Bio Unit 3- Molecular Genetics (Chapter 5)
1/84
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
85 Terms
Nucleotide
One individual unit/building block of DNA or RNA, made up of
one nitrogenous base of any kind
a sugar
phosphate group (on the sugar)
Erwin Chargaff’s experiment (Chargaff’s Rule)
Chargaff discovered the Chargaff’s Rule (base pairing) by looking at the chemical make up of DNA. He found that:
Adenine is always found with Thymine (and vice versa)
Guanine is always found with Cytosine (and vice versa)
Complementary Base Pairing
The pairing of complementary bases between the two strands of DNA as shown by Chargaff’s rule
Anti-parallel
Refers to the orientation of the two DNA strands against each other. They are parallel to each other but face opposite directions, meaning that when one strand runs 5' to 3', the other strand runs 3' to 5'. Referring to the carbons on the sugar from the ether group, from top to bottom of the strand.
genome
the total genetic material of an organism, the “Instruction book” on the entire body in DNA
gene
A distinct functional region within DNA that holds genomes.
Frederick Griffith’s Experiment
tested 2 forms of the same bacterium on mice: an S-strain that was pathogenic (carries the virus that would kill the mice) but could be made non-pathogenic by heating it, the other, R-strain, being non-pathogenic and harmless. When he injected the heated S strain and the R strain into the mice, the mice died. This indicated that there was something being transferred from the dead bacteria in the S-strain to the R-strain. This transfer is referred to as transformation.
Transformation
The movement of genetic material from a dead organism to a live one, altering their DNA. Found in Griffith’s Experiment
Initiation Phase
First phase of semi-conservative replication. A portion of the double helix is unwound to expose the bases for new base pairing
Elongation Phase
Phase 2 of semi-conservative replication. 2 new strands of DNA are put together using the individual parent strands as a template. The new DNA molecules (made of one parent and one new strand) wind back into helix shape.
Termination Phase
Last phase of semi-conservative replication. The new DNA molecules separate from each other completely. The replication machine is dismantled
Origin of Replication
The specific nucleotide sequence at which the process of replication starts.
Helicase
Type of enzymes that unwind and separate the double-stranded DNA during semi-conservative replication.
They cleave the h-bonds between the complementary base pairs to allow the strands to separate.
Semi conservative replication
The proved method of DNA replication in which each new DNA molecule consists of one original strand and one newly synthesized strand due to each new strand being made by using a parent strand as a guideline
DNA polymerase III
The enzyme that catalyzes the addition of new nucleotides one at a time to create a new strand based off of the parent strand.
only attaches to the 3’ hydroxyl end of pre-existing nucleotides
Can only synthesize a new strand from a parent strand in the 5’ to 3’ direction
primer
The short strands of RNA used to start the replication process by providing a free 3' hydroxyl group for the nucleotide addition done by DNA polymerase III
Okazaki fragments
Short DNA fragments created when a nucleotide is added to the various primers of the lagging strand
The use of primers means that the fragments always start with an RNA section
lagging strand
The second strand, ordered from 5’ to 3’ (forces the new strand to be made from 3’ to 5’)
needs several primers for replication which leads to the creation of Okazaki fragments
leading strand
needs only one primer for DNA replication
ordered 3’ to 5’ (allows the new strand to be from 5’ to 3’)
DNA polymerase I
enzyme that removes the RNA primers from the Okazaki fragments of the lagging strand, and fills that space by extending the neighboring DNA nucleotide. This causes the Okazaki fragments to be just made of DNA which are then joined together by DNA ligase
can also recognize mistakes in replication, cutting out the incorrect or extra base from the new strand and replacing it with the correct one (works with DNA polymerase II to do this) using the parent strand as a template
DNA ligase
the enzyme that catalyzes the joining together of Okazaki fragments that have only DNA nucleotides
Creates a continuous DNA strand
replication machine
protein-DNA complex at the end of each replication fork that carries out replication
DNA polymerase II
recognizes mistakes in replication and cuts out the incorrect base from the new strand, replacing it with the correct one using the parent strand as a template
Mismatch repair
A replication error fixing method in which specialized enzymes recognize the deformities in newly formed molecules that have mismatched base. These enzymes then bind to the DNA to remove the incorrect base from the new strand and replace it with the correct one
Errors in replication that pass the DNA polymerase and mismatch repair are called what?
Mutations
telomere
A repetitive DNA sequence at the end of a strand added to avoid the loss of important genetic information when the RNA section of the very last Okazaki fragment is removed and cannot be replaced
one gene/one polypeptide hypothesis
The theory that each gene only holds the information needed to produce a single polypeptide.
mRNA
“Messenger RNA”
RNA that contains the genetic information of a gene and carries it to the protein synthesis machinery
provides information that determines the amino acid sequence of a protein
acts as an intermediary
genetic code
a set of rules for determining how genetic information in the form of nucleotides are transferred into amino acid sequences in proteins.
a code specifying the relationship between a nucleotide codon and an amino acid
Triplet Hypothesis
Only 4 nucleotides in RNA, but 20 different amino acids. To produce 20 different combination, a minimum of three nucleotides needs to be used (4 times 4 times 4 = 64 vs 4 times 4 = 16) leading to the name
States that genetic code consists of 3 different nucleotides
gene expression
the transfer of genetic information from DNA to RNA to protein, leading to it determining the phenotype (and being expressed)
transcription
the transferring of genetic information from DNA template to RNA
the mRNA is synthesized using the DNA as a template
translation
the transferring of genetic information from mRNA to protein
the protein is synthesized using mRNA as a template
Alfred Hershey and Marsha Chase experiment
proved that DNA is the hereditary material not protein by using a virus called T2 bacteriophage
They tracked the DNA and protein molecules by using radioisotopes (35S for proteins and 32P for DNA).
When the 35S containing virus infected the bacteria, they found after agitation and separation that the radioactive material was only in the liquid medium, and not in the bacteria
When the 32P containing virus infected the bacteria, they found that the radioactive material was inside the bacteria, confirming that DNA is the genetic material as it was the only material to be able to enter the bacteria in order to alter it
What is T2 bacteriophage made out of?
protein coating over nucleic material
Rosalind Franklin’s experiment
used X-ray diffraction to study the structure of DNA
took X-Ray images of DNA molecules, and used their background with mathematics to calculate the diffractions in the image, and figured out what it would look like without the diffraction: arriving at the shape of the DNA that is known today
However they were not credited for this discovery as Watson and Crick stole their work to use for their own model and had it published before them
James Watson and Francis Crick
Put together the information and discoveries of all of the experiments (stolen from franklin) and created the first 3D DNA model that could be seen and understood by the general public
Proposed the double helix formation that allows for difference in DNA shape among species
Oswald Avery’s experiment
Used the process of elimination to provide further proof of Frederick Griffith’s conclusion
He subjected three separate identical heat killed S-strain bacteria to different enzymes (one for protein, one for DNA and one for RNA)
The enzyme treated extracts were then mixed with non-pathogenic R-strains
If the R-strains were transformed into pathogenic S-strains, then this meant that the hereditary material had not been destroyed by the enzyme, and that the target was wrong
The only R-strain that did not transform into a pathogenic S-strain was the one afflicted by the DNA enzyme, proving that the hereditary material that determines whether the strain is pathogenic is made of DNA
Therefore all genetic material is made of DNA
Spontaneous mutation
type of mutation that takes place when something in the replication process goes wrong (ex: strand slippage)
Induced mutation
a mutation that takes place due to the environment (ex: cigarettes, radiation)
Two types:
chemical mutagen
physical mutagen
Point Mutation
type of single gene mutation that involves the SUBSTITUTION of one nucleotide for another.
If a substitution changes the amino acid, it’s called a MISSENSE mutation
If a substitution does not change the amino acid, it’s called a SILENT mutation
If a substitution changes the amino acid to a “stop,” it’s called a NONSENSE mutation
Missense Mutation
A point mutation that changes the amino acid
Silent Mutation
A point mutation that does not change the amino acid
mutagen
substance that causes mutations
Two types: chemical and physical
mutation
Nonsense mutation
A point mutation that changes the amino acid to a “stop”
Frameshift
a type of single gene mutation that involves INSERTION or DELETION of a single base pair.
Single gene mutation
mutation that involves the change in nucleotide sequence of one gene
Chemical mutagen
Type of induced mutation
Molecules
enters the nucleus and reacts with DNA to induce a mutation
Physical mutagen
Type of induced mutation
an event or a substance (ex: high energy radiation like UV rays or X-rays )that changes the physical structure of DNA, causing a mutation
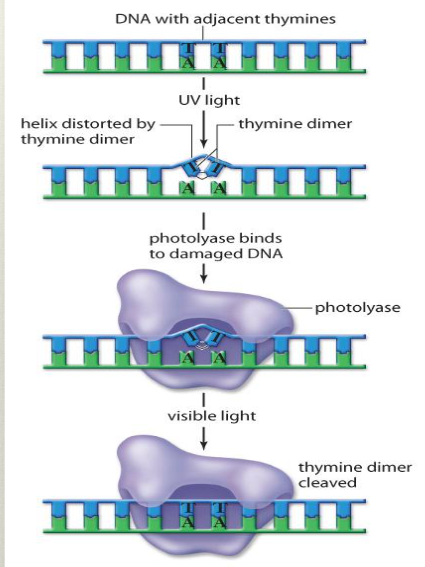
Photorepair
specific type of DNA repair specialized for fixing damage caused by UV radiation (binds thymines together covalently into a dimer, altering the shape of DNA)
photolyase enzyme recognizes the damage, binds to the dimer, and uses visible light to cleave the thymine dimer such that the covalent bond is broken
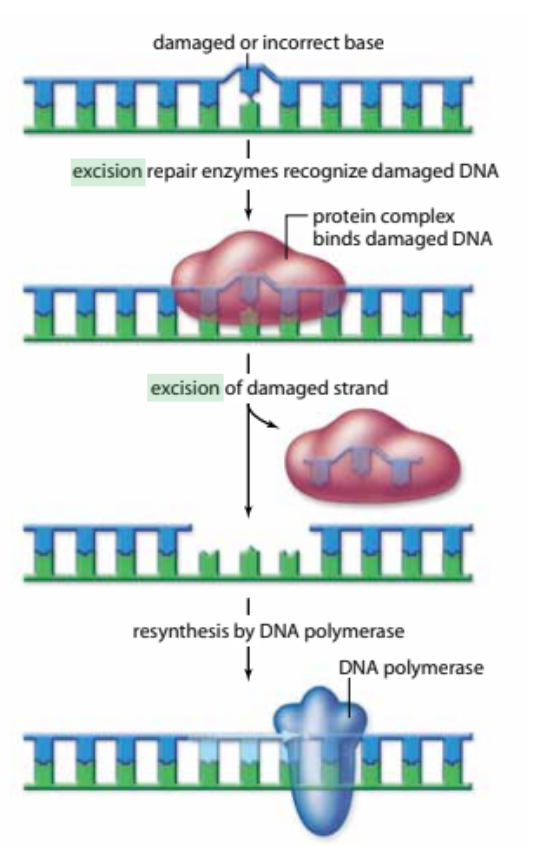
Excision Repair
a non-specific type of DNA repair that can fix a variety of damage through excision (cutting out) of the damaged or incorrect base to later be replaced by DNA polymerase
Specific Repair Mechanism
a DNA repair mechanism that is specialized to repair one specific type of damage
Ex: photorepair
Non-specific Repair Mechanism
A DNA repair mechanism that is versatile and capable of fixing multiple kinds of damage done to the DNA molecule
Ex: Excision Repair
Gene Regulation
the control of how much each gene is expressed
determines which genes are active or inactive and how active they are (how much protein is produced)
Constitutive Genes
Also called “housekeeping” genes
Essential for survival and therefore always active unlike most genes
What are the three different levels of DNA replication of prokaryotes that allows for gene regulation?
Transcription
Translation
After a protein has been synthesized
Operon
Found in prokaryotes
a region in which a cluster of genes are under the control of one promoter
Contains:
a promoter
an operator
structural genes
lac operon
where the genes that encode the enzymes necessary to break down the sugar lactose are located
Found in E.coli chromosome
inducible operon: activated when lactose is present
The Coding Region
the portion of DNA that contains structural genes required for protein synthesis
in lac operon, the structural genes for lactose are found here
Regulatory Region
the portion of DNA that contains genes and compartments that allow gene regulation to take place
the specific compartments and genes depend on the chromosome
What does the coding region of a lac operon contain?
structural genes that allow the synthesis for 3 different enzymes that break down lactose
What does the regulatory region of a lac operon contain?
A promoter
An operator
A CAP binding site
Promoter
a sequence of special nucleotides that indicate where RNA polymerase should bind to initiate transcription
Operator
a DNA sequence that a repressor protein binds to to inhibit transcription initiation
CAP site
a DNA sequence that a CAP (catabolize activation protein) binds to to increase the rate of transcription not necessary but useful when a lot is needed fast)
How and when is the lac operon deactivated?
when there is no lactose, and therefore the enzymes needed to break it down are not required
Deactivated by repressor that binds to the operator, preventing RNA synthase from doing so and initiating transcription
How and when is the lac operon activated?
when there is lactose which needs to be broken down, and allolactose is made
Activated through the allolactose binding to the repressor, preventing the repressor from binding to the operator
the operator becomes available for RNA synthase to bind to it
The binding of a CAP protein can further increase the rate at which transcription takes place
Inducible operon
an operon that can be activated when a product is present
trp (tryptophan) operon
always active unless a repressor is present
synthesizes enzymes that are required to produce tryptophan
What is a trp operon made of structurally?
a coding region
contains 5 different genes that produce enzymes that make tryptophan
a regulatory region that contains
a promoter
an operator
When and how is a trp operon activated?
under normal conditions a trp operon is always active because tryptophan is always necessary and must be synthesized
since the synthesized tryptophan is being used, it does not bind a repressor protein and the synthesis continues
When and how is a trp operon deactivated?
when there is a high concentration of tryptophan
Because there is a lot of tryptophan, it is more likely that the tryptophan binds to a repressor protein. Once it does, this repressor binds to the operator of trp operon to prevent RNA synthase from moving to the genes from the promoter by binding to it, thus deactivating the operon
Structural Genes
genes in an operon that code for the product protein
Restriction enzyme/endonuclease
An enzyme found in bacteria that recognizes a target sequence in the DNA of a virus that tries to inject its DNA inside the bacteria to take it over, and cuts it off
Used by scientists to cut up DNA such that only the desirable portions remain and can be added to other strands of DNA to create transgenic organisms
Restriction Site
The specific point of the DNA at which the restriction endonuclease cuts staggerdly , separating all except the base pairs at each end
What are the characteristics of restriction endonuclease that make it useful for scientists
Specificity
can target specific portions of the DNA to cut, which usually has the same base pattern but inverted between the strands
Staggered Cuts
cuts in a staggered pattern that leaves behind sticky ends which are unpaired bases that are useful in bonding the fragment with a fragment of DNA from a different organism
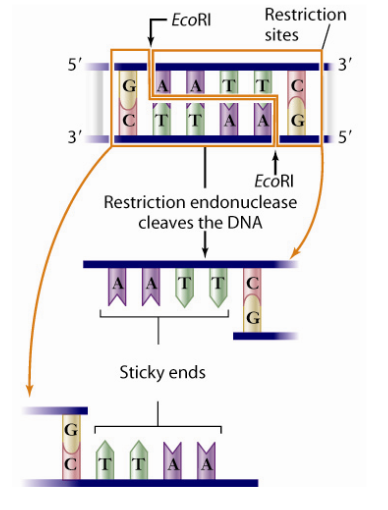
Sticky ends
unpaired bases at the end of DNA fragments caused by the staggered cut of restriction endonuclease
How is a recombinant DNA molecule made?
Two DNA molecules from different organisms are cut with the same restriction endonuclease
the endonuclease being the same causes them to have the same sticky ends, complimenting each other
the endonuclease is chosen specifically so that it can cut both DNA molecules
Base pairing between the sticky ends brings the two DNA molecules together
DNA ligase covalently joins the two strands
Repressible operon
an operon that is always active, and can only be deactivated by a high amount of the protein it produces
Enhancer
a DNA sequence that promotes transcription
made up of distal control elements
What are the binding sites on the large sub-unit of a ribosome called in order from left to right?
Site E, Site P, Site A
In what direction do ribosomes read?
5’ to 3’
What is the difference between a regulatory sequence and a regulatory gene?
A regulatory sequence is a sequence on the DNA molecule that a regulatory protein can bind to in order to control how much the genes that follow are expressed. ex:
promoter
A regulatory gene is a gene on the DNA molecule that codes for a regulatory protein (can also be enzyme)