biochem - FFP1
1/216
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
217 Terms
what are amino acids
• They are the basic building unit of proteins
• They contain short hydrocarbon chains, oxygen atoms and nitrogen
• Amino acids are the precursors of nucleosides, neurotransmitters, haem and many other molecules
• They can be converted to carbohydrates through the process known as gluconeogenesis
• There are 20 of them
contains;
R group
carboxylic group
amino group
central carbon
H
characteristics of side chains
**don’t need to learn all of the amino acids, need to learn ones on following slides
polar AAs found on surface of proteins
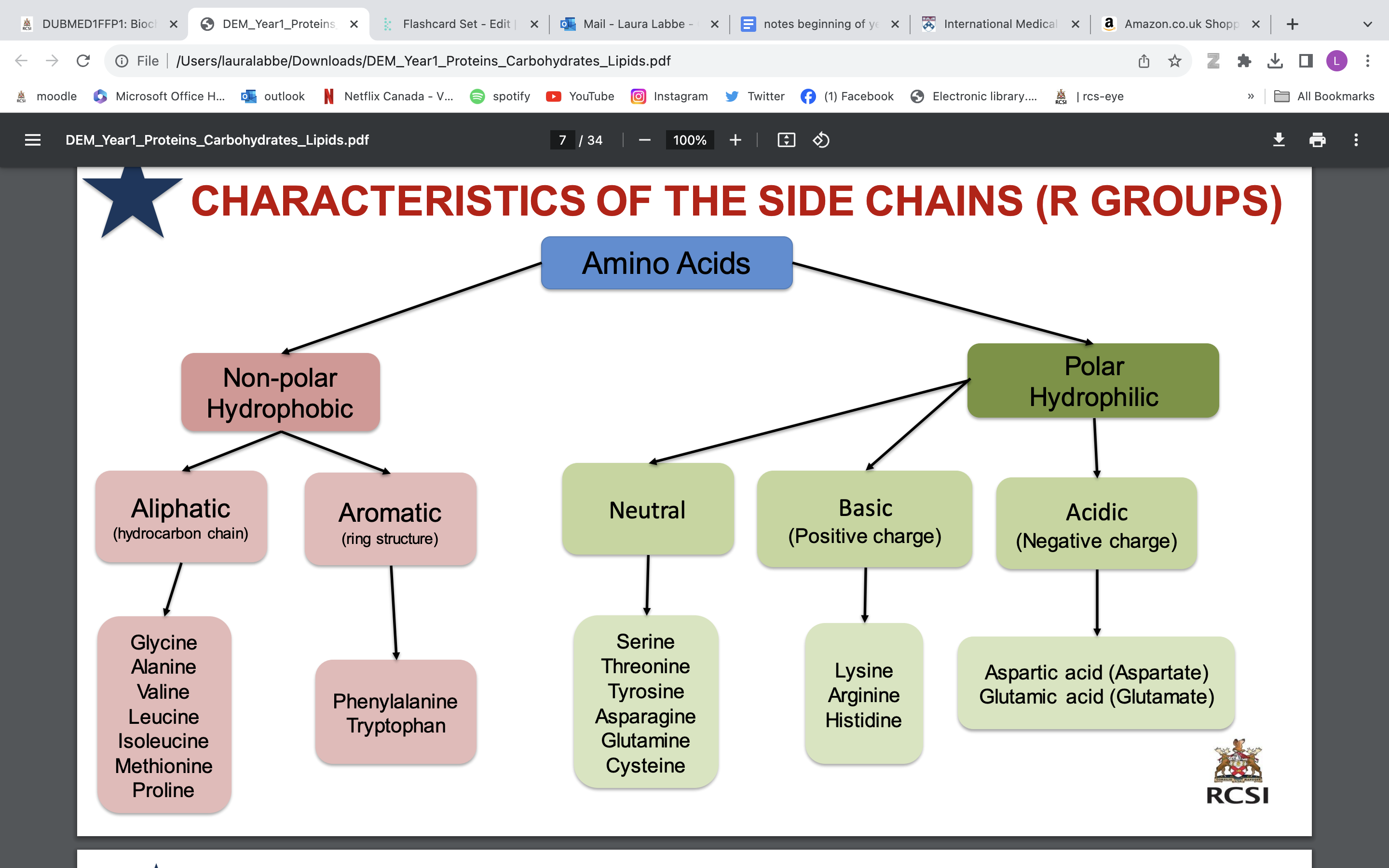
small amino acids
glycine and alanine
branched amino acids
valine, leucine, isoleucine
come up in clinic quite a bite, especially younger children (unusual colour and smell to urine)
sulphur containing amino acids
cysteine and methionine
methionine; often first aa in aa chain
The amino acid found at a bend in a protein
proline
Amino acids that can be phosphorylated: (functional change)
serine, threonine and tyrosine
signalling proteins
Amino acids that can be glycosylated:(functional change)
asparagine, serine and threonine
Amino acid that can be nitrosylated:(functional change)
cysteine
primary structure of amino acids
• Amino acids formed into a polypeptide chain
• Amino acids linked together with peptide bonds
• This bond is formed between the carboxyl group of one amino acid and the amino group of next amino acid
• The peptide bond is C(O)NH
• Chain has direction:
– Start = amino terminus = N terminus
– End = carboxyl terminus = C terminus
secondary structures of amino acids
• This is the spatial arrangement of the primary structure
• It is determined by hydrogen bonding
• The amino acid sequence controls folding
• These structures have a regular repetitive folding pattern
• Most important are the alpha helix and beta pleated sheet
in alpha helix= 3.6 AAs by 360 degrees
tertiary structure amino acids
• This is further folding of the polypeptide chain
• Folding into a globular form
• Compact folded structure
hydrophobic AAs on the inside
hydrophilic AAs on the outside
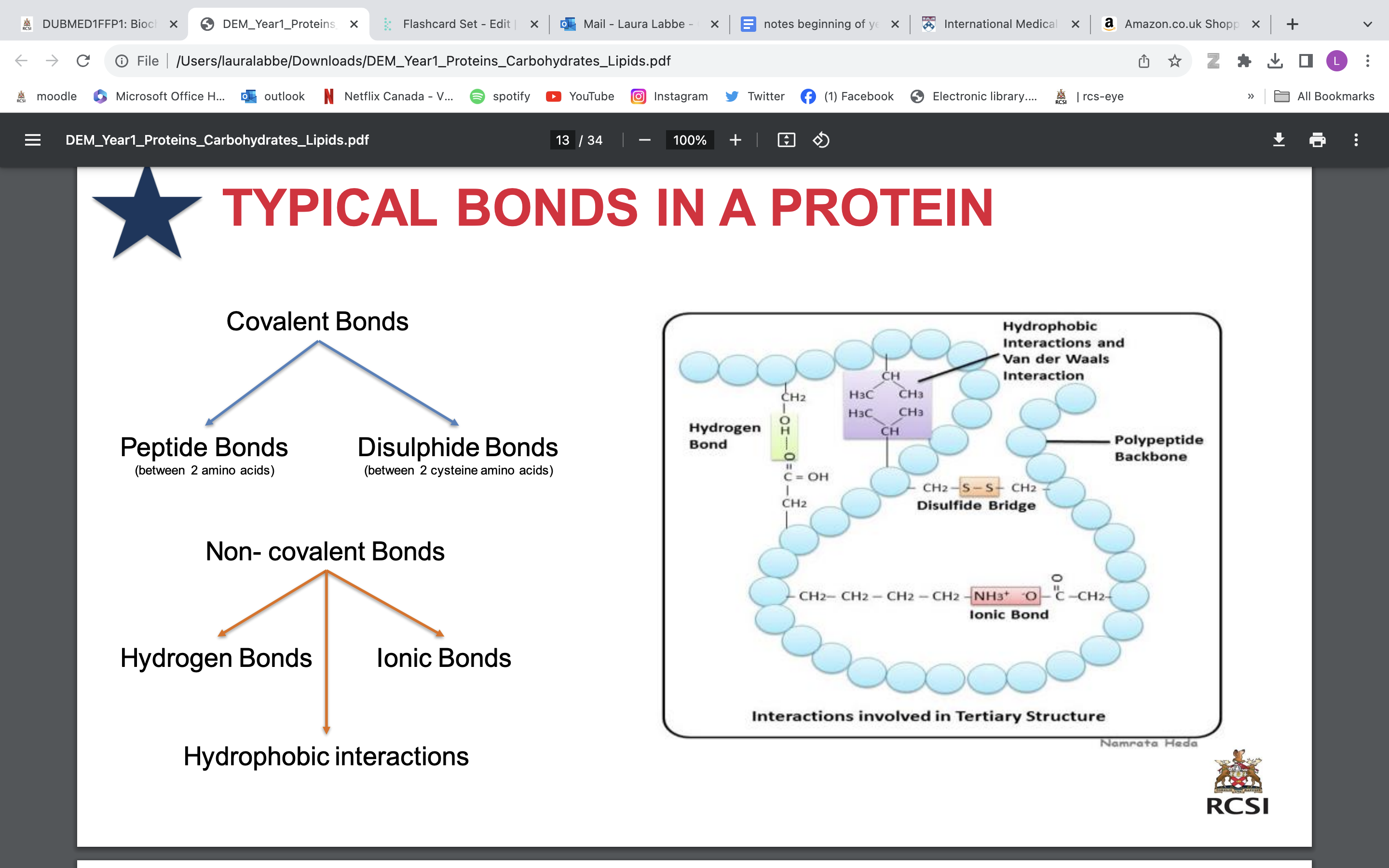
• It stabilised by a wide range of bonds and interactions between the side chains of amino acids:
Disulphide bonds (between 2 cysteines)
Hydrophobic interactions
Ionic bond
Hydrogen bonds
QUATERNARY STRUCTURE amino acids
• It is the arrangement of protein subunits in a multi-meric protein
• The 3D arrangement of more than one tertiary polypeptide
• Consist of 2 or more polypeptide chains
• Polypeptide may be the same or different
• Held together by
Non-covalent interactions
Inter-chain disulphide bonds
not necessary for a protein to get there
typical bonds in protein
Une liaison covalente est une liaison chimique dans laquelle deux atomes se partagent deux électrons d'une de leurs couches externes afin de former un doublet d'électrons liant les deux atomes.
NATIVE CONFORMATION protein
• It is the functional fully folded protein structure
• It is a unique three dimensional structure
• It determines the biological function of the protein
Enzymatic
Protection
Regulation
Signal transduction
Storage
Transport
POST TRANSLATIONAL MODIFICATIONS (PTM)
• Chemical modification of a protein after translation
• A functional group is attached to an amino acid
• Results in a change in protein function
• Increases the diversity of the proteome
• Some common PTMs
Phosphorylation: + phosphate = phosphoprotein
Glycosylation: + sugar group = glycoprotein
Ubiquitination: + ubiquitin = death signal- get rid of things
Nitrosylation: + NO (nitric oxide)
lipids
• Lipids are a heterogeneous group of water-insoluble (hydrophobic) organic molecules Functions
• Major source of energy in the body
• Structural components of cells and organelles
• Involved in cellular signaling events e.g.
Steroids
Prostaglandins
Leukotrienes
classification of lipids
• Fatty acids and their derivatives
• Lipids containing glycerol
Neutral lipids: Mono-, di-, tri-acylglycerol (triglycerides)
Charged lipids: Phospholipids
Lipids not containing glycerol
- Steroids
- Sphingolipids
Lipoproteins and lipopolysaccharides
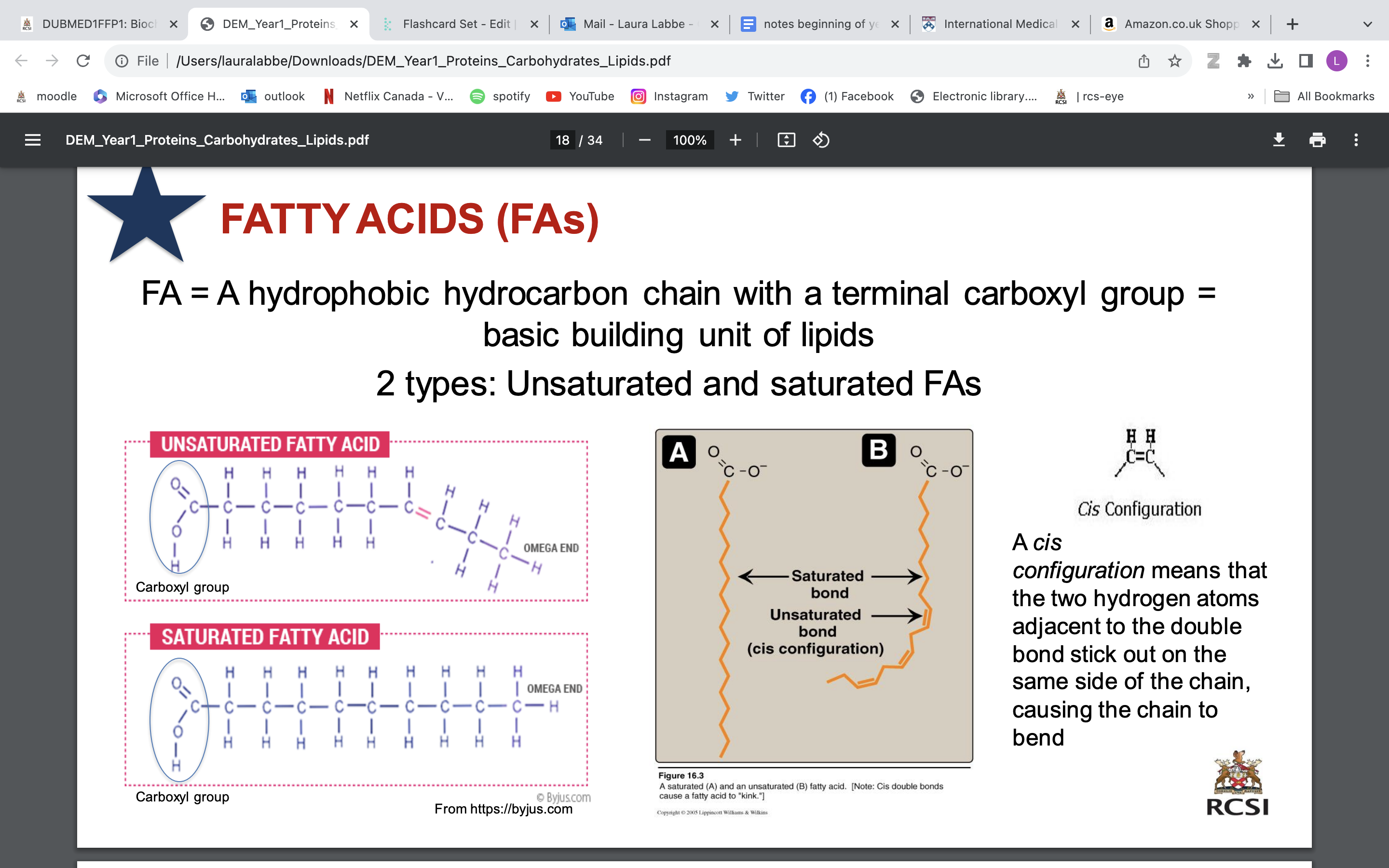
fatty acids
FA = A hydrophobic hydrocarbon chain with a terminal carboxyl group = basic building unit of lipids
2 types: Unsaturated (double bonds) and saturated (no double bond) FAs
omega fatty acids
• They are polyunsaturated fatty acids (PUFAs) = more than 1 double bond
• Omega refers to the position of the double bond from the omega (w) end of the FA chain
• Omega 3 refers to the position of the final double bond (from the carboxyl end) which is three carbons from the "omega" or tail end of the FA
• Omega 6 refers to the position of the final double bond (from the carboxyl end) which is six carbons from the "omega" or tail end of the FA
omega carbon; last carbon
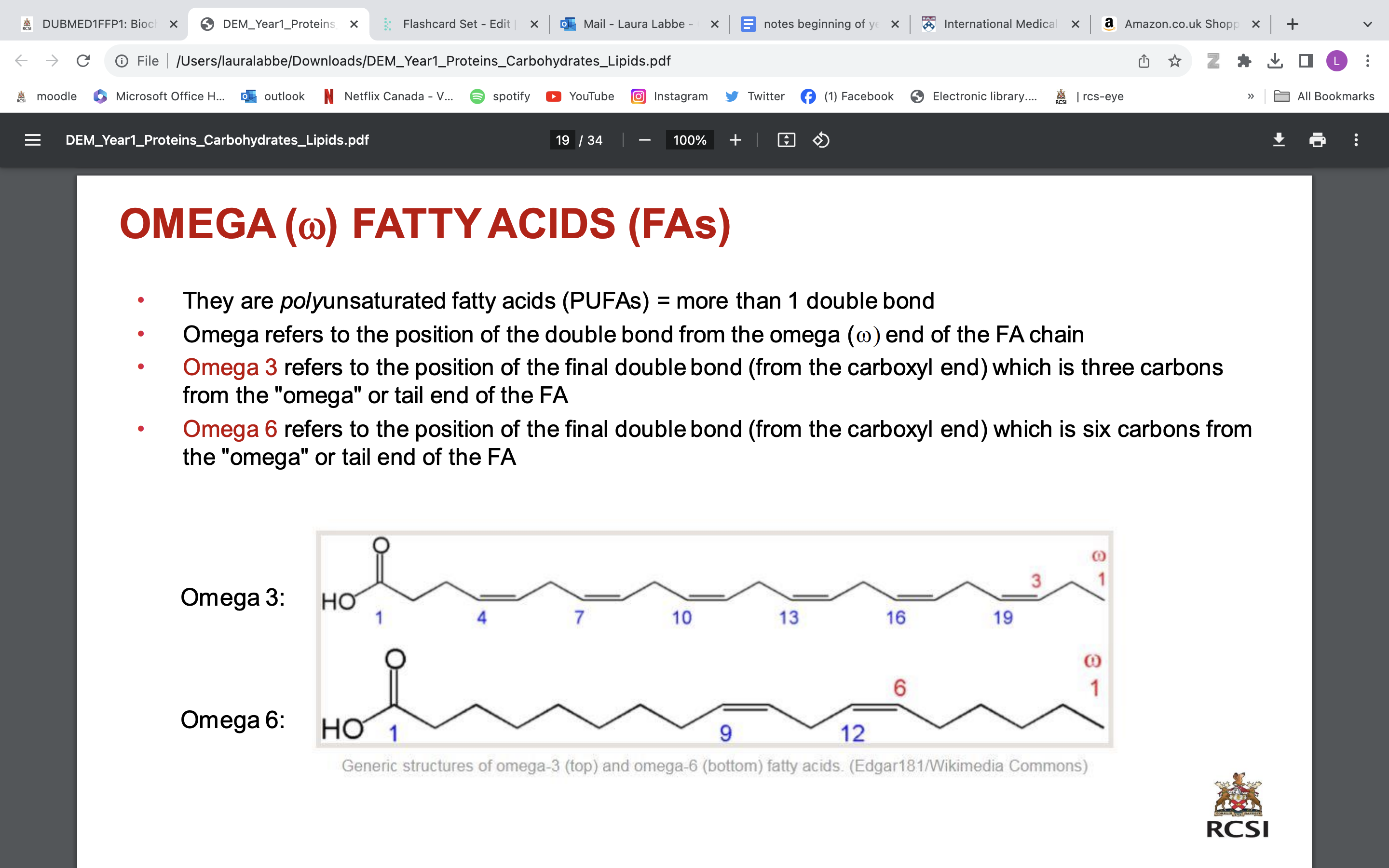
CHAIN LENGTHS OF FAs AND DOUBLE BOND POSITIONS
The carbon atoms are numbered, beginning with the carboxyl carbon as carbon 1
• Number before colon (:) = number of carbons in chain
• Number after colon = numbers and positions of double bonds relative to carboxyl carbon
• E.g. Arachidonic acid = 20:4(5,8,11,14)
20 carbon chain
4 double bonds between carbons 5-6, 8-9, 11-12 and 14-15
SIGNALLING FAs
prostaglandins and leukotrienes
prostaglandins
• Eicosanoids (PUFA -20)
• Synthesis: Arachidonic acid (a 20:4 FA) via COX (cyclooxygenase)
• Potent
• Short half-life (seconds)
• Multiple roles
Inflammation
Platelet homeostasis
leukotrienes
• Part of eicosanoid family
• Synthesis: arachidonic acid via LOX (lipoxygenase)
• Longer half-life (up to 4 hours)
• Multiple roles
Inflammation
Neutrophil adhesion
DIACYLGLYCEROL (DAG) AND MONOACYLGLYCEROL
DAG
Potent intracellular signaller
Mobilisation of calcium
monoacyglycerol
Breakdown product of TAG in fat digestion
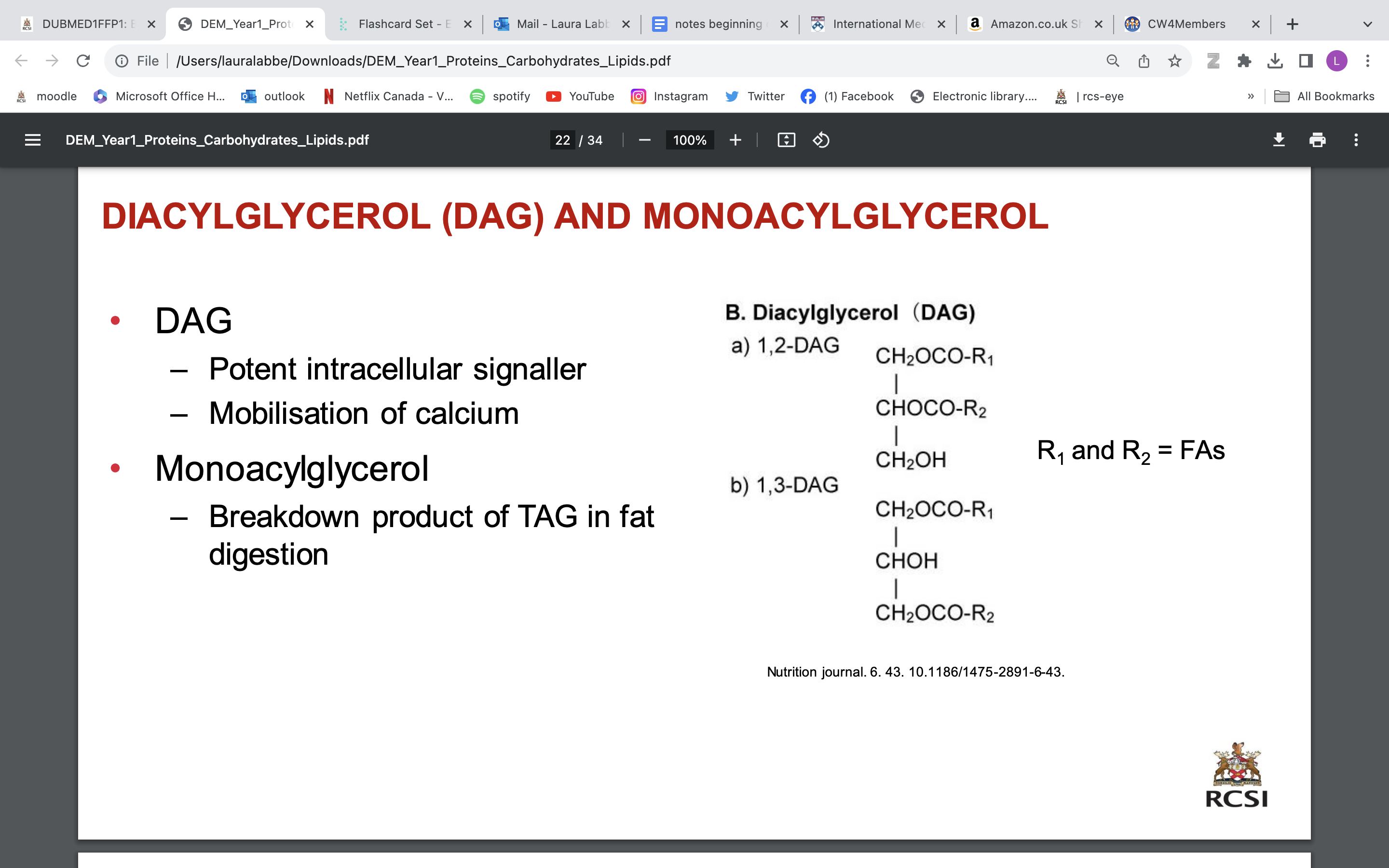
TRIGLYCERIDES (TRIACYLGLYCEROLS: TAG)
Made up of 3 FAs and glycerol
• The principal storage form of energy in the body
• Stored in adipose tissue
PHOSPHOLIPIDS
(PHOSPHOGLYCERIDES OR GLYCEROPHOSPHOLIPID)
• component of cell membrane
• They are amphipathic: hydrophilic head and hydrophobic tails
• The hydrophilic head is negatively charged
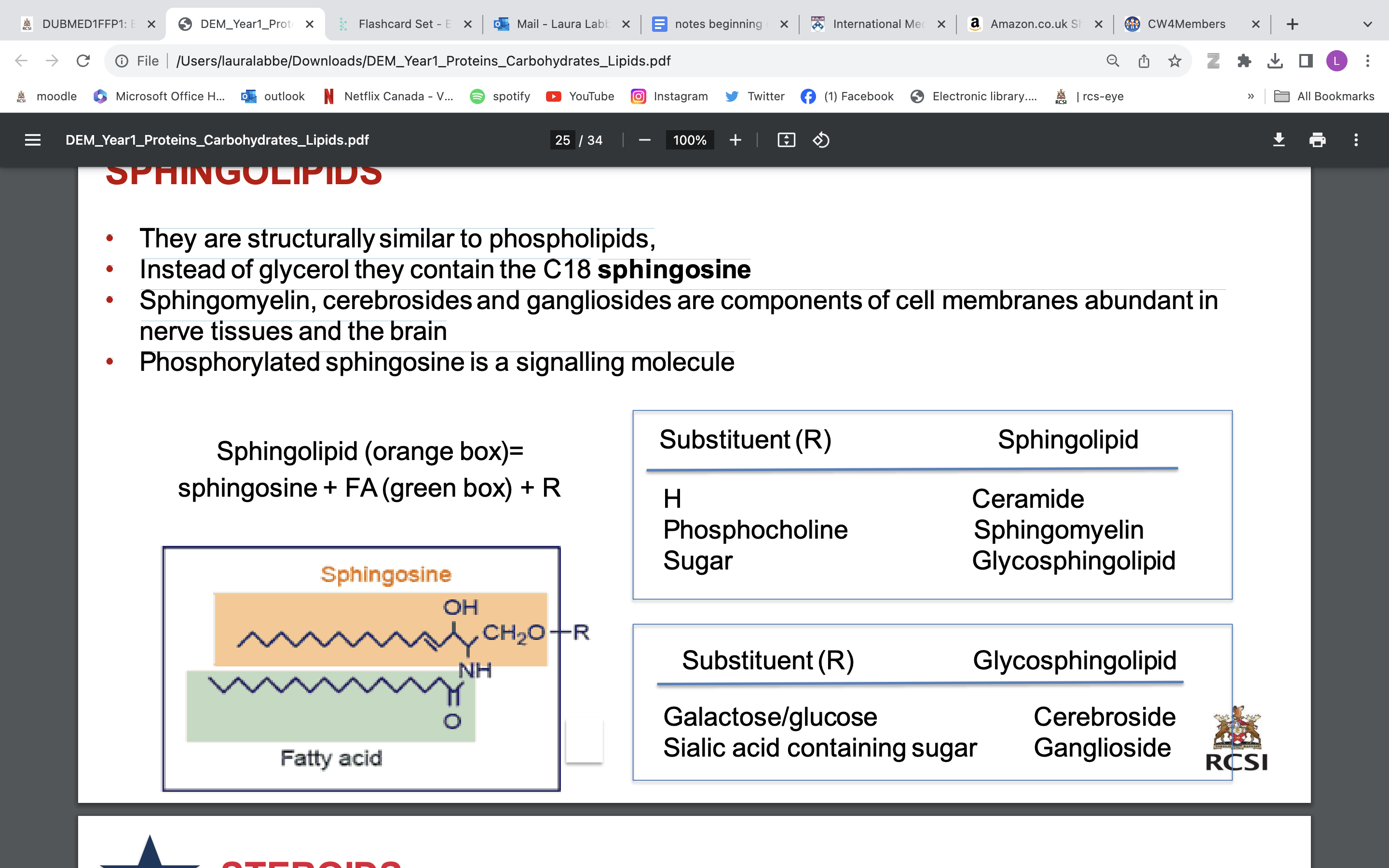
SPHINGOLIPIDS
• They are structurally similar to phospholipids,
• Instead of glycerol they contain the C18 sphingosine
• Sphingomyelin, cerebrosides and gangliosides are components of cell membranes abundant in nerve tissues and the brain
• Phosphorylated sphingosine is a signalling molecule
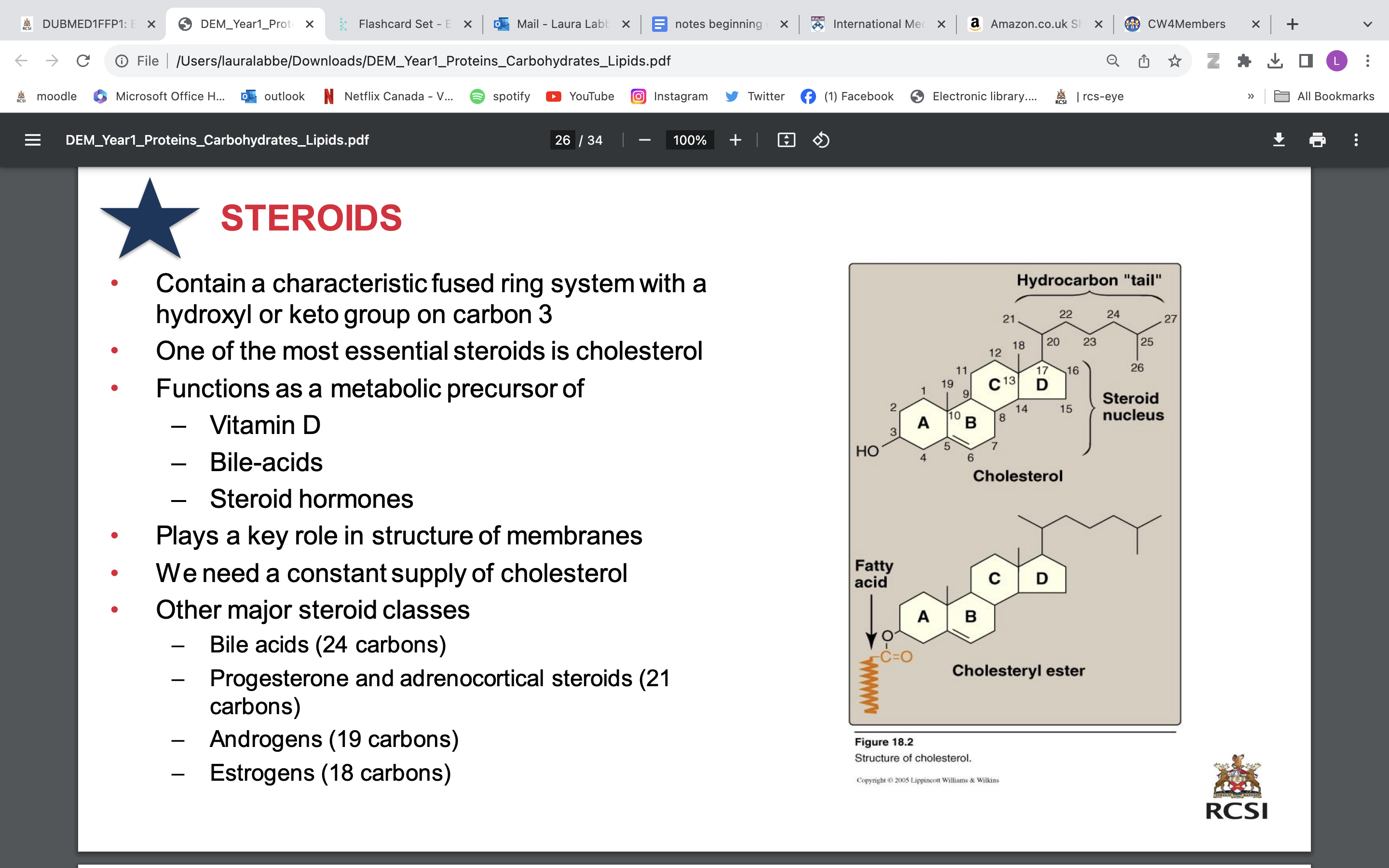
steroids
• Contain a characteristic fused ring system with a hydroxyl or keto group on carbon 3
• One of the most essential steroids is cholesterol
• Functions as a metabolic precursor of
Vitamin D
Bile-acids
Steroid hormones
• plays role in structure of membrane
• We need a constant supply of cholesterol
• Other major steroid classes
Bile acids (24 carbons)
Progesterone and adrenocortical steroids (21 carbons)
Androgens (19 carbons)
Estrogens (18 carbons)
lipoproteins
• Spherical particles found in plasma that transport lipids including cholesterol
• They are chylomicrons, very low density lipoproteins(VLDL), low density lipoprotein (LDL) and high density lipoprotein (HDL)
• Hydrophobic core of triacylglycerols and cholesteryl esters
• Phospholipid layer associated with cholesterol and protein
what are carbohydrates
• They are molecules that contain carbon (C), hydrogen (H), and oxygen (O) atoms
• Also known as a saccharide
• A single saccharide is called a monosaccharide = basic building unit of carbohydrates
• Two monosaccharides linked together is a disaccharide
• An oligosaccharide is a few linked monosaccharides
Can be associated with proteins (glycoproteins) or lipids (glycolipids)
• Polysaccharides consist of many monosaccharides eg starch or glycogen
monosaccharides
• They are simple sugar units
• They have the empirical formula = (CH2O)n where n = 3 – 7 carbons
• The ratio of hydrogen to oxygen is approximately 2 : 1
n = 3 carbons: triose, e.g. glyceraldehyde
• n = 5 carbons: pentose, e.g. ribose
• n = 6 carbons: hexose, e.g. glucose
• They are poly-hydroxy aldehydes (aldose) or ketones (ketose)
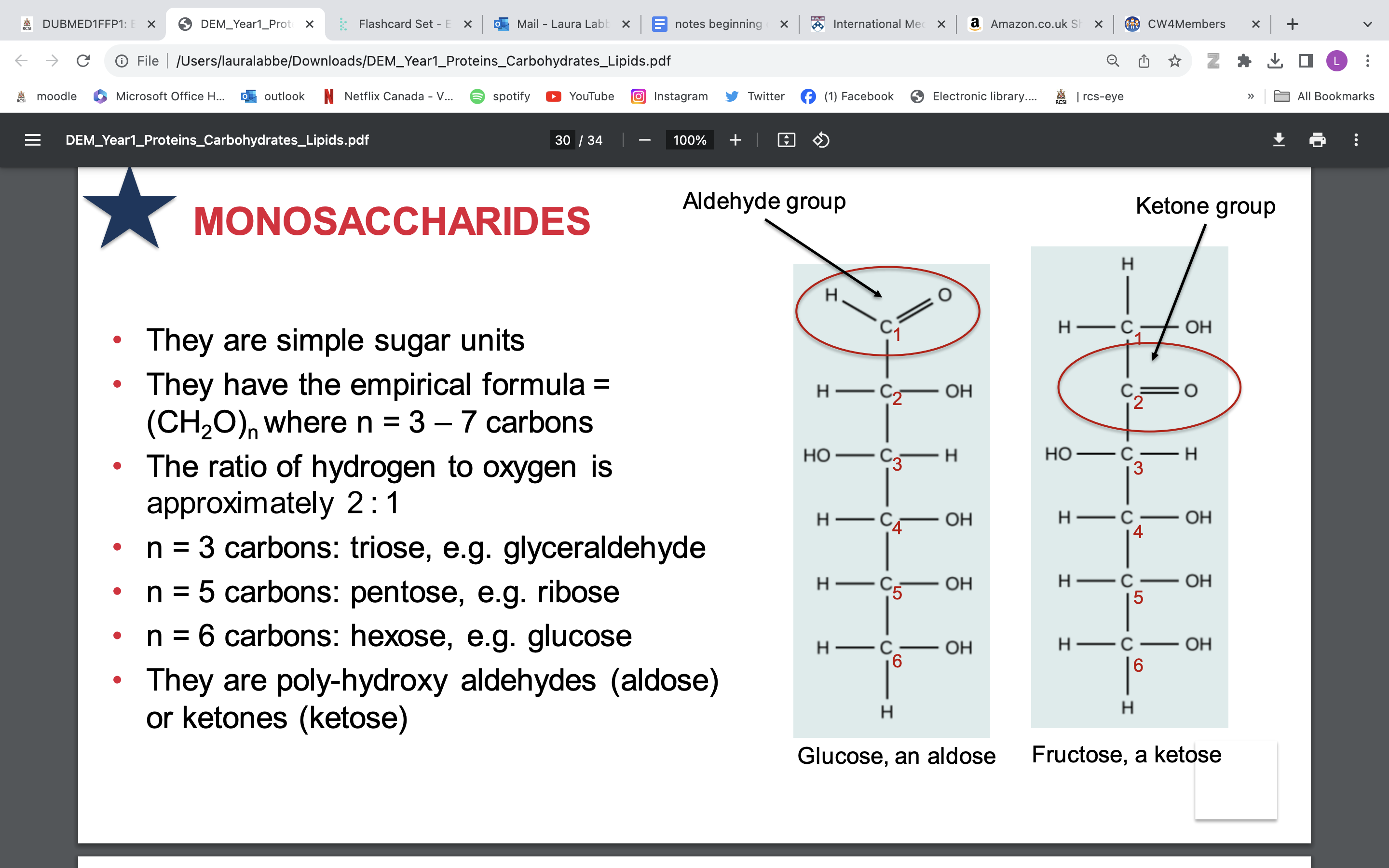
CYCLISATION of monosaccharides
monosaccharides like to form ring when watery environment
• In an aldose its all happening at carbon 1 = C1
• C1 in aldose = carbonyl carbon
• C1 in cyclised aldose = anomeric carbon
• Have an alpha anomeric carbon (OH points down)
• Have a beta anomeric carbon (OH points up)
• In a ketose it all happens at carbon 2 = C2
THE GLYCOSIDIC BOND: BOND BETWEEN 2 SUGARS = DISACCHARIDE
• The name or type of bond depends on
– The numbers of the connected carbons
– The position of the anomeric hydroxyl group
If the –OH group is in the alpha configuration it is an alpha (a) bond
If the –OH group is in the beta configuration it is a beta (b) bond
Example: Lactose = b-galactose + glucose
– Bond is between carbon 1 of b-galactose and carbon 4 of glucose: condensation event
– Bond (linkage) is b(1→ 4) glycosidic bond
• Carbohydrates can also bind to noncarbohydrate structures
✓ Purines and pyrimidine bases in nucleic acids
✓ Aromatic rings eg in steroids
✓ Proteins: glycoproteins
✓ Lipids: glycolipids
polysaccharides
• n > 12 – hundreds
• Variations can occur in the chain
Monosaccharides
Glycosidic bonds
Branch points
Structure
functions;
• Storage in animals:
- Glycogen: A homopolymer of glucose. Branched every 12-14 residues (a1- 4, a1-6)
• Storage in plants:
– Starch: A homopolymer of glucose; composed of:
Amylopectin (80-85%) branched every 24- 30 residues (a1-4, a1-6)
Amylose (15-20%) non branched helical structure (a1-4)
• Structure in plants:
– Cellulose: Homopolymer of glucose. Long straight chains (β1-4)
fed and fasting states
digestion, absorption, transport- while eating
triglyceride synthesis
lipolysis
beta oxidation→ energy
ketogenesis (liver only)→ ketone bodies→ energy
biological membrane components
• Lipid bilayer: structural backbone
• Phospholipids (most abundant)
– Glycolipids
– Cholesterol
• Proteins are:
– Transporters
– Enzymes
– Signal transductors
• Carbohydrates attached to proteins and lipids
dynamic structure! not static
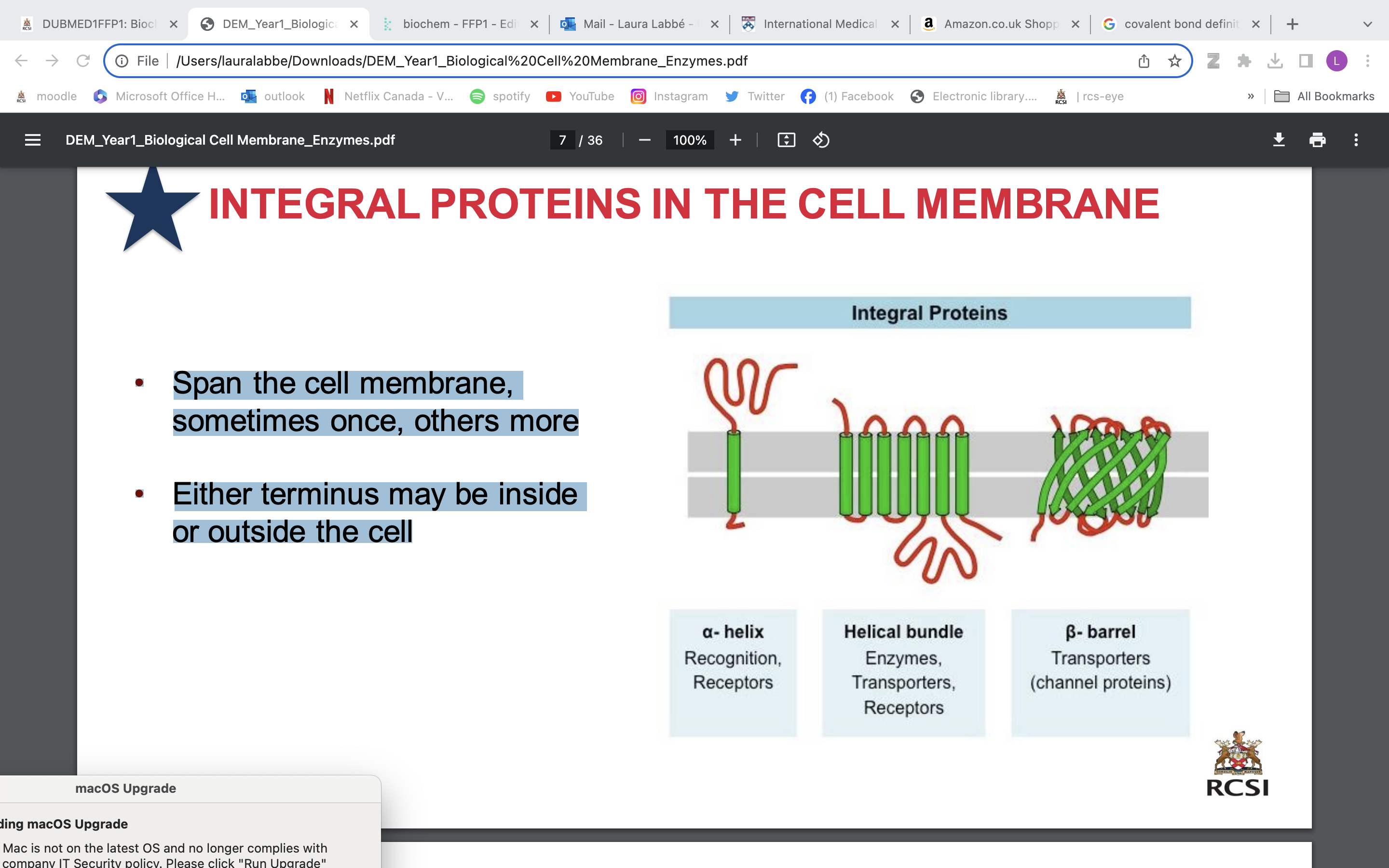
alpha helix; recognition, receptors
beta barrels; transporter (channel proteins)
helical bundle ; enzymes, transporters, receptors
lipid bilayer
• Polar head group of phospholipids has an affinity for water
• Their hydrocarbon tails avoid water
• Therefore, the favoured structure is a bimolecular sheet
• Formation is a self-assembly process
• Hydrophobic interactions are the major driving forces
• Electrostatic and hydrogen bonding attractions between polar head and water
• It is self-sealing
• Majority are phospholipids
• They are amphipathic: hydrophilic and hydrophobic components
• These lipids spontaneously form bilayers in aqueous solution:
– vesicles
– micelles
• Also have cholesterol
integral proteins in cell membrane
• Span the cell membrane, sometimes once, others more
• Either terminus may be inside or outside the cell
peripheral membrane proteins
• These proteins adhere sometimes, temporarily to the biological membrane
• They do not enter into the hydrophobic space within the cell membrane but can push into the peripheral regions of the lipid bilayer
• They attach to integral membrane proteins
membrane carbohydrates
• Components of complex molecules on the cell membrane
– glycoproteins
– glycolipids
– proteoglycans
• The carbohydrates forms a layer around the cell on the outside of the membrane only
• It is referred to as the Glycocalyx
• This layer functions as a barrier between a cell and its surrounding
• It also serves as a mediator for cell-cell interactions
• It protects a cell membrane from the direct action of physical forces and stresses allowing the membrane to maintain its integrity.
• It is involved in development and progression of many diseases
function of cell plasma membrane
• It is a sheet-like structure that forms closed boundaries
• Physical isolation, it acts as a barrier
• Holds molecules inside in the cell
• It allows for regulation of exchange into and out of the membrane
• It creates an environment that responds to changes in the cell’s environment
– unique intracellular pH (redox)
– response of its receptors
• It provides structural support for the cell
fluid mosaic model of membrane
• Lipid bilayer
– Solvent for integral proteins
– Permeability barrier
• Small amount of lipids interact specifically with particular membrane proteins
• Proteins can diffuse laterally but are not free to rotate
TRANSPORT ACROSS THE MEMBRANE: PASSIVE DIFFUSION
• The unaided spontaneous movement of solute molecules down their concentration gradient, from high to low, until solutes equilibrate across the bilayer
• Entropically driven
• Maximum entropy at equilibrium
TRANSPORT ACROSS THE MEMBRANE: FACILITATED DIFFUSION
• Carrier mediated diffusion (Uses an integral protein)
• Occurs down concentration gradient
• Dependent on integral proteins
• Does not 'use' energy
antiport; 2 molecules move in opposite directions simultaneously
uniport; 1 molecule one way
symport; 2 molecules same way
active transport across membrane
• Movement against a concentration gradient
• Does use energy
• 3 forms
– Primary active transport
– Secondary active transport
– Group translocation
secondary active transport
• Carrier protein that moves a specific substance against its concentration gradient
• Carrier protein can also move another substance at the same time
• The concentration gradient for one substance provides the driving force for the carrier protein
• Second substance gets a ‘free ride’
requires energy in form of ATP
movement of melecules via second protein depend on energy made by sodium-potassium pump (primary)
REVIEW VIDEO
primary active transport
requires energy in form of ATP (phosphate)
exp- sodium potassium pump, potassium goes in while sodium goes out of cell
3 sodium receptors for 2 potassium
when phosphate binds to protein- allows Na to leave and then potassium to bind.
when phosphate released, potassium released in cell
protein change shape to allow molecules to go in and out
since all molecules involved are + and not in equal quantities- electrical gradient is created
work against concentration gradient
endocytosis
substance moved into cell
exocytosis
substance moved out of cell
enzymes
• Enzymes are protein catalysts
• They increase the speed of chemical reactions in the body by 106 – 1012 fold
• They are not consumed during the reaction, can be used again after reaction
• Enzymes are specific:
– each enzyme binds just one (or a few) substrates (keys) – each enzyme catalyses one reaction type
• Some enzymes require cofactors
REVIEW VIDEO
enzyme terms
• Enzymes (E) have an active site
• This is formed by folding of the enzyme
• It is the place where the substrate(s) (S) bind
• It is lined with amino acid side-chains that bind the substrate non-covalently
• It forms the enzyme-substrate [ES] complex
• The interaction results in the creation of a product (P)
lock and key model
E+ S → ES → E+ P
nomenclature and naming of enzymes
The naming of enzymes is based on:
• The type of reaction catalysed (followed by the suffix –ase), e.g. Dehydrogenase, protease, transferase, reductase
• The nature of the substrate, e.g. Glucokinase, hexokinase
• The source of the enzyme, e.g. Pancreatic lipase, salivary amylase
• Its regulation, e.g. Hormone specific lipase
• There are other names that are not obvious enzyme names, e.g. Trypsin, pepsin, thrombin
effect of temperature and ph enzymes
• Each enzyme has an optimum temperature and pH for its biological activity
• Velocity of the enzymatic reaction increases with temperature up to a point and then starts to decrease due to protein denaturation
• Each enzyme has an optimum pH at which it functions most effectively
• Changes in pH may affect
– the function of active site amino acid sidechains
– may cause denaturation
enzyme kinetics
• Another major factor that affects enzyme rate is substrate concentration [S]
• As [S] increases, the rate of the reaction increases
• The increase is linear until it reaches a plateau, where further increase in [S] does not produce any increase in the reaction rate.
• This occurs because at low [S] the rate of reaction is limited by the number of substrate molecules available to bind to the active site
• As the number of molecules increases further, the enzyme becomes saturated, i.e. all the binding sites are constantly occupied by the substrate
• The maximum reaction velocity is designated V max
THE MICHAELIS MENTEN EQUATION
• A plot of the initial rate of the reaction (v) against different [S] results in a curve described by the Michaelis–Menten equation
• The maximum reaction rate (Vmax) can be estimated from the plateau of the curve
• The other parameter characteristic for an enzyme-substrate pair is the Michaelis constant (Km)
• This is the substrate concentration at which the rate is half Vmax
• The Km reflects the affinity of the enzyme for its substrate, expressed in molar concentration
• A low Km indicates a high affinity
• This indicates only a low concentration of substrate is required to saturate the enzyme
![<p>• A plot of the initial rate of the reaction (v) against different [S] results in a curve described by the Michaelis–Menten equation</p><p>• The maximum reaction rate (Vmax) can be estimated from the plateau of the curve</p><p>• The other parameter characteristic for an enzyme-substrate pair is the Michaelis constant (Km)</p><p>• This is the substrate concentration at which the rate is half Vmax</p><p>• The Km reflects the affinity of the enzyme for its substrate, expressed in molar concentration</p><p><strong>• A low Km indicates a high affinity</strong></p><p>• This indicates only a low concentration of substrate is required to saturate the enzyme</p>](https://knowt-user-attachments.s3.amazonaws.com/4ec467c7-cf90-48fd-98ba-b4c068f05949.jpeg)
enzyme inhibition
• An inhibitor is any substance that can diminish the velocity of an enzyme-catalysed reaction
• A reversible inhibitor binds to the enzyme with non-covalent bonds
• A dilution of the enzyme-inhibitor complex leads to dissociation and recovery of activity
• An irreversible inhibitor may form covalent bonds
• The enzyme cannot regain activity
COMPETITIVE AND NON COMPETITIVE INHIBITION
• In the presence of a competitive inhibitor (competing for same site) :
• The reaction velocity reaches the same Vmax observed in the absence of the inhibitor.
• At sufficiently high [S] the effect can be reversed by increasing [S].
• Increases the apparent Km: more substrate is needed to reach 1/2 Vmax
in the presence of non competitive inhibitor
• The Vmax is decreased
• Effect cannot be overcome by an increase in [S].
• The Km remains the same
enzyme cofactors
• Cofactors are non-protein substances required for an enzymatic reaction to proceed
• They are often metal ions, such as copper or magnesium
• Organic cofactors are known as coenzymes
• Vitamins often act as coenzymes
• They are frequently involved in oxidation–reduction reactions: acting as electron donors or acceptors
metabolism
a set of chemical reactions essential for life. These reactions can be divided into: catabolic and anabolic reactions
• Each step in a metabolic pathway is controlled by enzymes which enable energetically unfavourable reactions to proceed.
• By a process of enzyme inhibition or activation, metabolic pathways respond to changes to cellular or body environment and need.
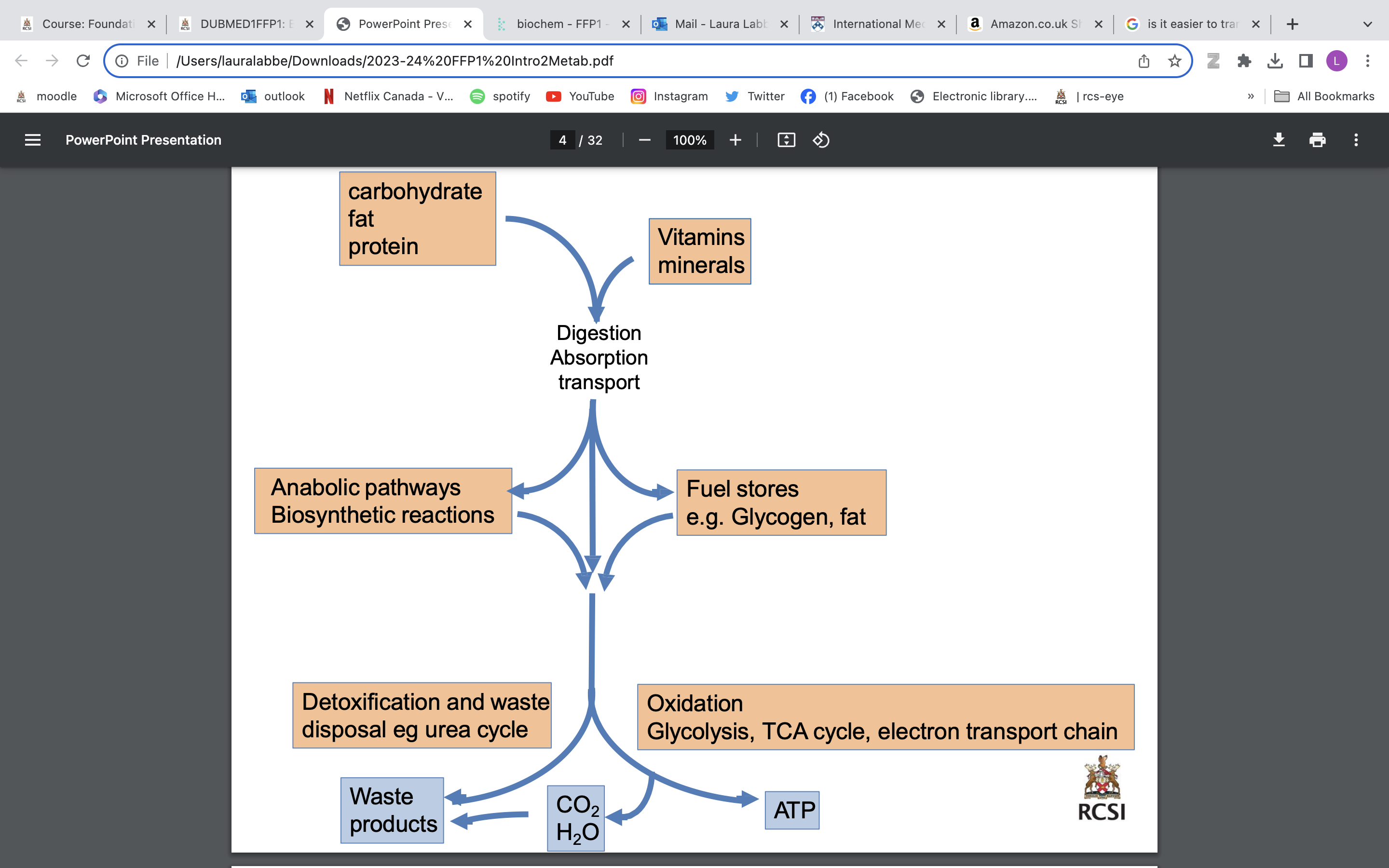
anabolic reactions
involve the synthesis of complex molecules - essential for life but which consume energy. “building up”
catabolic reactions
involve the breakdown of organic matter, ultimately to produce energy by cellular respiration, “breaking down”
anabolism vs catabolism
Anabolism example: Glucose used as a building block to make glycogen← energy input
Catabolism example: Carbohydrate (starch) → glucose (monosaccharide)→ energy out
Energy generated from catabolic pathways can be used in anabolic pathways
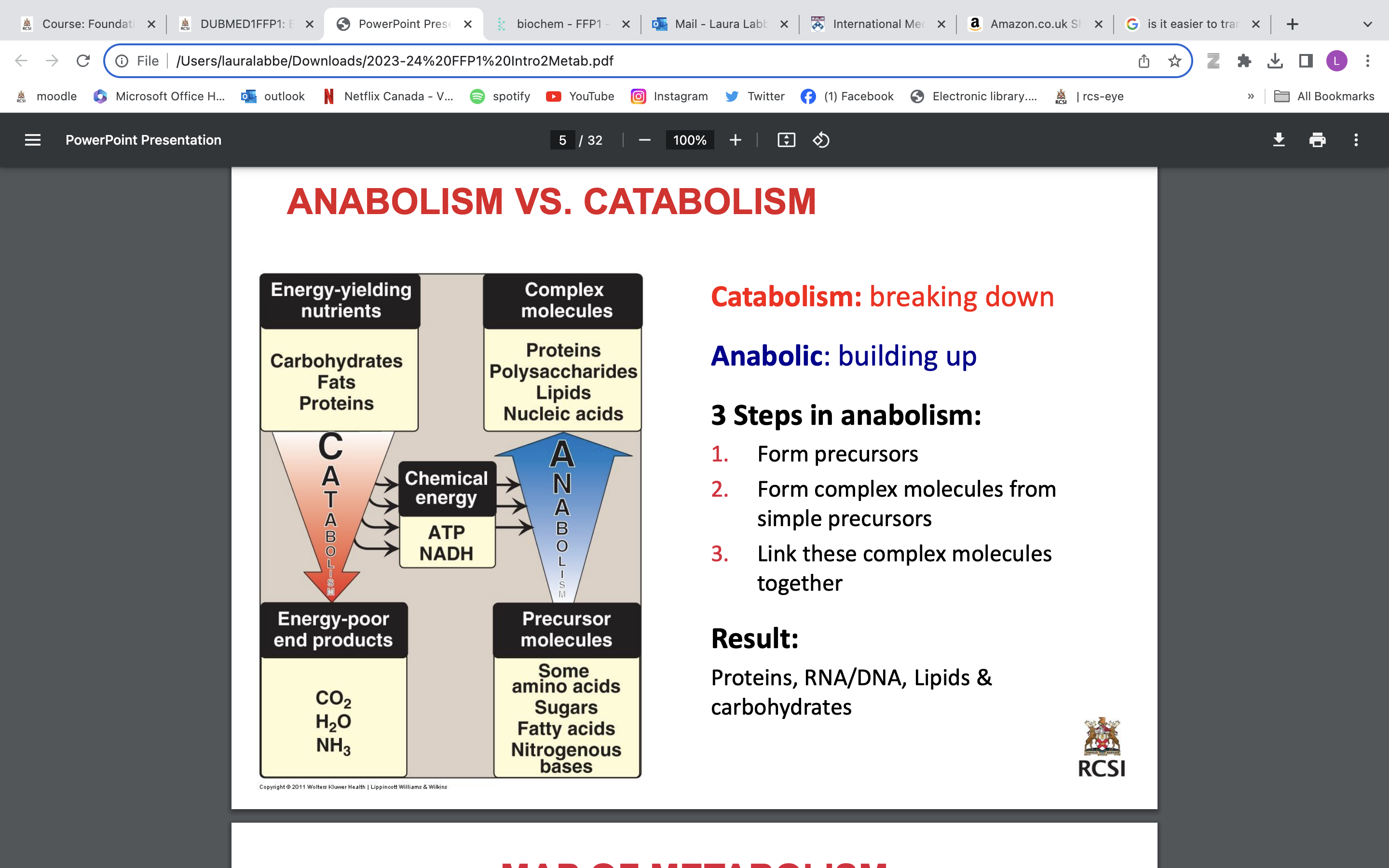
3 steps in anabolism
1. Form precursors
2. Form complex molecules from simple precursors
3. Link these complex molecules together
Result: Proteins, RNA/DNA, Lipids & carbohydrates
energy sources
• ATP – high energy phosphate group
• NADH – high energy electrons – reducing power
• NADPH - high energy electrons – reducing power
• FADH2 - high energy electrons – reducing power
• Where do ATP, NADH, NADPH, FADH2 come from?
• Catabolic reactions: e.g. Glycolysis & TCAcycle
energy stored in ATP
energy in phosphate bonds can be used to do “work” in cell and in anabolic reactions Cleavage of ATP to ADP releases energy
ATP→ ADP + Pi. DG= -8kCal/mol
Free energy released put to work in cell:
• Mechanical work – muscle contraction
• Transport work – K+ ATPase
• Biosynthetic work – amino acids, FAs, urea
ATP-ADP cycle:
• Energy requiring processes in the body use energy from the hydrolysis of ATP.
• To regenerate ATP need to oxidise fuels (breaking down foodstuffs)
Energy production: Catabolism Carbohydrate Lipids (FAs) Protein
Energy utilisation: Biosyn. Macromol Muscle contraction ion transport Thermogenesis etc
ATP→ ADP + Pi and vice versa! vicious cycle
change of energy aka delta G
• All processes in the body/cell involve changes in energy
• ΔG is the energy available to do work
• Predicts whether a reaction is favourable or not
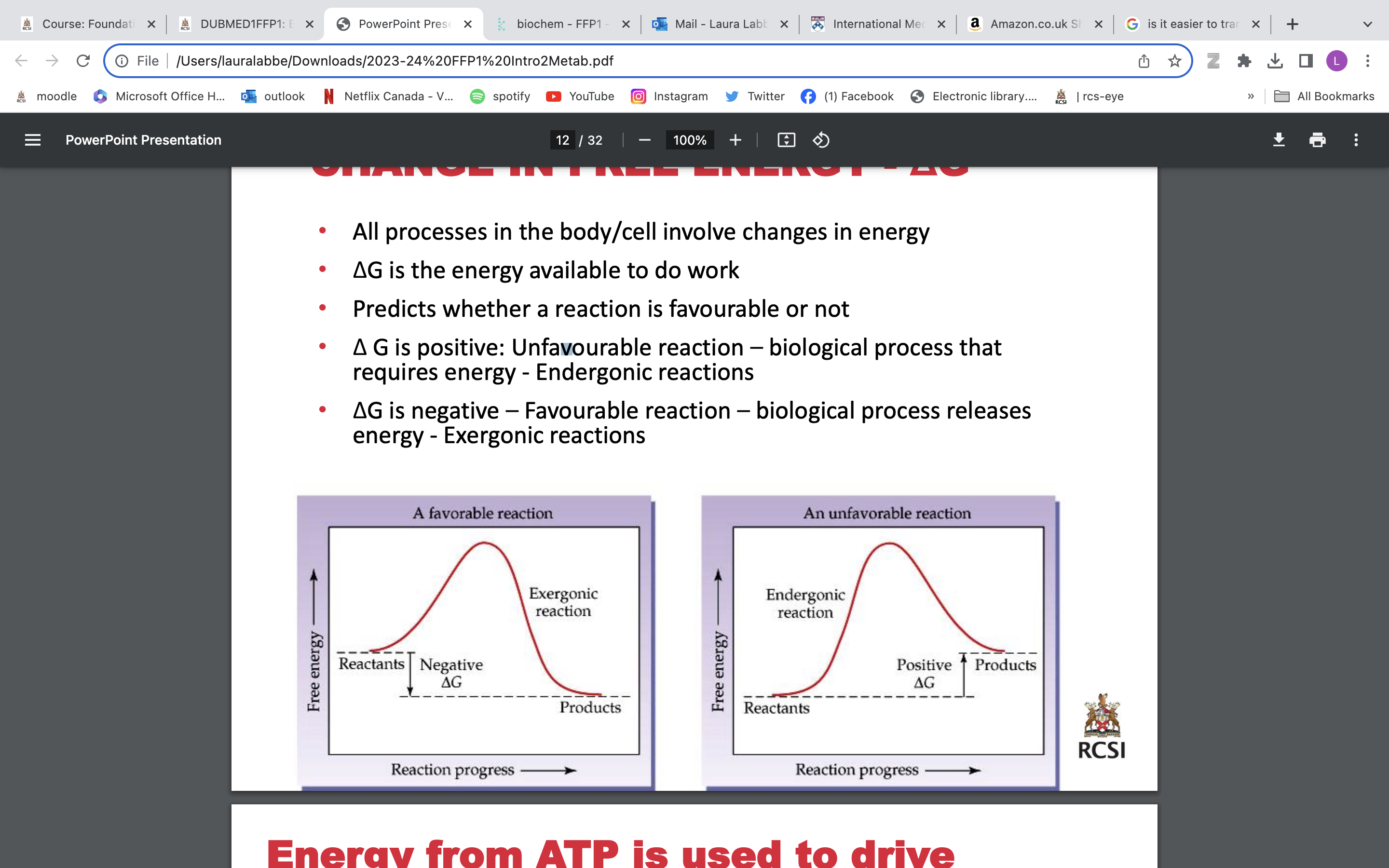
• Δ G is positive: Unfavourable reaction, biological process that requires energy - Endergonic reactions
Energy from ATP is used to drive unfavourable reactions
Goal for metabolism of food = Generate energy in form of ATP
-ΔG is negative – Favourable reaction – biological process releases energy - Exergonic reactions
example glucose metabolism
coenzymes:
NAD+ + 2H+ + 2e → NADH + H+
FAD + 2H+ + 2e → FADH2
electrons shown above contain a lot of energy
can exist in reduced or oxidized form
Oxidation - the loss of electrons or hydrogen
Reduction - the gain of electrons or hydrogen
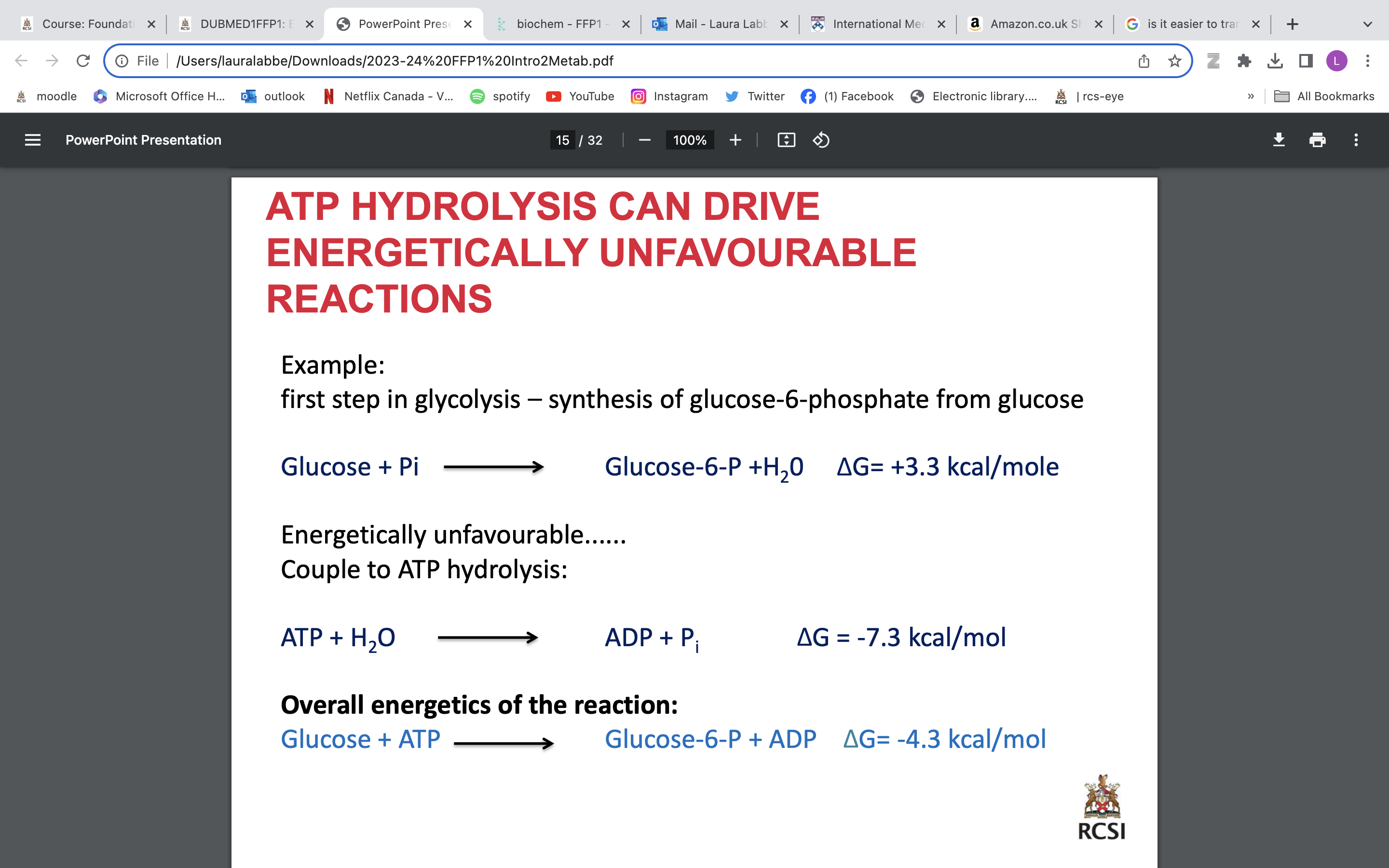
coupled redox reaction
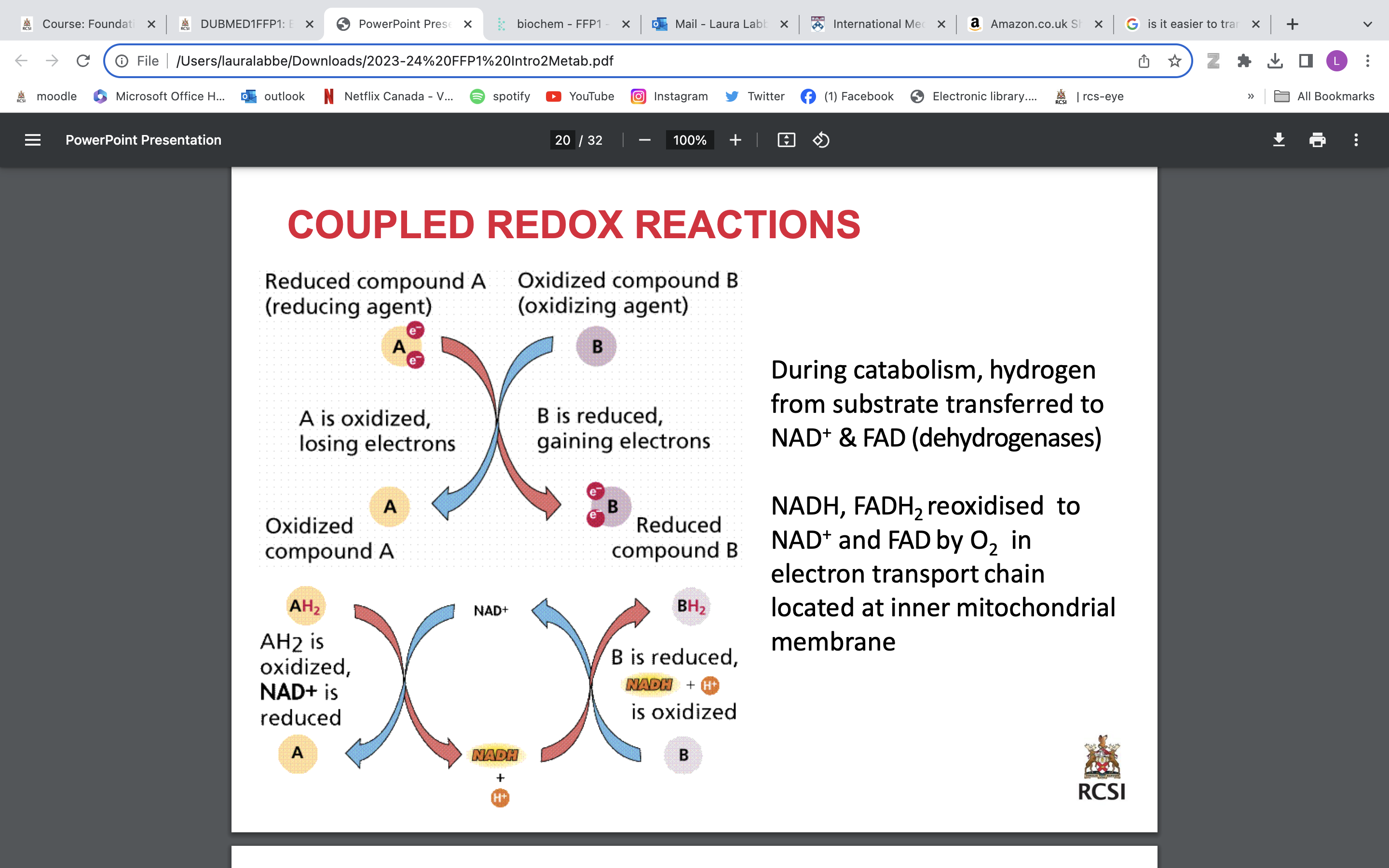
mitochondrion
Functions of the mitochondria
• Production of energy – ATP (cellular respiration)
• Cell signalling - mitochondria store calcium in the matrix and can release it
• Mitochondrially-encoded genes
• Most cells have 100’s-1000’s of mitochondria (except RBCs; none)
cytosol
Electron transport chain inner mito membrane Energy from NADH, FADH2generated during glycolysis and TCA cycle is used to make ATP
Oxygen is final acceptor of electrons
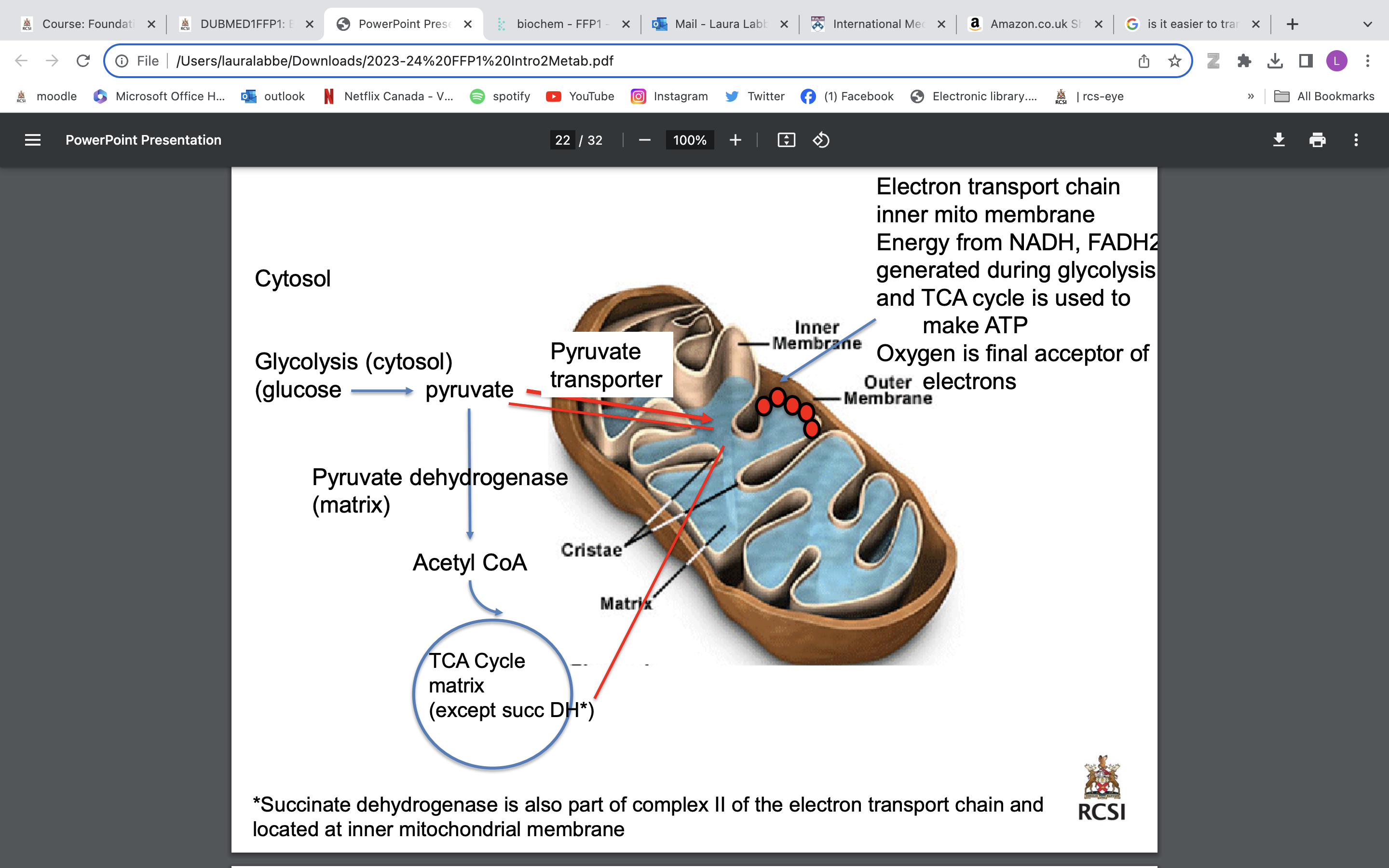
aerobic atp production
3 molecules of ATP per NADH+H+
2 molecules of ATP per FADH2
Conversion in the electron transport chain
1 glucose molecule results in either 36 or 38ATP depending which shuttle system is used to transfer e from the NADH generated in glycolysis into the mitochondrion (ETC lecture)
Under anaerobic conditions, only 2 ATP/molecule of glucose as pyruvate converted to lactate in a reaction that consumes 2 NADH.
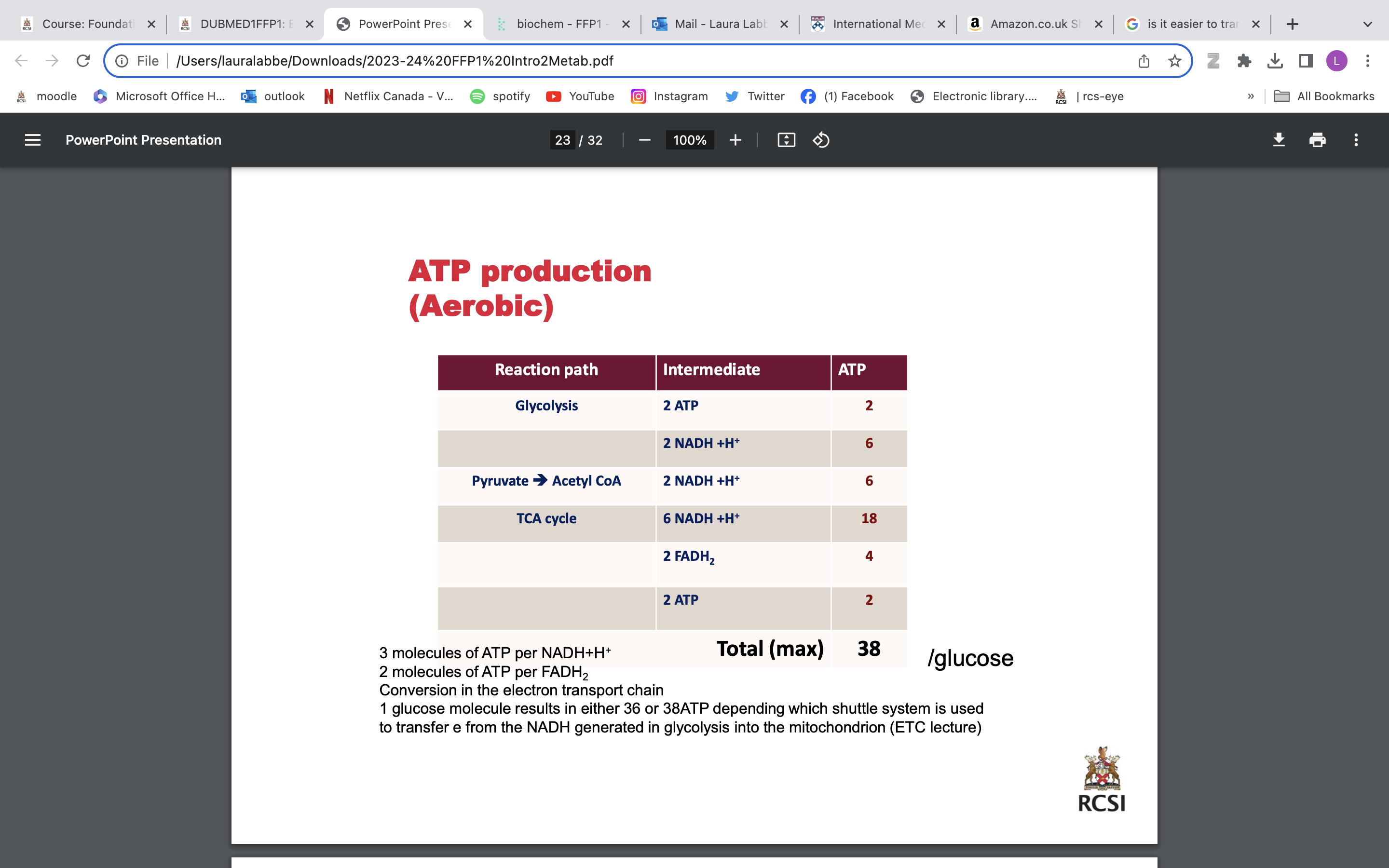
ELECTRON TRANSPORT CHAIN (ETC) OXIDATIVE PHOSPHORYLATION
Final common pathway for coenzymes NADH and FADH2 produced in catabolism and the TCA cycle. Reduced Coenzymes donate a pair of electrons to electron carriers. Electrons passed down ETC losing energy & generating ATP. Energy not converted to ATP used to transport Ca2+ & generate heat (extra energy, not necessary) TGs in adipose tissue are bodys principal storage form of energy TG breakdown yields large amounts of acetyl CoA and NADH Amino acids can be used to replenish TCA cycle intermediates Some Aas can be converted to pyruvate
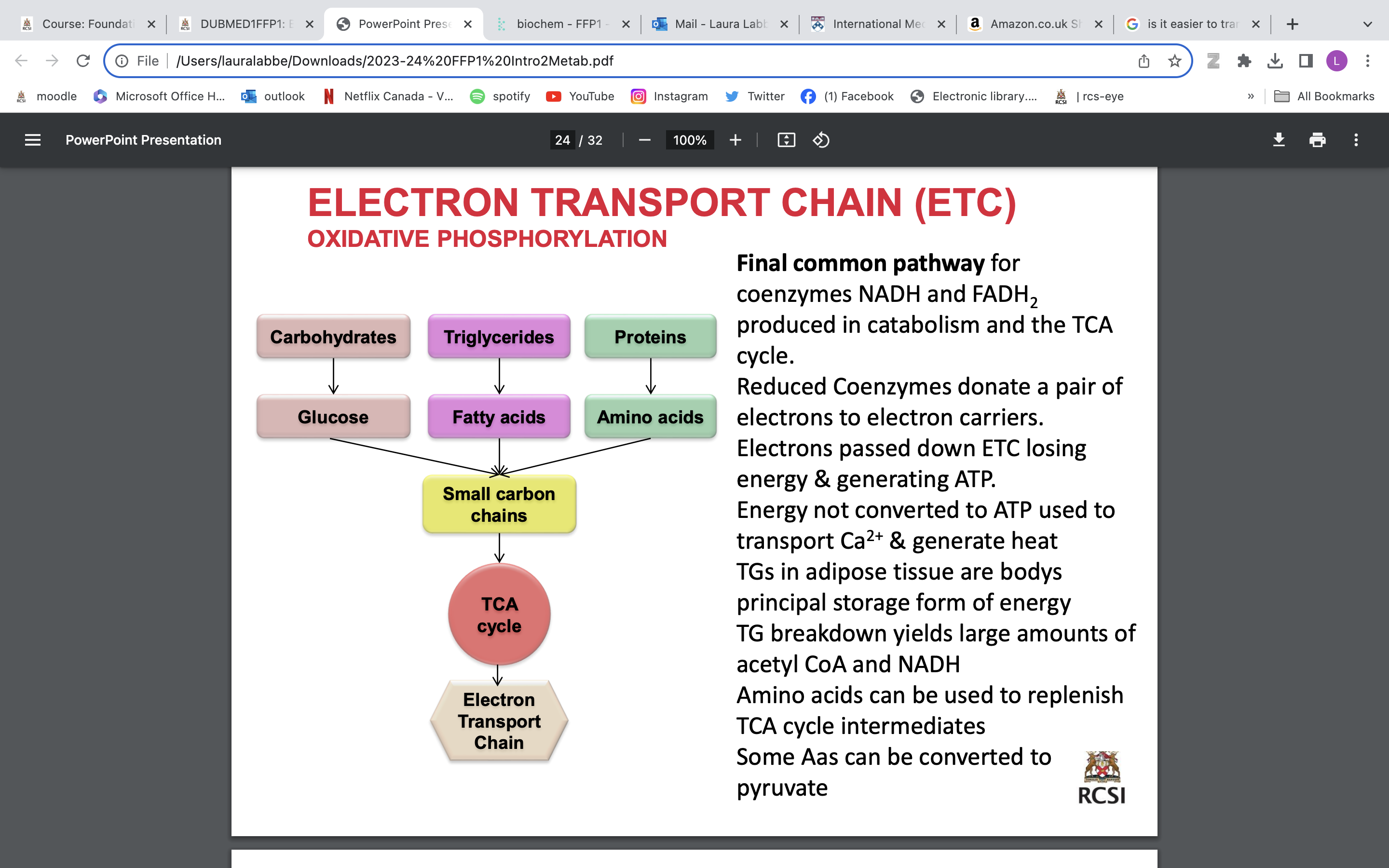
REDOX AND THE ELECTRON TRANSPORT CHAIN (ETC)
REDOX:
Addition of electrons = reduction
Removal of electrons = oxidation
• Energy released is harvested as ATP by oxidativephosphorylation e.g. transport of 2e - from NADH to O2 via ETC produces 52.6 kCal
• Energy required to produce 3 ATPfrom ADP + Pi = 3 X 7.3 = 21.9 kCal (balance released as heat)
• Transport of 2 e - from FADH2 to O2 produces 2 XATP
intermediates for anabolic reactions
Catabolic reactions provide intermediates for anabolic reactions.
The TCA Cycle provides a ‘pool’ of metabolic intermediates eg: Oxaloacetate is used in Gluconeogenesisto make Glucose
Acetyl CoA is also a substrate for biosynthetic reactions eg: synthesis of fatty acids, cholesterol & ketone bodies
The TCA cycle is amphibolic ie: it is involved in anabolic & catabolic pathways
Gluconeogenesis: synthesis of glucose from non-carbohydrate precursors, maintains blood glucose during “fasting”
metabolisms regulating metabolic pathways
Feedback Inhibition e.g. NADH can inihibit enzymes involved in its production, NAD+ can stimulate
Phosphorylation/dephosphorylation e.g. pyruvate dehydrogenase, glycogen synthase, glycogen phosphorylase
Hormonal regulation eg insulin, glucagon
Allosteric regulation eg ATP, acetyl CoA, Ca2+
Availability of substrates e.g. availability of oxaloacetate regulates citrate synthase activity
Oxygen availability
In the absence of oxygen glucose catabolism ends with conversion of pyruvate to lactate. No energy generation by TCA cycle or ETC -
Hypoxia/anoxia – failure of oxidative phosphorylation
Ischaemia – no ATP synthesis, shutdown of energy-dependent processes, osmotic imbalance due to failure of ATP-dependent ion pumps, pyruvate dehydrogenase inhibited, increased lactate, decreased pH
protein expression and inborn errors of metabolism
usually:
dna transcribed to rna and translated to protein
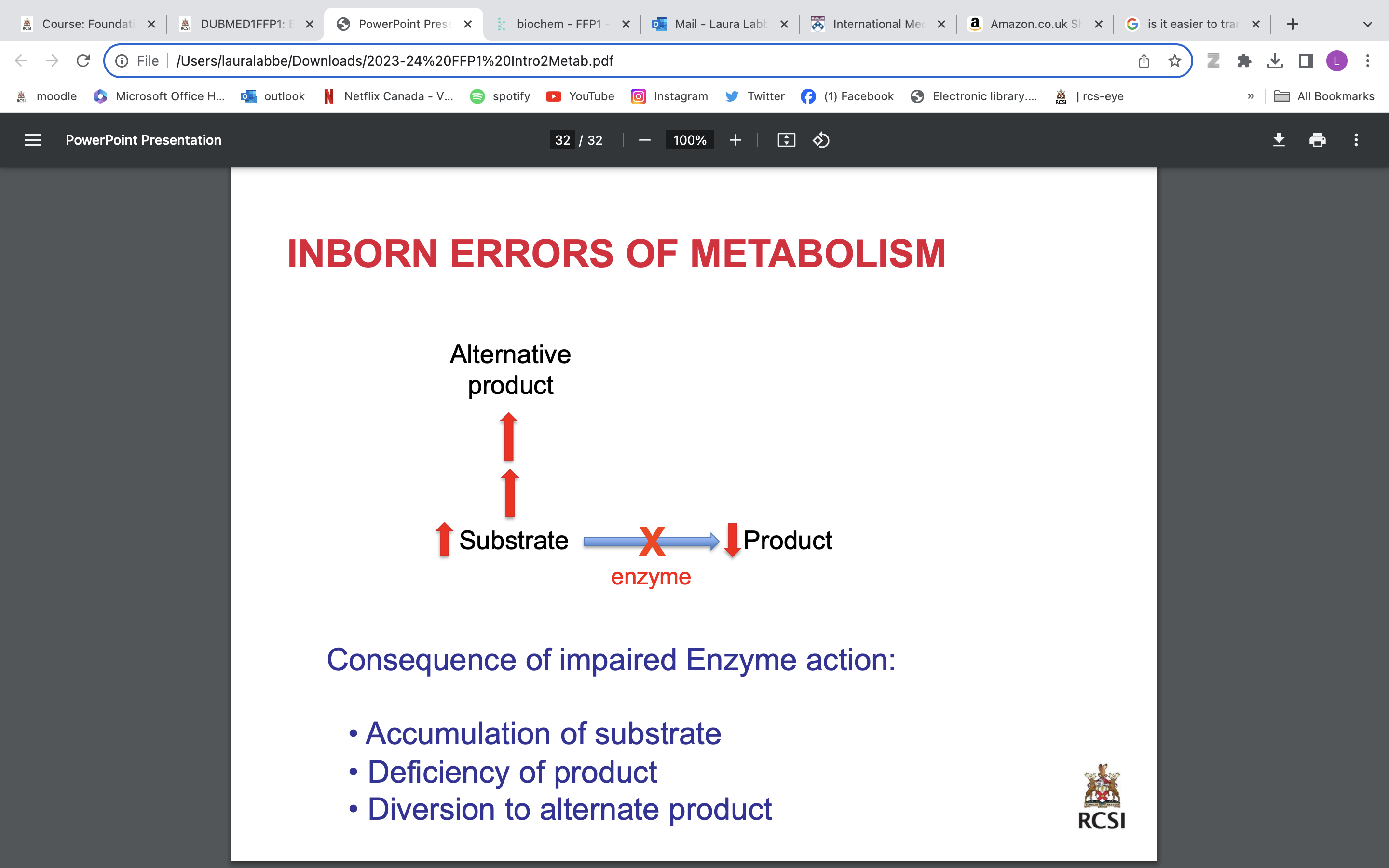
inborn errors of metabolism
mutation in gene→ defective protein→ impaired function→ disorder/ disease
Consequence of impaired Enzyme action:
• Accumulation of substrate
• Deficiency of product
• Diversion to alternate product
what is blood
•One of the largest organs distributed throughout the entire body
•Blood circulates through the body’s heart, arteries, veins, capillaries
•70kg man = 5.6L
–7- 8% body weight
•Temperature = 38°C
•Slightly alkaline
•pH 7.35-7.45
Carries TO the body tissues…
•oxygen
•nutrients
•hormones
•water
•solutes
•heat
Carries AWAY from the tissues…
•waste matter
•carbon dioxide
blood composition
•Plasma 55 % (~ 3.5 L)
–Liquid component of blood in which the cells are suspended
–Complex aqueous solution
–Gases, salts, proteins, carbohydrates & lipids
•Formed elements 45%
–Red cells (erythrocytes) 99%
–Platelets < 1%
–White cells (leukocytes) < 1%
•Whole blood allowed to clot
–clot removed
–remaining fluid is SERUM
–serum does not contain coagulation
factors
Haemopoiesis
production & development of new blood cells
•Two-thirds white cell production:
–‘Leucopoiesis’
•One-third red cell production
–‘Erythropoiesis’
•>500 times more RBCs than WBCs in circulation
•Differentiated cells lose their capacity for self-renewal
•Single stem cell produces >106 mature cells, and accounts for < 0.1% of all cells in bone marrow
•Stem cells grow and divide in bone marrow
•Lose Cell Adhesion Molecules (CAMs)
–Allow cells to leave marrow & enter circulation
•Require Growth factors:
–Erythropoietin
–colony stimulating factors
–Interleukins
–thrombopoietin
In children, haematopoiesis occurs in the marrow of the long bones such as the femur and tibia.
In adults, it occurs mainly in the pelvis, cranium, vertebrae, and sternum.
In some cases, the liver, thymus, and spleen may resume their haematopoietic function, if necessary. This is called extramedullary haematopoiesis.
Maturation, activation, and some proliferation of lymphoid cells occurs in the spleen and lymph nodes.
Erythropoiesis
rbc production
Typically one proerythroblast gives rise to about 16 mature RBCs
Anucleate, discoid shape
~120 day life-span
1% destroyed per day
anemia
Anaemia:
A decrease in Hb concentration below the reference range for the age & sex:
11.5 – 16.0 g/dL (female)
13.5 – 17.5 g/dL (male)
Inherited haemolytic anaemias:
• Glucose-6-phosphate dehydrogenase (G6PD) deficiency
• Sickle cell anaemia
life cycle of RBCs
•The life span of an RBC is ~120 days
•Senescent RBCs removed by macrophages
•Haemoglobin components are recycled:
•globin - amino acids reutilized
• iron reutilised
•Haem excreted in bile
aerobic metabolism
uses O2 while anaerobic does not→ aerobic more efficient
Multi-cellular organisms need to transport O2 to all tissues for aerobic metabolism and to store O2
Specialised proteins for this: Myoglobin and Haemoglobin
All tissues in the body undergoing aerobic respiration require O2 to function normally, aerobic metabolism being the most efficient way to generate energy.
Humans aren’t unique in this regard and all multicellular organisms need to find ways to shuttle O2 to all tissues within a body.
Similarly, there is a key advantage to being able to not only transport O2, but also store 02.
haem group
Oxygen binding by proteins depends on the haem group.
• Not part of the polypeptide chain
• Tightly bound to the protein
• Essential for haemoglobin activity [Fe2+/ferrous]
• Iron is held in position by 4 N’s
• Fe2+ can make 2 more bonds…
common feature of both myoglobin and haemoglobin is the presence of a heme group, which enables the oxygen-binding properties of each protein.
i.e. depending on the number of haem groups a protein has will determine how many oxygen molecules a given protein can carry…haemoglobin has 4, whereas myoglobin only has 1.
Haem is a prosthetic organic compound that isn’t part of a protein, i.e. it isn’t genetically encoded and doesn’t consist of amino acid residues
Haem groups are characterized by capacity to bind Fe2+ which sits right in the middle of the haem porphyrin ring and has free bonds that enable it to capture O2.
structure of myoglobin
O2 reservoir within heart & skeletal muscle cells
153 aa 17kDa compact protein
Structure:
• 75% a- helix
• 8 helices (labelled A-H)
• Non-helical regions (AB, BC etc.)
• Exterior hydrophilic
• Interior hydrophobic except histidines E7 & F8
Haem sits in a crevice near the surface lined with non-polar residues
E7 = distal His
F8 = proximal His
Oxygenation moves the haem iron which moves His F8
Helix F moves & causes other elements of the structure to move
Hb vs Mb
•Mb is a storage protein
•binds O2 avidly, dissociates slowly
•Mb is not co-operative
•Mb is 1 polypeptide
hemoglobin
2 Major functions:
•Transports O2 to tissues.
•Transports CO2 and protons away from tissues.
Structure:
•4 polypeptide chains
•Each chain has a haem group
= can bind 4 oxygens
•Subunits held together by non-covalent interactions
Gene expression of the a & b chains
The composition of the 4 protein sub-units of haemoglobin can change between individuals and in different points in your life and this is mediated by differential gene expression of distinct functional sub-units.
For example there are haemoglobin proteins that predominate during embryonic development and also a well-characterized fetal haemoglobin which has increased oxygen binding capacity for reasons we’ll discuss later.
In adults, the Hb A predominates in approximately 85% of adults, whereas a reduced number of adults express Hb A2.
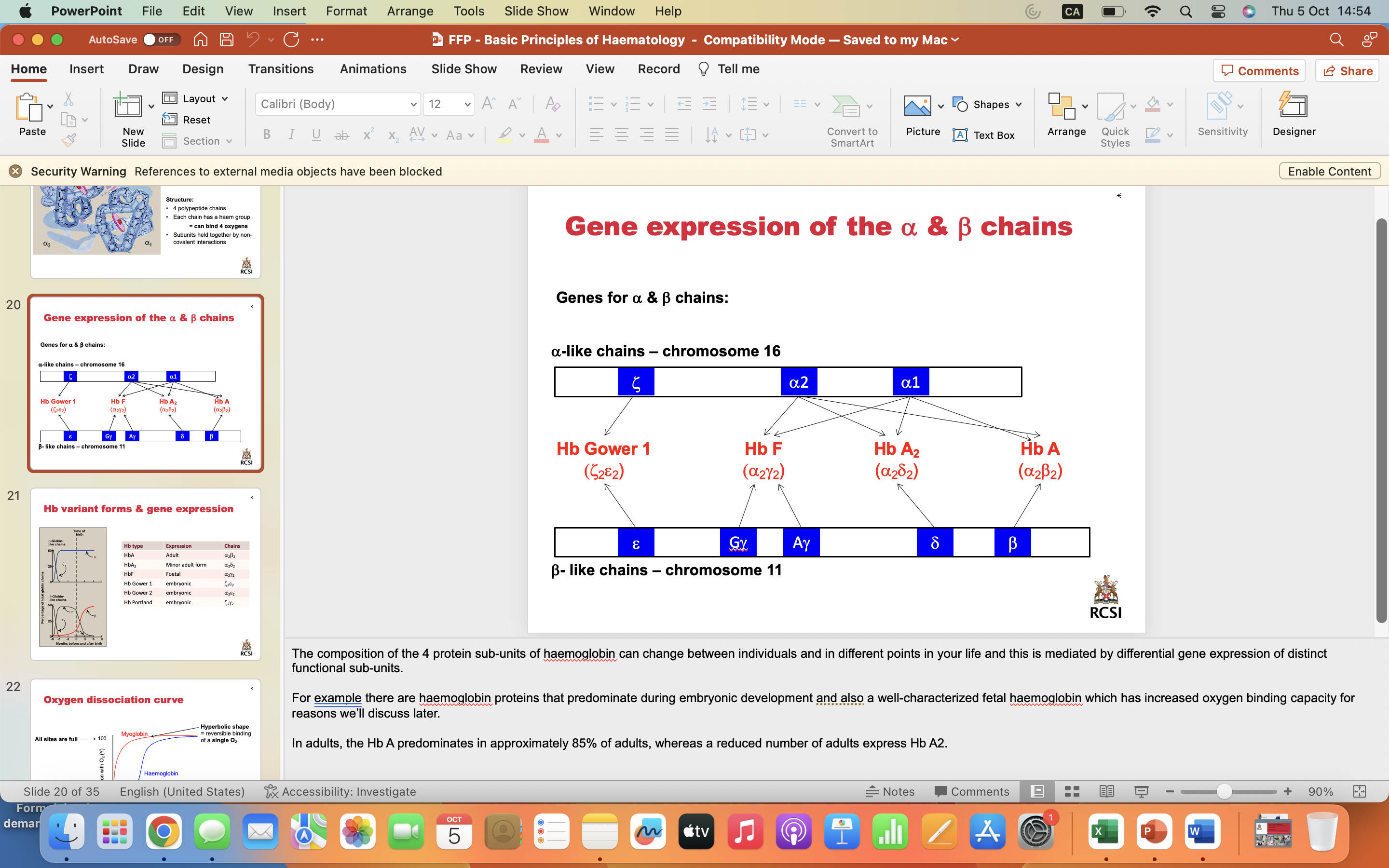
Hb variant forms & gene expression
These graphs illustrate how the alpha and beta chains of haemoglobin change during development
Alpha chain embryonic form of haemoglobin utilize zeta-form of alpha chain, which exists until fetal formation at which point the alpha chain predominates
Beta chains vary most during development, with the epsilon and gamma chains being expressed during embryonic development, which switches to the featl gamma form during fetal development. In the time after childbirth, the beta form slowly replaces the fetal gamma form until mature Hb A is formed.
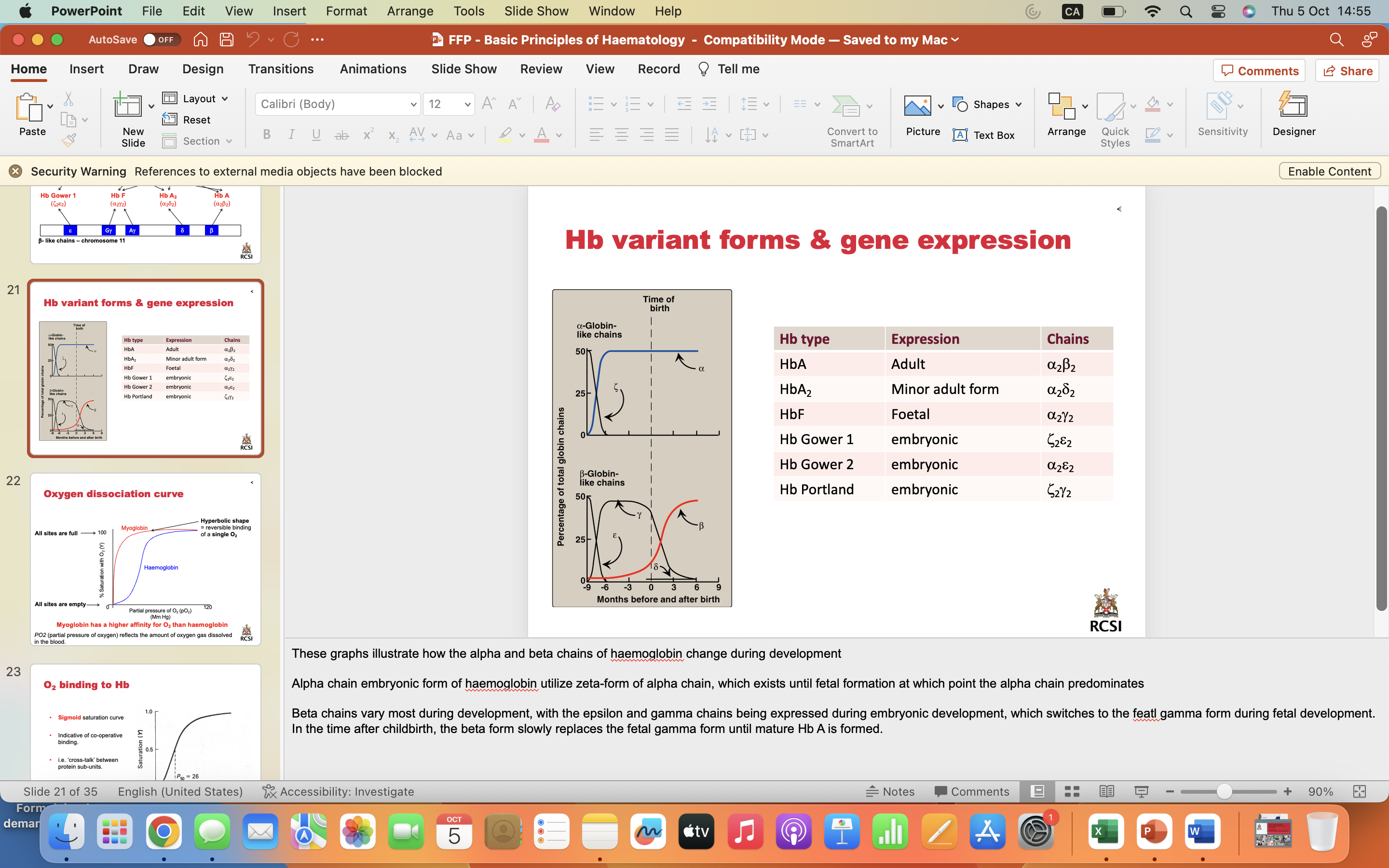
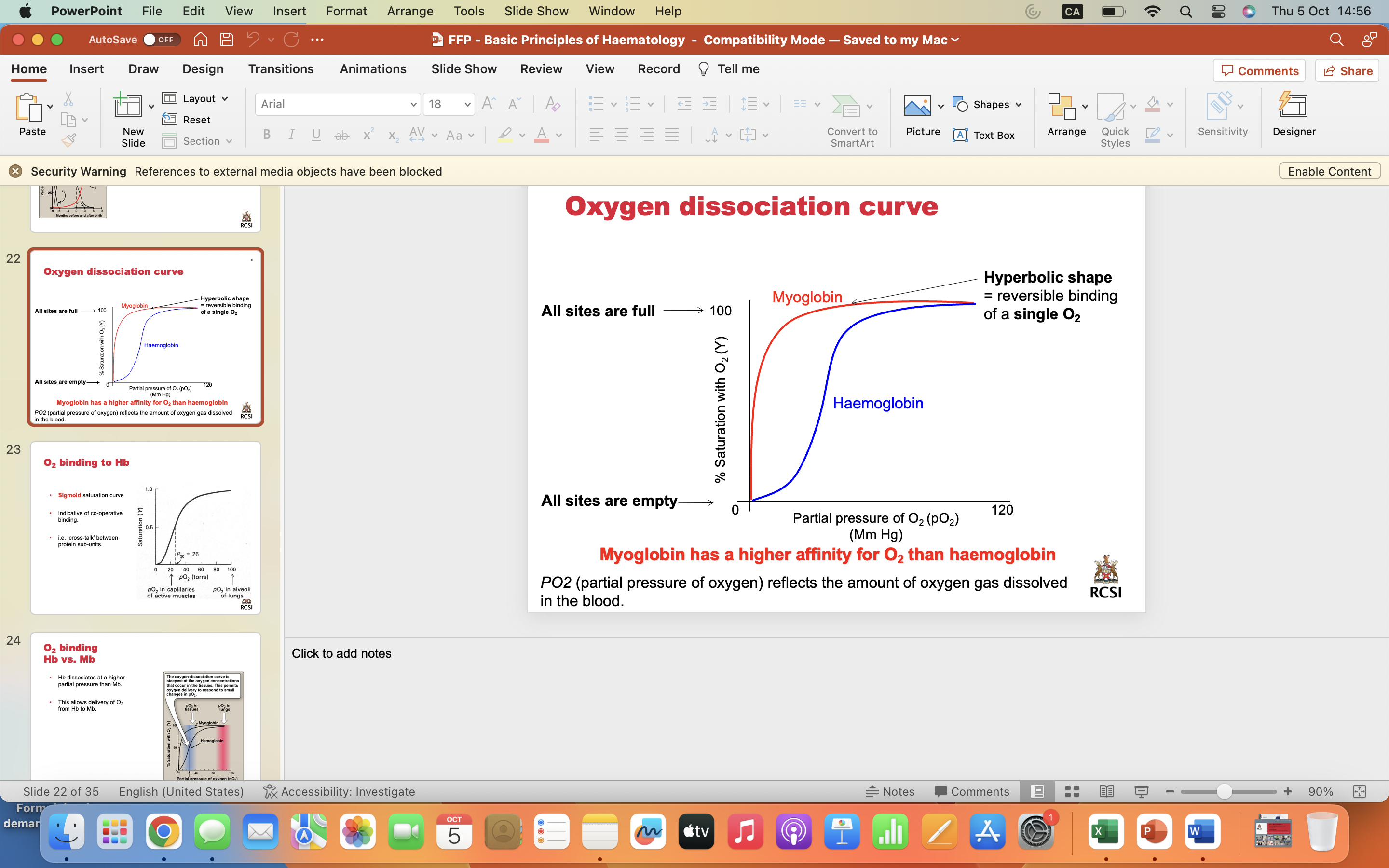
Oxygen dissociation curve
Myoglobin has a higher affinity for O2 than haemoglobin
PO2 (partial pressure of oxygen) reflects the amount of oxygen gas dissolved
in the blood.