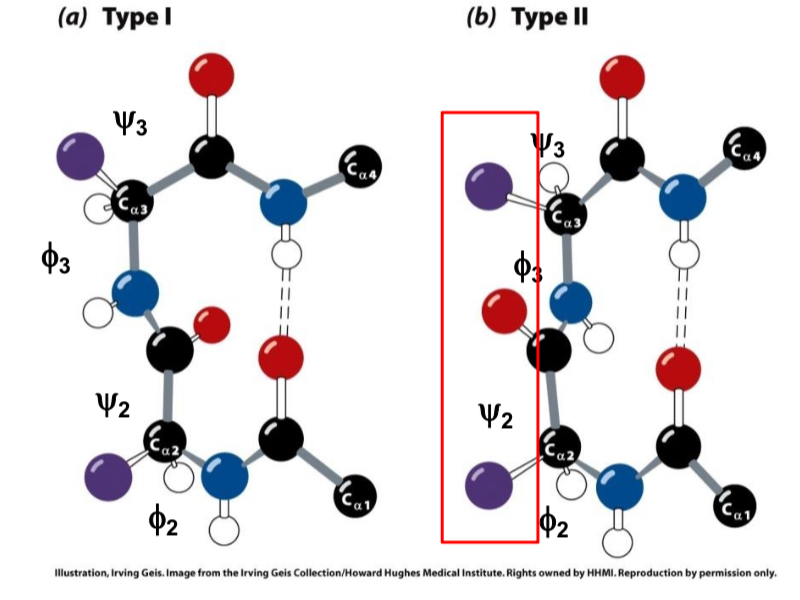
What type is when the carbonyl is on the same side as sidechains?
Type II
Fibrous proteins
Highly elongated molecules whose shapes are dominated by a single type of secondary structure.
No significant tertiary structure arrangements.
Usually insoluble in water.
Examples: Keratin and Collagen
Globular proteins
More “compacted” molecules, containing several types of regular secondary structures.
Usually arranged in domains with unique tertiary structure. May also contain irregular, more flexible, segments (coils).
Usually water soluble.
Examples: Globin, Hemoglobin, Cytochrome c…
About 98% of proteins are globular. But some Fibrous proteins are extremely abundant.
Collagen is the most abundant protein in vertebrates!
Keratin: A Quasi α-Helical Protein
Nails, hair, horns and feathers α or β-forms
Over 50 variants, tissue specific
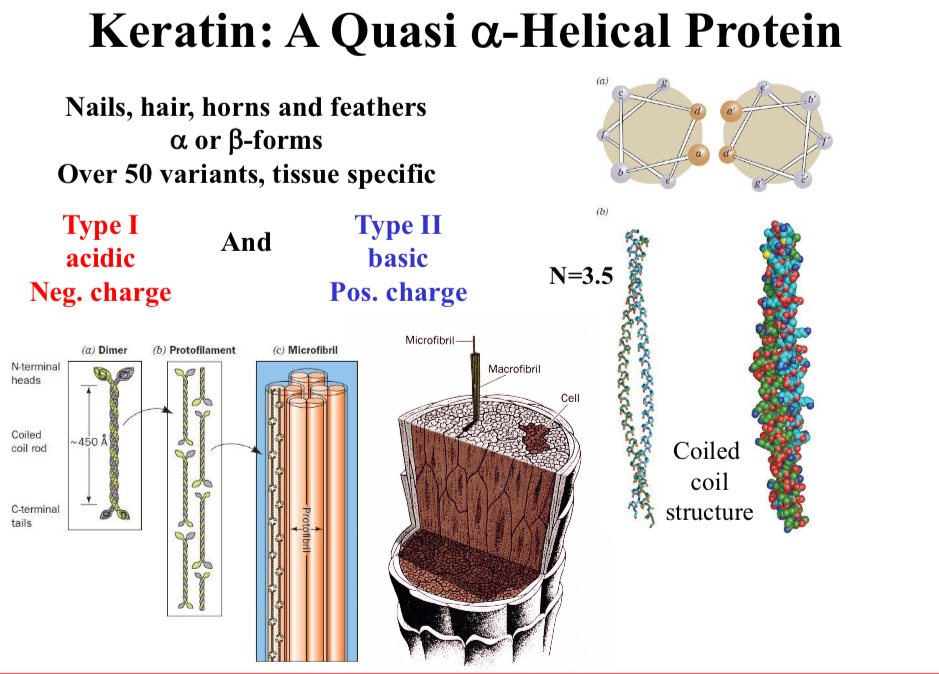
2 types of keratin
Type I: acidic, negative charge
Type II: basic, positive charge
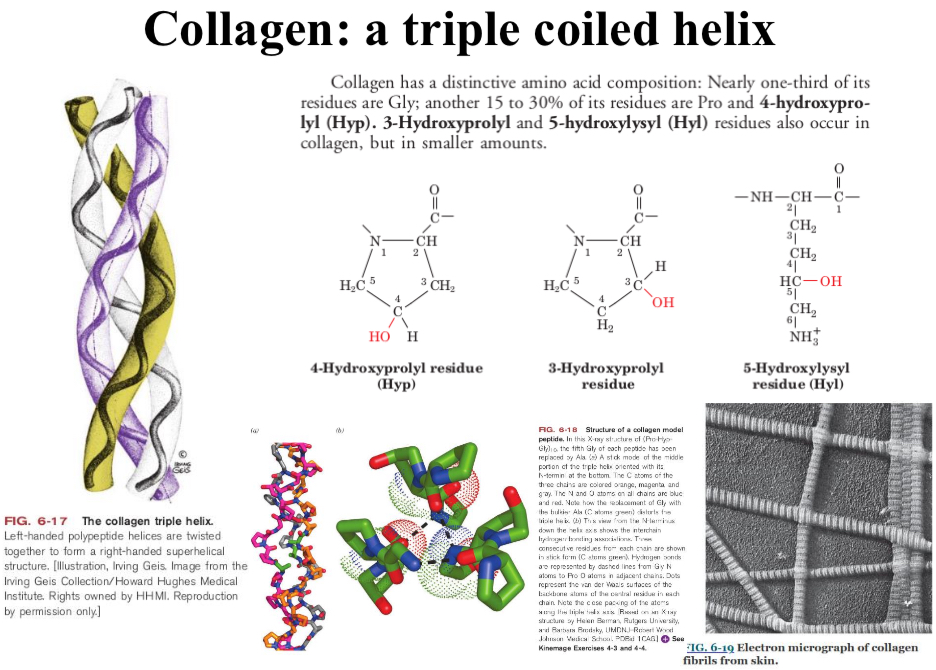
Collagen
A triple coiled helix
Sidechain Locations in Proteins; Non-polar: (Val, Leu, Ile, Met, and Phe)
Occur mostly in the interior of a protein keeping them out of the water (hydrophobic effect)
Sidechain Locations in Proteins; Charged polar: (Arg, His, Lys, Asp, and Glu)
Normally located on the surface of the protein in contact with water
Sidechain Locations in Proteins; Uncharged polar: (Ser, Thr, Asn, Gln, and Tyr)
Usually on the protein surface but also occur in the interior of the protein.
Usually involved in hydrogen bonds with neighbors
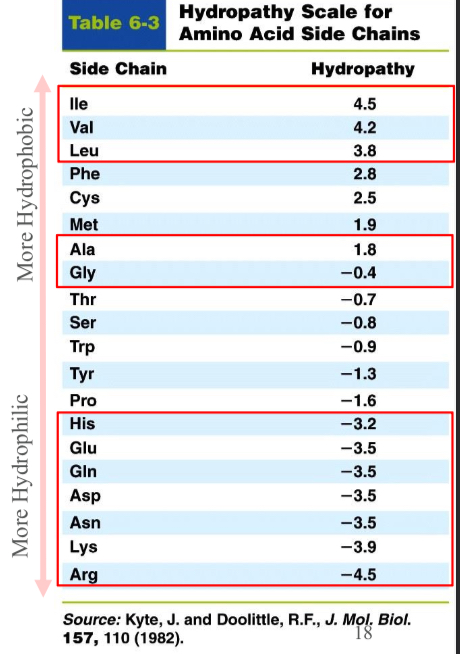
Hydrophaty scale
Allows an assessment of which amino acid residues would point towards the interior (high numbers) or towards the surface (low numbers)
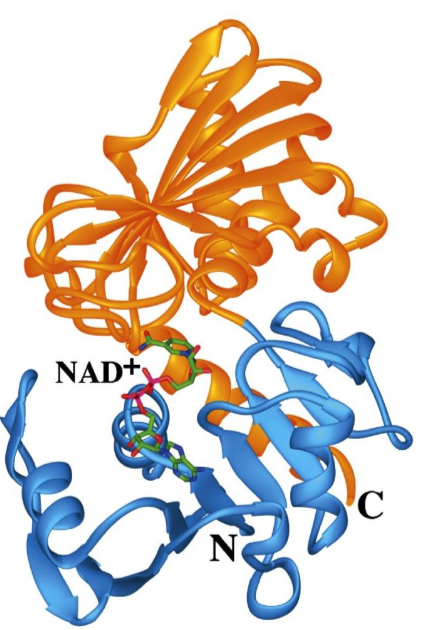
Protein Domains
Many single polypeptide chains fold into multiple structural domains, each with their own function
glyceraldehyde-3-phosphate (GAP) dehydrogenase
The blue domain binds NAD+
The orange domain binds GAP
A glycolysis pathway enzyme
Note: both domains are part of the tertiary structure of this polypeptide chain!
Supersecondary structural motifs
Small, recognizable combinations of a few secondary structure elements that occur repeatedly in different proteins.
They are usually smaller then protein domains and usually don’t have an independent function by themselves, but serve as building blocks for more complex structures
Protein domains
Larger, compact and independently folding units of proteins’ tertiary structure.
Often corresponds to a protein unit that is functional (e.g., specific biochemical function) and “evolutionary” (i.e., conserved over the evolution)
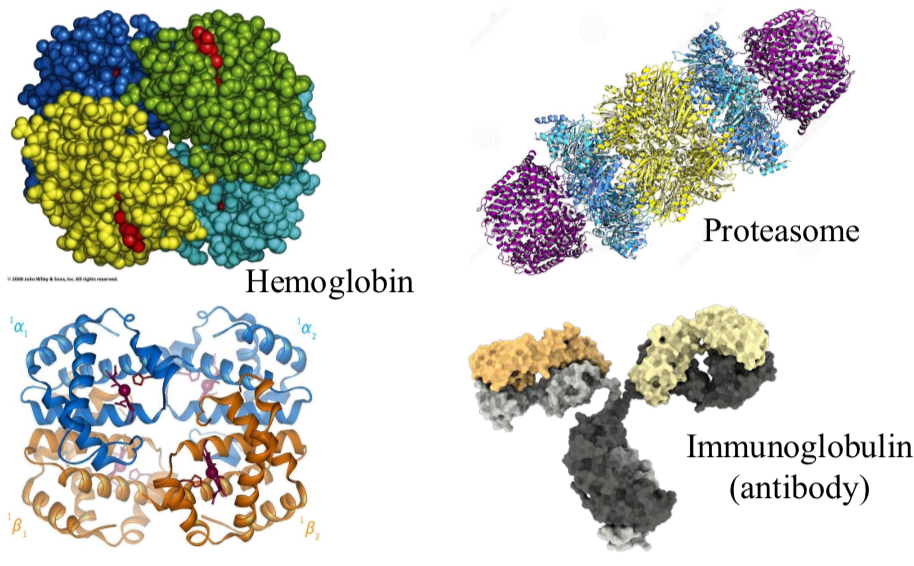
Quaternary Protein Structure
4° structure is the relative placement of different polypeptide segments
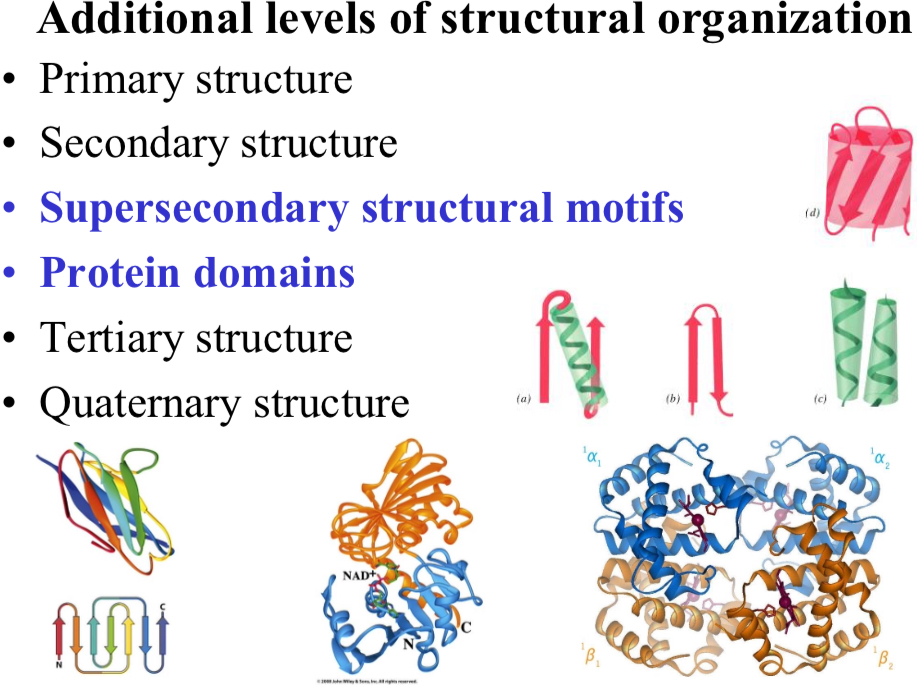
Additional levels of structural organization
Primary structure
Secondary structure
Supersecondary structural motifs
Protein domains
Tertiary structure
Quaternary structure
Protein structures are stabilized by…
Several different forces
Electrostatics, hydrogen bonds and van der Waals forces hold…
A protein together (tertiary and quaternary structs.)
Hydrophobic effects force global protein conformation and…
Has the greatest effect on structure and stability.
In transmembrane proteins the most hydrophobic residues are found…
In membrane spanning regions.
Peptide chains can be cross-linked by…
Disulfides, salt-bridge networks, metal ions, prosthetic groups, or other ligand compounds
Proteins refold very rapidly and generally…
In only one stable conformation.
Heating disrupts protein structure thermal vibrations and disrupt weak bonding forces.
Note that there are heat stable proteins, which have a few more hydrogen bonds and salt bridges — networks of “weak” interactions
pH changes lead to denaturation.
Protonation of amino acids leads to loss of charge and H-bonding
Detergents associate with the…
Nonpolar amino acids blocking water interactions
Chaotropic agents (e.g., Urea and Guanidinium ions)
Disrupt hydrophobic interactions by increasing the solubility of nonpolar groups (most commonly used protein denaturants, but mechanism of action not well understood).
What is the first step in protein folding?
Secondary structure formation
What is the driving factor in protein folding?
Hydrophobic affect
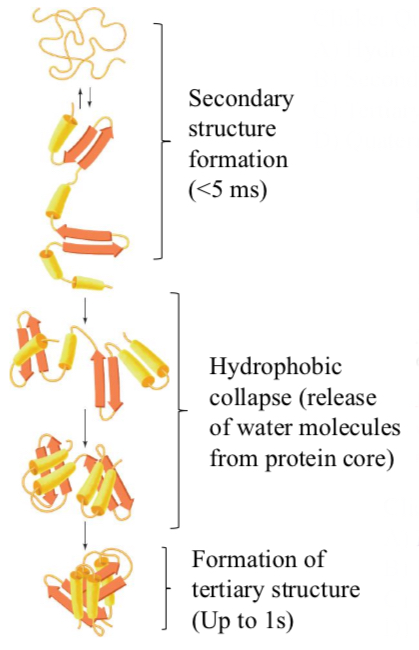
Proteins fold in a hierarchical way
Secondary structure formation (<5 ms)
Hydrophobic collapse (release of water molecules from protein core)
Formation of tertiary structure (Up to 1 s)
Folded state
Lower protein entropy, but much higher water entropy due to the release of waters from “cages” around exposed hydrophobic surfaces.
Proteins can (usually) fold spontaneously into their native states via directed pathways rather than random conformational searches.
Proteins appear to fold in a hierarchical manner, with small local elements of structure forming and then coalescing to yield larger elements, which coalesce with other such elements to form yet larger elements.
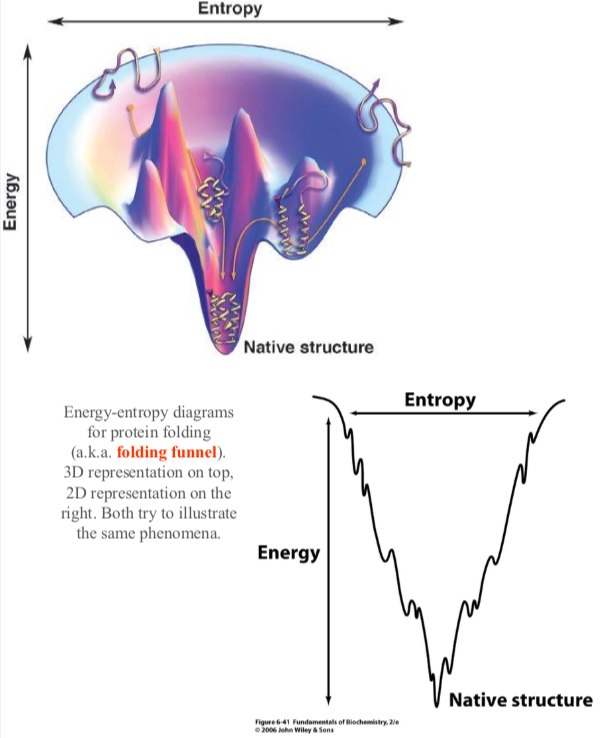
There are several local minimums that could trap a protein however the dynamic nature of a protein allows folding to a global energy minimum.
However certain proteins, and larger complexes, require help in folding
Protein disulfide isomerase (PDI)
Either reforms disulfide bonds as a shuffle mechanism or oxidizes new disulfide bonds
Molecular Chaperones
Bind unfolded proteins and prevent improper folding.
Especially important in multi-subunit complexes
Hsp70
Present in both eukaryotic and procaryotic cells
Trigger Factor
Prokaryotic ribosome-associated chaperon
Chaperonins
Form large multisubunit assemblies, both in eukaryotic and prokaryotic cells.
Hsp90
Among the most abundant proteins in eukaryotic cells
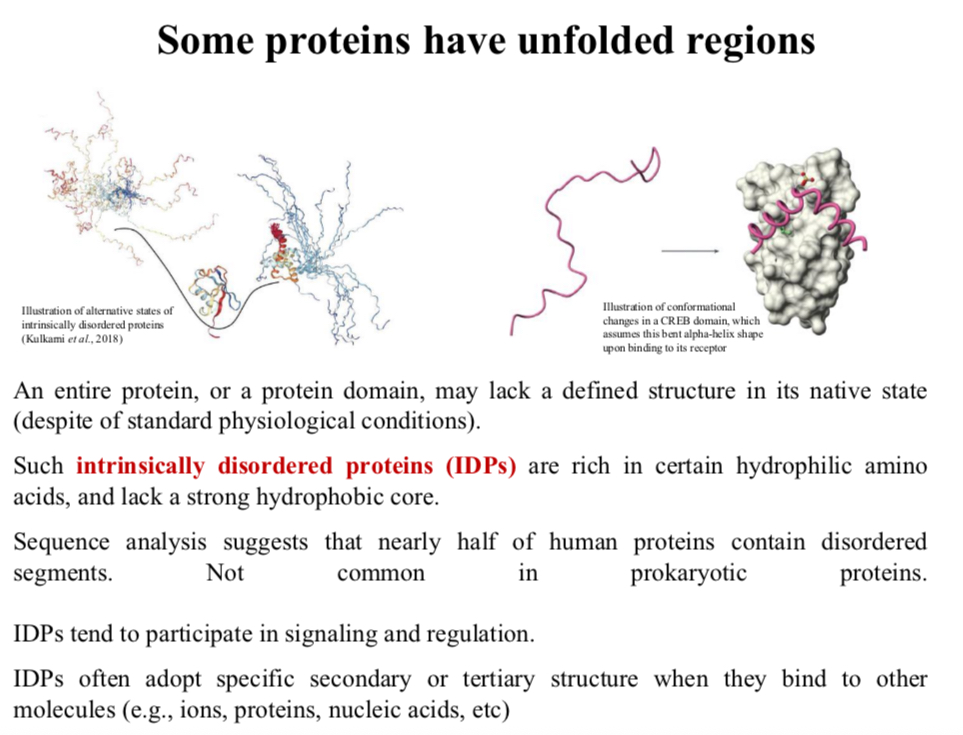
Some proteins have unfolded regions
An entire protein, or a protein domain, may lack a defined structure in its native state (despite of standard physiological conditions).
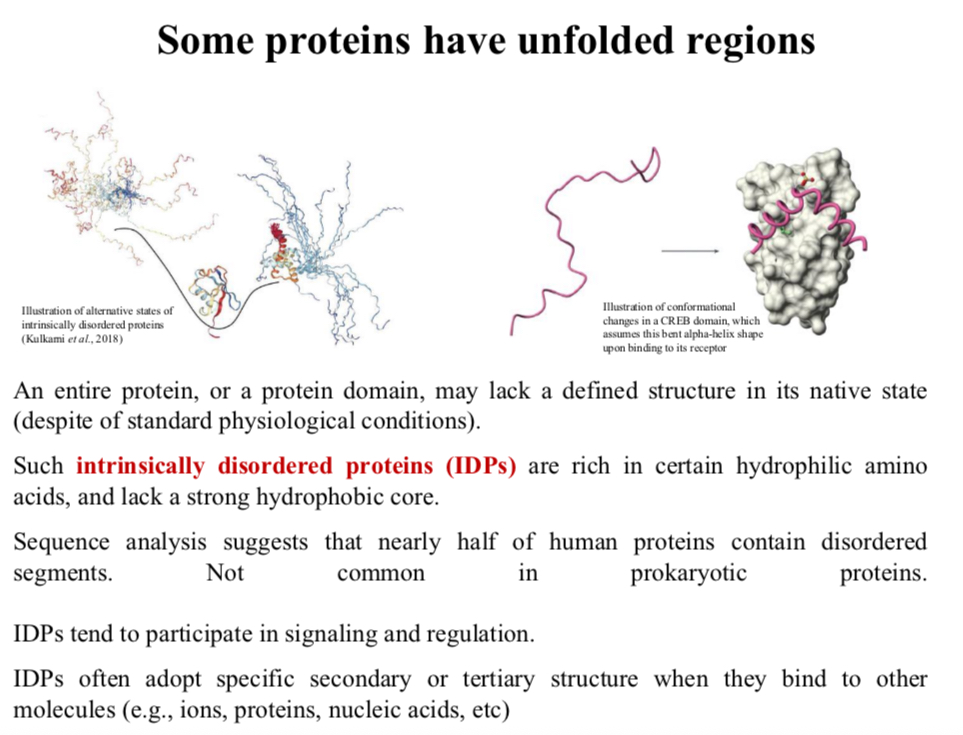
Such intrinsically disordered proteins (IDPs) are rich in certain hydrophilic amino acids, and lack a strong hydrophobic core.
Sequence analysis suggests that nearly half of human proteins contain disordered segments.
Not common in prokaryotic proteins.
IDPs tend to participate in signaling and regulation.
IDPs often adopt specific secondary or tertiary structure when they bind to other molecules (e.g., ions, proteins, nucleic acids, etc)
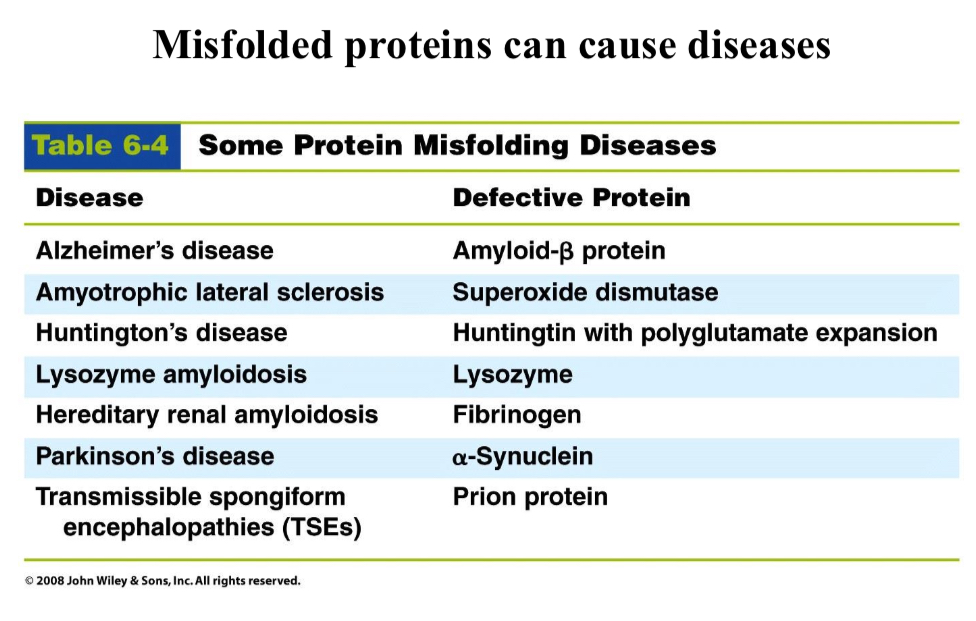
Misfolded proteins can…
Cause diseases
Transmissible spongiform encephalopathies (TSE) are not caused by virus or bacteria but…
Are associated with an “infectious” protein (prion)
The protein PrP has two different folds
The “normal” fold (PrPC) is essential to neuritogenesis, neuronal homeostasis, cell signaling, cell adhesion, and a protective role against stress.
However, the misfolded protein (PrPSC) causes the disease.
Contact between the (PrPC) and (PrPSC) leads to…
(PrPSC)
Eating nerve tissue of infected humans or cows but not sheep leads to the disease.
The incorrect fold leads to amyloid fibrils that causes mental regression
Sheep TSE
Scrapie
Cow TSE
Bovine spongiform encephalopathy (mad cow disease)
Human TSE
Creutzfeldt-Jakob Disease
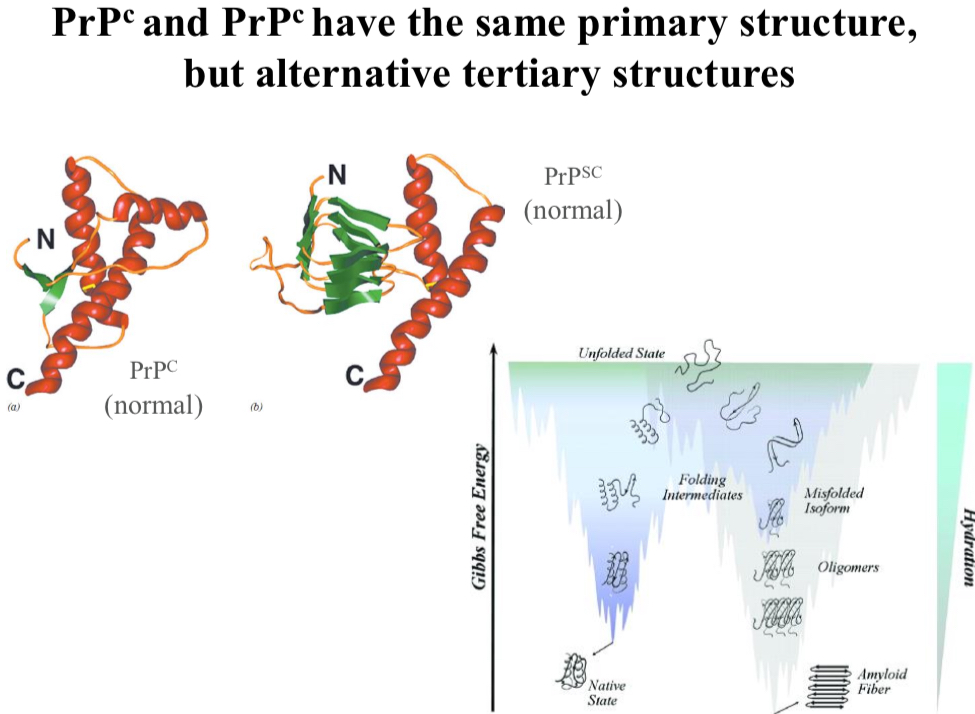
PrPC and PrPSC have the same primary structure, but…
Alternative tertiary structures
1958
John Kendrew, 3D model of sperm whale myoglobin from X-ray crystallography
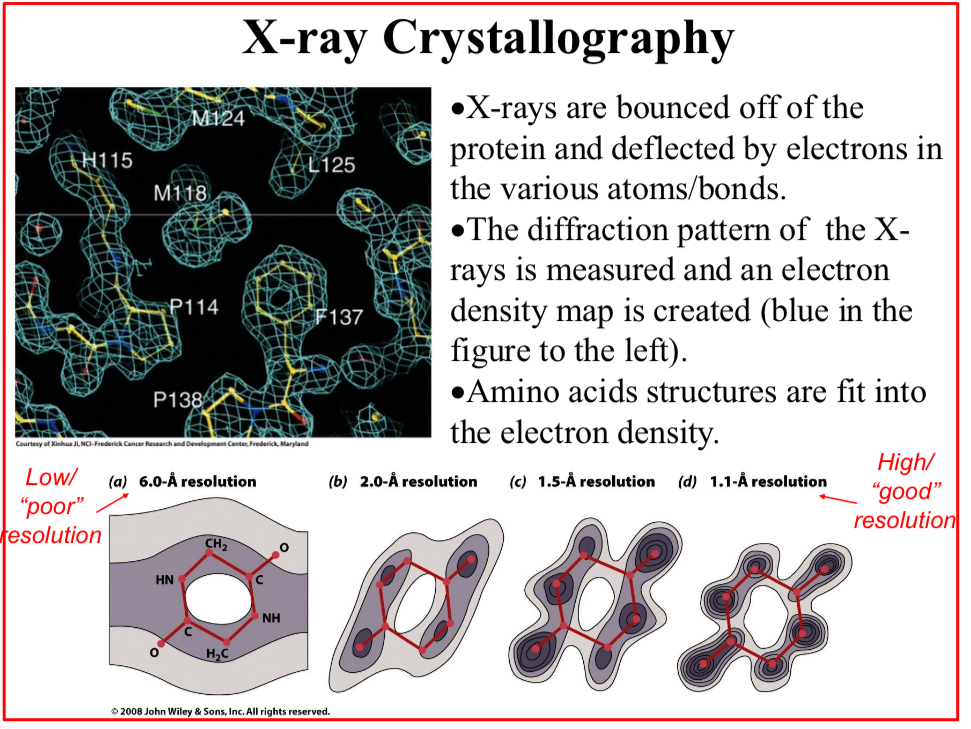
X-ray Crystallography
X-rays are bounced off of the protein and deflected by electrons in the various atoms/bonds.
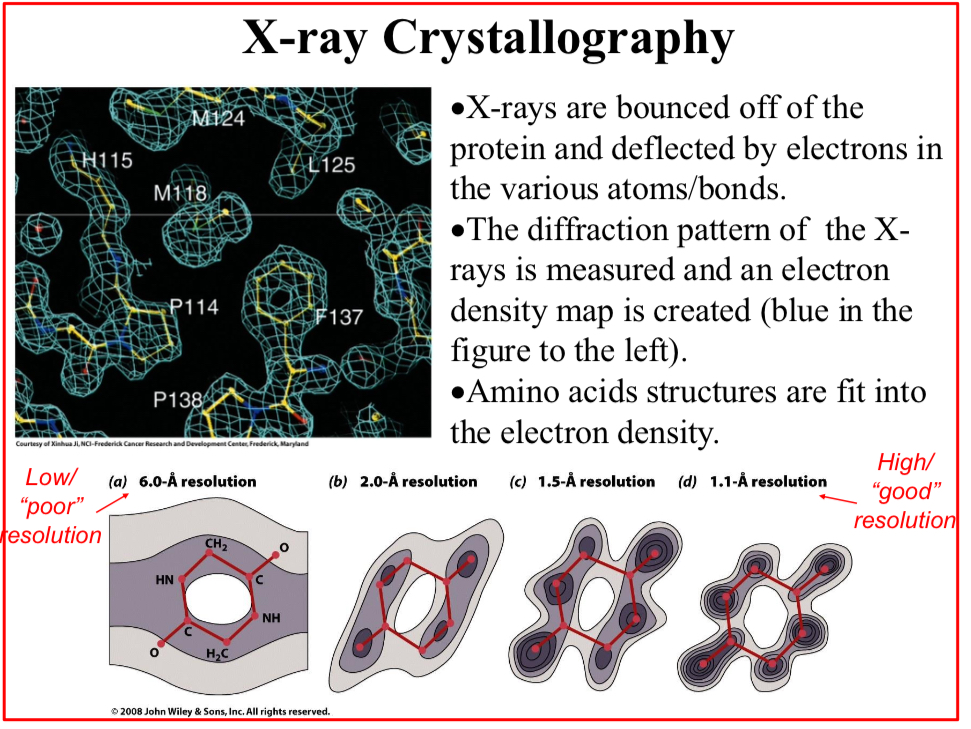
The diffraction pattern of the X-rays is measured and an electron density map is created
Amino acids structures are fit into the electron density.
Given the crystal consistency, the density maps are not as precise as they could, and…
The crystal is said to have a “resolution limit”.

X-ray Crystallography advantages:
Crystalline proteins assume conformations that are very similar to…
That of the protein in solution (near-native structure)

X-ray Crystallography advantages:
Most independent X-ray crystallography experiments describe…
The same conformation for the same structure (consistency/reproducibility)

X-ray Crystallography advantages:
Many enzymes are catalytic active in the crystalline state.
Since activity is highly dependent on structure, this is strong evidence that the crystalline conformations must indeed be near-native.
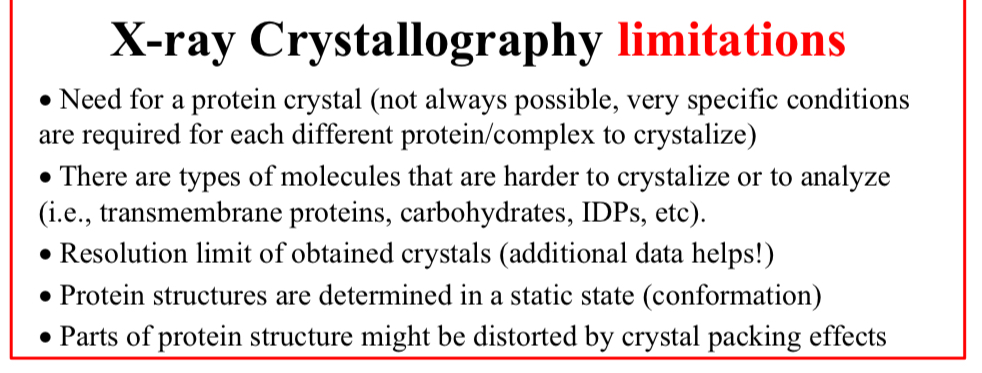
X-ray Crystallography limitations:
Need for a protein crystal
Not always possible, very specific conditions are required for each different protein/complex to crystalize
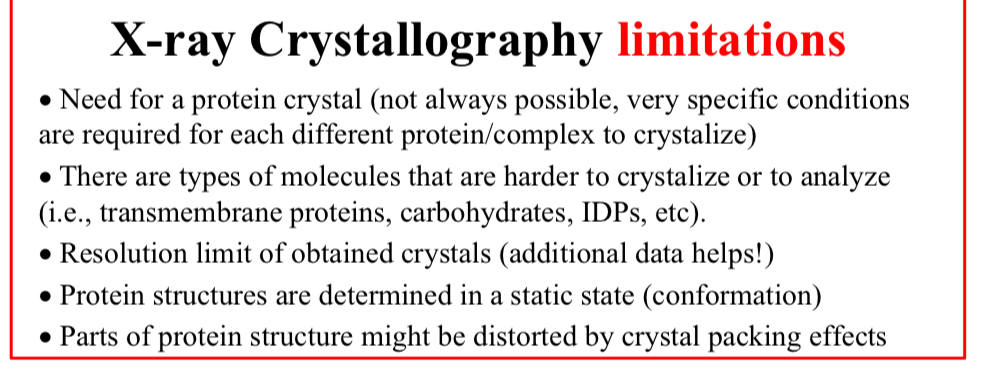
X-ray Crystallography limitations:
There are types of molecules that are harder to crystalize or to analyze
i.e., transmembrane proteins, carbohydrates, IDPs, etc
X-ray Crystallography limitations:
Resolution limit of…
Obtained crystals (additional data helps!)
X-ray Crystallography limitations:
Protein structures are determined in…
A static state (conformation)
X-ray Crystallography limitations:
Parts of protein structure might be…
Distorted by crystal packing effects
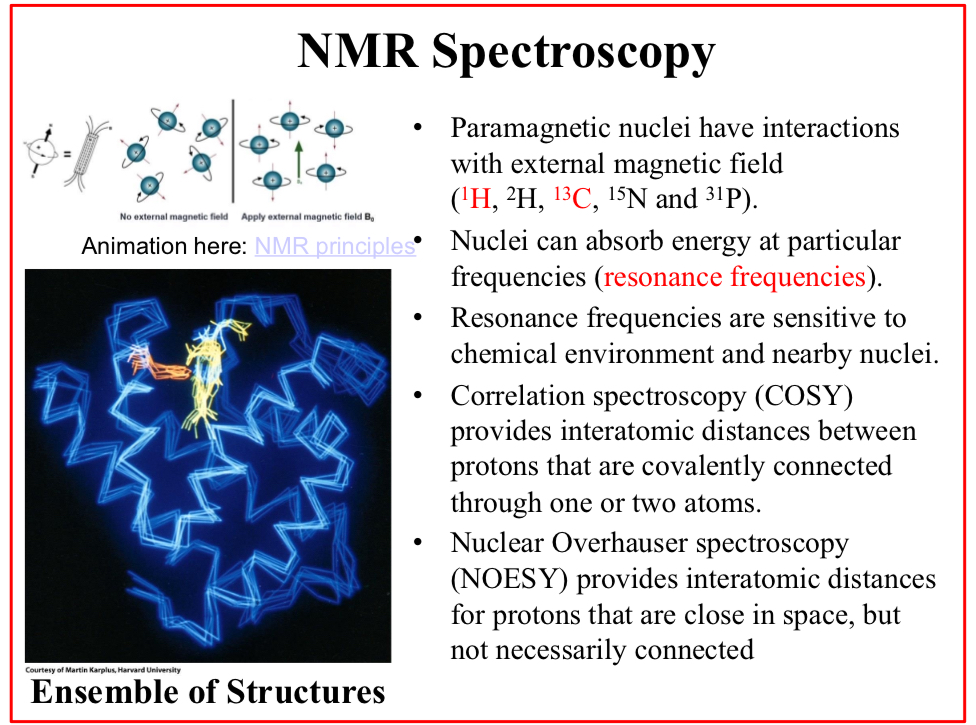
NMR Spectroscopy
Paramagnetic nuclei have interactions with external magnetic field (1H, 2H, 13C, 15N and 31P).
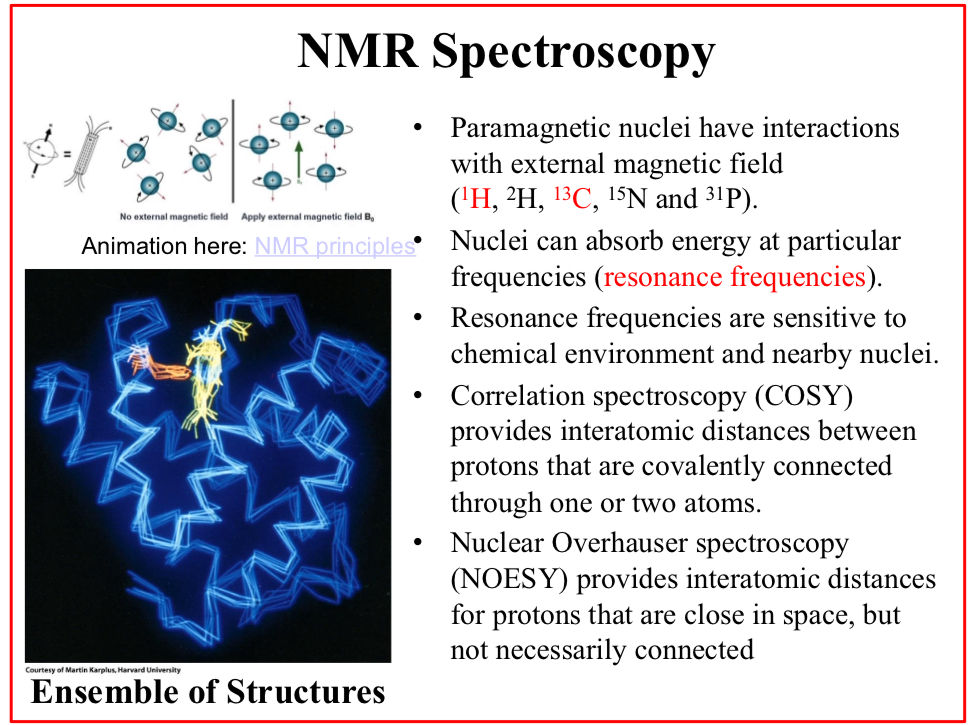
Nuclei can absorb energy at particular frequencies (resonance frequencies).
Resonance frequencies are sensitive to chemical environment and nearby nuclei.
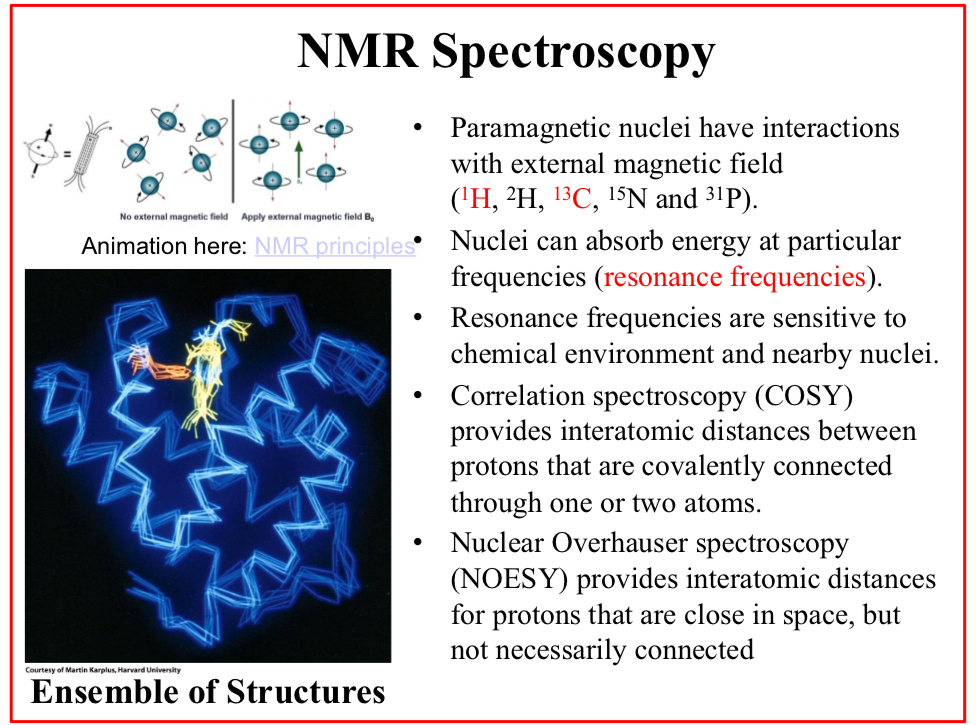
Correlation spectroscopy (COSY)
Provides interatomic distances between protons that are covalently connected through one or two atoms.
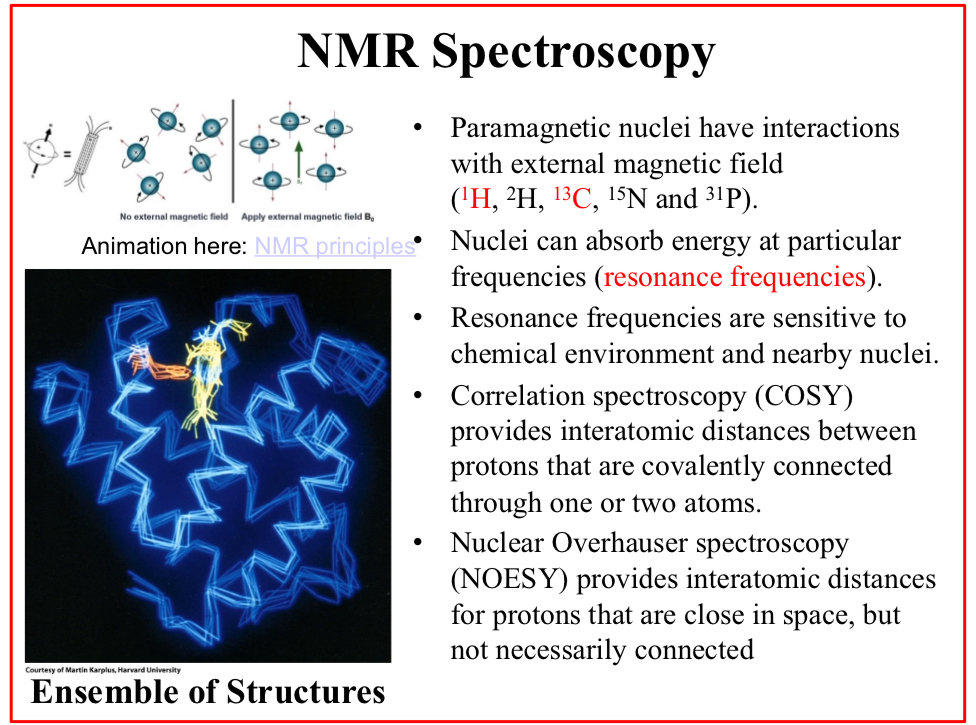
Nuclear Overhauser spectroscopy (NOESY)
Provides interatomic distances for protons that are close in space, but not necessarily connected
NMR spectroscopy advantages:
No need for crystallization and…
Can be used to determine the protein structure in solution
NMR spectroscopy advantages:
Provides not a single conformation, but…
An ensemble of conformations
NMR spectroscopy advantages:
Can probe motions over time scales spanning…
10 orders of magnitude
NMR spectroscopy advantages:
Resolution can be comparable with X-ray crystallography
NMR spectroscopy results are usually consistent with crystallographic data