*path bac exam 3
0.0(0)
Card Sorting
1/136
Earn XP
Description and Tags
Last updated 1:25 PM on 4/2/23
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
137 Terms
1
New cards
constitutive vs non-constitutive
**constitutive:** genes that are always expressed
* typically perform housekeeping functions
* tibosomal RNAs and proteins involved in protein synthesis
\
**non-constitutive:** genes that are only expressed when their products are required
* subject to regulation
* virulence genes
* typically perform housekeeping functions
* tibosomal RNAs and proteins involved in protein synthesis
\
**non-constitutive:** genes that are only expressed when their products are required
* subject to regulation
* virulence genes
2
New cards
why are some genes regulated
avoid host defenses - antigenic + phase variation - turn off genes to evade immune system
energy - don’t make components that aren’t needed
energy - don’t make components that aren’t needed
3
New cards
central dogma
DNA - transcription - RNA - translation - protein
DNA - gene expression - protein
DNA - gene expression - protein
4
New cards
regulation of gene expression
transcriptional level: DNA → RNA + post-transcriptional levle
\
translational level: RNA → protein + post-translational level
\
translational level: RNA → protein + post-translational level
5
New cards
transcriptional regulation - 5
transcriptional regulators
two component systems
sigma factors
supercoiling
nucleoid associated proteins
two component systems
sigma factors
supercoiling
nucleoid associated proteins
6
New cards
gene structure 6
**Promoter** – Where RNA polymerase binds to DA
**Transcription start site** – Where transcription begins
**Shine and Dalgarno site** – AKA RBS – Where the ribosome binds the mRNA
**Translation start site** – Where translation begins – ATG start codon
**Translation stop site** – Where translation stops – 3 possible codons
**Transcription stop site** – Where transcription terminates
**Transcription start site** – Where transcription begins
**Shine and Dalgarno site** – AKA RBS – Where the ribosome binds the mRNA
**Translation start site** – Where translation begins – ATG start codon
**Translation stop site** – Where translation stops – 3 possible codons
**Transcription stop site** – Where transcription terminates
7
New cards
gene structure - ecoli and RNA pol binding
TTGACA = -35 box
TATAAT = -10 box
the optimal inter-base distance cannot be more than 1+/- WHY? RNA pol binds 3 dimentionally and if there is more the 1 bp change then TATAAT will be on the other side of the DNA helix and RNA cannot bind
TATAAT = -10 box
the optimal inter-base distance cannot be more than 1+/- WHY? RNA pol binds 3 dimentionally and if there is more the 1 bp change then TATAAT will be on the other side of the DNA helix and RNA cannot bind
8
New cards
operon
group of 2+ genes together on the same transcription unit
9
New cards
transcription activator v repression
activator: required to turn on the expression of one or more genes
repressor: required to turn off the expression of one or more genes
repressor: required to turn off the expression of one or more genes
10
New cards
transcriptional activators
typically bind to the DNA in the vicinity (mostly upstream) of the promoter to help recruit RNA polymerase
can overlap with the promoter
can overlap with the promoter
11
New cards
transcriptional repressors
most common mechanism of action by repressions is to bind to promoter DNA and prevent RNA polymerase binding
can also compete with activators for the same binding site
can also compete with activators for the same binding site
12
New cards
bacterial virulence gene regulation
regulon definition
regulon definition
transcription regulator proteins often activate transcription from more than one promoter
regulon: all of the genes under transcription control of the same regulator
regulon: all of the genes under transcription control of the same regulator
13
New cards
two component system
what does it typically do
signals 4
what does it typically do
signals 4
two component signal transduction systems
stimulus-response coupling mechanism
two components:
* sensor protein: bacteria use to sense their environment
* response protein: bacteria use to carry out a response
response is typically alterations in gene expression - response protein is typically a transcription regulator
signals: pH, osmolarity, AMPs, cation concentration
stimulus-response coupling mechanism
two components:
* sensor protein: bacteria use to sense their environment
* response protein: bacteria use to carry out a response
response is typically alterations in gene expression - response protein is typically a transcription regulator
signals: pH, osmolarity, AMPs, cation concentration
14
New cards
two component signal transduction steps
signal received by histidine kinase (sensor)
autophosphorylation of His
phosphate is transferred to Aspartic acid on response regulator
conformational change → cellular response
reaction: altered gene expression
autophosphorylation of His
phosphate is transferred to Aspartic acid on response regulator
conformational change → cellular response
reaction: altered gene expression
15
New cards
histidine kinase
composed of: HK domain, variety of other domains
sense a variety of stimuli from a variety of sources
* extracellular: lots of complex loops outside the membrane
* intramembrane: lots of transmembrane domains
* intracellular: transmembrane domains and short external loops
sense a variety of stimuli from a variety of sources
* extracellular: lots of complex loops outside the membrane
* intramembrane: lots of transmembrane domains
* intracellular: transmembrane domains and short external loops
16
New cards
activation of HK
activation of HK: dimerization, autophosphorylation
autophosphorylation ALWAYS occurs on a conserved histidine AA residue
autophosphorylation ALWAYS occurs on a conserved histidine AA residue
17
New cards
response regulator
typically have 2 domains: N-term receiver domain, c-term output domain
N-term receiver domain: ALWAYS contains as Aspartic Acid residue, receives a phosphate group
C-term output domain: usually has a DNA binding domain, most common is HTH (helix-turn-helix)
\
receiver domain is phosphorylated, helix-turn-helix region is exposed, dimerizes, then can bind DNA
N-term receiver domain: ALWAYS contains as Aspartic Acid residue, receives a phosphate group
C-term output domain: usually has a DNA binding domain, most common is HTH (helix-turn-helix)
\
receiver domain is phosphorylated, helix-turn-helix region is exposed, dimerizes, then can bind DNA
18
New cards
NarX-NarL system
simple 2-component system
nitrate signal binds NarX, autophosphorylates NarL, dimerizes, transcriptional activation
nitrate signal binds NarX, autophosphorylates NarL, dimerizes, transcriptional activation
19
New cards
Rcs phosphorelay system
complex multi-component system
not all “two component systems” have just TWO components
Rcs system has 5 components
not all “two component systems” have just TWO components
Rcs system has 5 components
20
New cards
BvgS/R system
some histidine kinases have multiple domains (HK and RR domains)
phosphate group is always transferred ….
H → D and D → H NEVER H → H or D →D
check slide 30 lec 13
phosphate group is always transferred ….
H → D and D → H NEVER H → H or D →D
check slide 30 lec 13
21
New cards
what is a sigma factor
sigma factor is a protein that interacts with RNA polymerase and DIRECTS it to promoters
not considered a part of RNA polymerase
not directly required for transcription
once open complex forms sigma factor is released
sigma factor + core RNA polymerase = holoenzyme
bacterial use different sigma factors to direct RNA polymerase to different genes
turns on regulatory networks at different times
* sporulation
* stress response
* virulence
not considered a part of RNA polymerase
not directly required for transcription
once open complex forms sigma factor is released
sigma factor + core RNA polymerase = holoenzyme
bacterial use different sigma factors to direct RNA polymerase to different genes
turns on regulatory networks at different times
* sporulation
* stress response
* virulence
22
New cards
sigma factors
direct RNA polymerase to different subsets of genes depending on environmental conditions
oS = stationary phase genes, starvation
oE = envelope stress genes, something wrong with membrane
oF = flagella genes
oH = heat shock genes
having different sigma factors = use the same RNA pol and direct it to genes that need to be expressed
sigma factors allow the cell to reuse core RNA polymerase = converses energy
\# sigma factors varies greatly depending on the bactera
oS = stationary phase genes, starvation
oE = envelope stress genes, something wrong with membrane
oF = flagella genes
oH = heat shock genes
having different sigma factors = use the same RNA pol and direct it to genes that need to be expressed
sigma factors allow the cell to reuse core RNA polymerase = converses energy
\# sigma factors varies greatly depending on the bactera
23
New cards
genome condensation
to fit the nucleoid (genome) inside the cell bacteria use a combination of
* nucleoid associated proteins
* supercoiling
* nucleoid associated proteins
* supercoiling
24
New cards
DNA supercoiling
DNA helix underwound
introduces negative (right handed) supercoils
condenses DNA
degree of negative superhelicity changes according to environmental/physiological conditions and stimuli
introduces negative (right handed) supercoils
condenses DNA
degree of negative superhelicity changes according to environmental/physiological conditions and stimuli
25
New cards
how do bacterial control supercoiling levels
DNA gyrase and Topoisomerase 1
Gyrase introduces negative supercoils
Topoisomerase 1 introduces positive supercoils (relaxes DNA)
underwinding/negative supercoiling
overwinding/ positive supercoiling
?? slide 41 lec 13
Gyrase introduces negative supercoils
Topoisomerase 1 introduces positive supercoils (relaxes DNA)
underwinding/negative supercoiling
overwinding/ positive supercoiling
?? slide 41 lec 13
26
New cards
DNA supercoiling during infection
as salmonella bacteria enters macrophage, salmonella DNA becomes more relaxes (less negatively supercoiled)
DNA relaxation? = signal to activate genes require for survival inside macrophages
promoters become activated by DNA relaxation
slide 42 lec 13
DNA relaxation? = signal to activate genes require for survival inside macrophages
promoters become activated by DNA relaxation
slide 42 lec 13
27
New cards
nucleoid associated proteins
functions 3
participate in DNA transactions 4
four major NAPs
regulation of gene expression? 2
functions 3
participate in DNA transactions 4
four major NAPs
regulation of gene expression? 2
class of small DNA binding proteins found associated with the genome (nucleoid)
bind DNA
bend DNA
participate in DNA transactions
* transcription
* recombination
* replication
* integration and excision
functions:
* condense DNA
* DNA structural organization (supercoiling)
* transcriptional regulators
four major nucleoid associated proteins
* H-NS
* HU
* IHF
* Fis
play a role in regulation of gene expression
* structural: i.e. positive interaction bends DNA so activator can bind RNA pol
* as transcription activators/ repressors: typically indirect (don’t contact RNAP), compete for binding with activator/repressors
bind DNA
bend DNA
participate in DNA transactions
* transcription
* recombination
* replication
* integration and excision
functions:
* condense DNA
* DNA structural organization (supercoiling)
* transcriptional regulators
four major nucleoid associated proteins
* H-NS
* HU
* IHF
* Fis
play a role in regulation of gene expression
* structural: i.e. positive interaction bends DNA so activator can bind RNA pol
* as transcription activators/ repressors: typically indirect (don’t contact RNAP), compete for binding with activator/repressors
28
New cards
virulence regulation 3
cofactors and ligands → DtxR
regulatory RNAs
quorum sensing
regulatory RNAs
quorum sensing
29
New cards
transcription regulators
positive regulation: activator > repressor
negative regulation: repressor > activator
negative regulation: repressor > activator
30
New cards
activation of transcription factors
function
function
transcription factors may be activated or deactivated
TCS: phosphorylation of response regulator activates protein
interaction with other transcription factors (homo or hetero dimerization)
ligands can bind to TF activating/deactivating the protein
ligand: a substance (usually a small molecule) that forms a complex with a protein to serve a biological purpose
* activate
* deactivate
* modify function
TCS: phosphorylation of response regulator activates protein
interaction with other transcription factors (homo or hetero dimerization)
ligands can bind to TF activating/deactivating the protein
ligand: a substance (usually a small molecule) that forms a complex with a protein to serve a biological purpose
* activate
* deactivate
* modify function
31
New cards
regulation of dtxR and tox
regulation of dipthereia toxin (DT) gene expression is controlled by a transcription regulator DtxR
DtxR is activated when Fe2+ levels are high (binds)
DtxR-Fe2+ binds to the tox gene promoter and represses transcription
DtxR regulates tox gene, binds Iron, becomes active,
lots of Fe2+ = tox not produced
low Fe2+ (like inside host)= tox produced, Fe2+ dissociates from DtxR, DtxR no longer binds to tox promoter → transcription of tox → make DT
DtxR is activated when Fe2+ levels are high (binds)
DtxR-Fe2+ binds to the tox gene promoter and represses transcription
DtxR regulates tox gene, binds Iron, becomes active,
lots of Fe2+ = tox not produced
low Fe2+ (like inside host)= tox produced, Fe2+ dissociates from DtxR, DtxR no longer binds to tox promoter → transcription of tox → make DT
32
New cards
regulatory RNAs
aka small RNAs (sRNAs)
heterogeneous class of RNA
diversity of regulatory mechanisms
regulate directly or indirectly multiple genes
can be part of the transcript they regulate (cis-acting)
regulate other transcripts (trans-acting)
regulatory activity typically depends on structure and base pairing
heterogeneous class of RNA
diversity of regulatory mechanisms
regulate directly or indirectly multiple genes
can be part of the transcript they regulate (cis-acting)
regulate other transcripts (trans-acting)
regulatory activity typically depends on structure and base pairing
33
New cards
regulatory RNAs types 3
riboswitches and antitermination
cis-acting RNA thermosensors
trans-acting RNAs
cis-acting RNA thermosensors
trans-acting RNAs
34
New cards
riboswitches and antitermination SAM
SAM (S-adenosylmethionine) riboswitch - example of a small molecule regulating its own production
long 5” UTR --------- CDS: gene for protein involved in methionine biosynthesis (pathway to generate SAM)
lots of SAM? binds to RNA structure and stabilizes it, it leads to a terminator stabilized and terminates sequence, does not transcribe CDS
low SAM? antiterminator creater, CDS is transcribed, CDS contains same gene → Low SAM makes more SAM
long 5” UTR --------- CDS: gene for protein involved in methionine biosynthesis (pathway to generate SAM)
lots of SAM? binds to RNA structure and stabilizes it, it leads to a terminator stabilized and terminates sequence, does not transcribe CDS
low SAM? antiterminator creater, CDS is transcribed, CDS contains same gene → Low SAM makes more SAM
35
New cards
riboswitches and antitermination T box
Tbox riboswitch
CDS encode tRNA synthetase to charge uncharged-tRNAs
when charged tRNA is present in the cell, does not bind UTR and terminator terminates sequence
when uncharged tRNA present in the cell, base pairs with in UTR and disrupts the terminator, antitermination, CDS made
a charged tRNA cannot disable the terminator, it can only work with uncharged tRNA (the AA gets in the way)
\
CDS encode tRNA synthetase to charge uncharged-tRNAs
when charged tRNA is present in the cell, does not bind UTR and terminator terminates sequence
when uncharged tRNA present in the cell, base pairs with in UTR and disrupts the terminator, antitermination, CDS made
a charged tRNA cannot disable the terminator, it can only work with uncharged tRNA (the AA gets in the way)
\
36
New cards
RNA thermosensors
translation regulation
cis acting
full length RNA transcript is made
RNA temperature sensitive
\
at low temp, RNA adopts a stable secondary structure
ribosome binding site (RBS = Shine Dalgarno) is masked or sequesteered
ribosome cannot get access to the mRNA → protein is not translated
\
at high temperature RNA secondary structure charges or destabilized
RBS no longer sequestered
binding of ribosome → translation
cis acting
full length RNA transcript is made
RNA temperature sensitive
\
at low temp, RNA adopts a stable secondary structure
ribosome binding site (RBS = Shine Dalgarno) is masked or sequesteered
ribosome cannot get access to the mRNA → protein is not translated
\
at high temperature RNA secondary structure charges or destabilized
RBS no longer sequestered
binding of ribosome → translation
37
New cards
trans-acting RNAs
mech of action 2
mech of action 2
very heterogenous group
2 main mechanisms of action
1. sRNAs that bind to RNA (usually by base pairing)
2. sRNAs that bind to proteins
2 main mechanisms of action
1. sRNAs that bind to RNA (usually by base pairing)
2. sRNAs that bind to proteins
38
New cards
sRNAs that bind to RNA
typically a method of post-transcriptional regulation
* no translation: sRNA binds RBS, ribosome cannot bind, = no translation
* mRNA degration: sRNA binds RBS, dsRNA made, body assumes it is a virus and degrades the RNA
* translation occurs: mRNA forms secondary structure where RBS is blocked, sRNA binds and releases the base pairing reaction and free’s the RBS? = translation
* no translation: sRNA binds RBS, ribosome cannot bind, = no translation
* mRNA degration: sRNA binds RBS, dsRNA made, body assumes it is a virus and degrades the RNA
* translation occurs: mRNA forms secondary structure where RBS is blocked, sRNA binds and releases the base pairing reaction and free’s the RBS? = translation
39
New cards
sRNAs that bind to proteins - Csr
CsrA is a translation repressor protein
binds to the RBS of glgCAP mRNA
prevents translation
crsB is a sRNA containing multiple CsrA binding sites
csrB can titrate CsrA away from glgCAP allowing translation to occur
binds to the RBS of glgCAP mRNA
prevents translation
crsB is a sRNA containing multiple CsrA binding sites
csrB can titrate CsrA away from glgCAP allowing translation to occur
40
New cards
quorum sensing
other name
other name
aka population density dependent gene regulation
cell to cell signaling mechanism that enables bacteria to collectively control gene expression
this type of bacterial communication is achieved only at **higher cell densities**
at low densities bacteria release molecules called __**autoinducers**__ into the extracellular medium
when the bacteria reach a high density (**quorum**) the concentration of autoinducers exceeds a particular threshold
autoinducer is transported back into the cell and activates expression of a particular set of genes
genes responsible for virulence, competence, light production
cell to cell signaling mechanism that enables bacteria to collectively control gene expression
this type of bacterial communication is achieved only at **higher cell densities**
at low densities bacteria release molecules called __**autoinducers**__ into the extracellular medium
when the bacteria reach a high density (**quorum**) the concentration of autoinducers exceeds a particular threshold
autoinducer is transported back into the cell and activates expression of a particular set of genes
genes responsible for virulence, competence, light production
41
New cards
quorum sensing controlled processes 5
bioluminescence
biofilm formation
sporulation
competence
virulence gene expression
biofilm formation
sporulation
competence
virulence gene expression
42
New cards
quorum sensing molecules
2 major types of molecules
\
N-acyl-homoserine lactones (AHLs) Gram-negative
\
Autoinducer peptides (AIPs) Gram-positive
\
N-acyl-homoserine lactones (AHLs) Gram-negative
\
Autoinducer peptides (AIPs) Gram-positive
43
New cards
N-acyl-homoserine lactones
synthesized by
recognized by
what does it do
what changes
synthesized by
recognized by
what does it do
what changes
mediates quorum sensing in G-
several types depending on their length of acyl side chain
able to diffuse through membrane
synthesized by autoinducer synthase = LuxI
recognized by autoinducer receptor/DNA binding transcriptional activator protein = LuxR
several types depending on their length of acyl side chain
able to diffuse through membrane
synthesized by autoinducer synthase = LuxI
recognized by autoinducer receptor/DNA binding transcriptional activator protein = LuxR
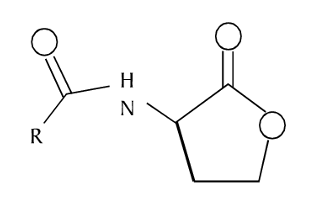
44
New cards
autoinducer peptides
often involves?
recognized by
regulates processes 3
shape
often involves?
recognized by
regulates processes 3
shape
G+ use small peptides as quorum sensing molecules
signaling often involves TCS
peptide is recognized by membrane bound histidine kinase
regulates processes such as: competence, sporulation, virulent gene expression
may be linear or cyclic peptides
signaling often involves TCS
peptide is recognized by membrane bound histidine kinase
regulates processes such as: competence, sporulation, virulent gene expression
may be linear or cyclic peptides
45
New cards
restriction enzymes
recognize and cut DNA at specific recognition sites
type 2 RE cleave DNA at/around the site
cleavage may produce sticky ends or blunt ends in the DNA target
hundreds are commercially available
very useful in cloning
most commercially available restriction endonucleases were identified in bacteria
restrictions enzymes are named based on the organism in which they were discovered
HindIII: isolated from *Haemophilus influenzaeI* strain Rd
old formatting: *Hin*d III
new formatting: HindIII
type 2 RE cleave DNA at/around the site
cleavage may produce sticky ends or blunt ends in the DNA target
hundreds are commercially available
very useful in cloning
most commercially available restriction endonucleases were identified in bacteria
restrictions enzymes are named based on the organism in which they were discovered
HindIII: isolated from *Haemophilus influenzaeI* strain Rd
old formatting: *Hin*d III
new formatting: HindIII
46
New cards
what prevents restriction enzymes from cutting u the host bacteria DNA
methylation via methylases
47
New cards
restriction-modification systems
restriction endonucleases are paired with a second enzyme - methylase
methylase recognizes the same DNA site as the restriction endonucleases
after DNA replication methylase adds a methyl group to the DNA
together the two enzymes represent a Restriction-Modification system
methylated DNA will NOT be recognized and cut by the corresponding restriction endonuclease
Restriction-Modification systems can help protect the bacterial cell from foreign DNA
methylase recognizes the same DNA site as the restriction endonucleases
after DNA replication methylase adds a methyl group to the DNA
together the two enzymes represent a Restriction-Modification system
methylated DNA will NOT be recognized and cut by the corresponding restriction endonuclease
Restriction-Modification systems can help protect the bacterial cell from foreign DNA
48
New cards
how does restriction-modification systems protect bacterial call from foreign DNA?
host DNA is methylated by methylase
foreign DNA is not methylated
restriction enzymes cut up the foreign DNA and kills it
host DNA is protected
foreign DNA is not methylated
restriction enzymes cut up the foreign DNA and kills it
host DNA is protected
49
New cards
CRISPR origins
name
name
originally identified as “phage-like” repetitive DNA sequences on bacterial genomes
unknown function
widespread, found in bacterial and archaeal genomes
NAME: Clustered Regularly Interspaced Short Pallindromic Repeats = CRISPR
unknown function
widespread, found in bacterial and archaeal genomes
NAME: Clustered Regularly Interspaced Short Pallindromic Repeats = CRISPR
50
New cards
CRISPR structure
consists of
1. direct repeats, varying in size from 21-37 bp
2. interspaced by similarly sized non-reptitive sequences (spacers)
repeats are pallindromic repeats: DNA sequence that reads the same forward and backward, can create hairpins
family of homologous genes were found associated with CRISPR
CRISPR associated genes (Cas)
number of Cas genes can vary
1. direct repeats, varying in size from 21-37 bp
2. interspaced by similarly sized non-reptitive sequences (spacers)
repeats are pallindromic repeats: DNA sequence that reads the same forward and backward, can create hairpins
family of homologous genes were found associated with CRISPR
CRISPR associated genes (Cas)
number of Cas genes can vary
51
New cards
CRISPR Cas array
Cas gene(s) + Repeats + spacers
52
New cards
CRISPR yogurt origins
researchers at DANISCO were investing practical questions in yogurt fermentation industry
phage contamination: most serious problem in fermentation industries
phage-resistant strains would emerge after phage pandemics
after viral challenge, bacteria integrated new spacers derived from phage genomic sequences, CRISPR + Cas provided resistance against phages
phage contamination: most serious problem in fermentation industries
phage-resistant strains would emerge after phage pandemics
after viral challenge, bacteria integrated new spacers derived from phage genomic sequences, CRISPR + Cas provided resistance against phages
53
New cards
CRISPR steps
acquisition: spacer acquisition
expression: CRISPR expression crRNA biogenesis
interference: or immunity
expression: CRISPR expression crRNA biogenesis
interference: or immunity
54
New cards
adaptation
invasion of the cell by foreign genetic elements (bacteriophages or plasmids)
spacers: corresponding to foreign DNA sequences are acquired and inserted into the CRISPR locus between repeat sequences
if the bacterial cell survives infection with bacteriophage it will recognize that phage in the future and be resistant
spacers: corresponding to foreign DNA sequences are acquired and inserted into the CRISPR locus between repeat sequences
if the bacterial cell survives infection with bacteriophage it will recognize that phage in the future and be resistant
55
New cards
CRISPR expression
CRISPR repeat-spacer arrays are transcribed into long primary transcripts
precursor CRISPR RNA = pre-crRNA
subsquently processed into a set of short crRNAs
each crRNA contains a single spacer and a repeat fragment
\
3 major types of CRISPR
type 1 and 3 are similar, type 2 is different
(end products are the same, mechanisms slightly different)
precursor CRISPR RNA = pre-crRNA
subsquently processed into a set of short crRNAs
each crRNA contains a single spacer and a repeat fragment
\
3 major types of CRISPR
type 1 and 3 are similar, type 2 is different
(end products are the same, mechanisms slightly different)
56
New cards
type 1 CRISPR + mech
CRISPR array: multiple Cas genes + repeats + spacer
\
promoter generates long pre-crRNA transcript, processed into individual crRNA’s (broken into 1 spacer + 1 repeat), palindromes form hairpins,
Cas genes encode a nuclease - an enzyme complex that cuts DNA, combines with crRNA, crRNA programs Cas nuclease to cut spacers, spacers come from viruses, so when virus infects again the spacer+nuclease can bind to viral DNA and cut + destroy virus
\
promoter generates long pre-crRNA transcript, processed into individual crRNA’s (broken into 1 spacer + 1 repeat), palindromes form hairpins,
Cas genes encode a nuclease - an enzyme complex that cuts DNA, combines with crRNA, crRNA programs Cas nuclease to cut spacers, spacers come from viruses, so when virus infects again the spacer+nuclease can bind to viral DNA and cut + destroy virus
57
New cards
type 2 CRISPR
CRISPR array: tracer RNA gene + 1 Cas gene + repeats + spacers
there is ONE Cas gene: nuclease
contains tracer gene
transcribes genes → tracer gene, pre-crRNA, Cas gene
pre-crRNA → crRNA into single spacer/repeat pieces
tracer + crRNA = guide RNA
guide RNA binds to Cas to program it
tracrRNA binds repeat
there is ONE Cas gene: nuclease
contains tracer gene
transcribes genes → tracer gene, pre-crRNA, Cas gene
pre-crRNA → crRNA into single spacer/repeat pieces
tracer + crRNA = guide RNA
guide RNA binds to Cas to program it
tracrRNA binds repeat
58
New cards
CRISPR interference
crRNAs bond with Cas proteins to form an effector complex
recognizes the target sequence in the invasive nucleic acid by base pairing
induces sequence-specific cleavage and degradation of foreign DNA
prevents proliferation and propagation of foreign genetic elements
recognizes the target sequence in the invasive nucleic acid by base pairing
induces sequence-specific cleavage and degradation of foreign DNA
prevents proliferation and propagation of foreign genetic elements
59
New cards
type 1 vs 2 CRISPR
type 1:
* only 1 RNA = crRNA
* Cas nuclease typically many protein subunits
type 2:
* 2 RNAs: tracrRNA + crRNA = guide RNA
* Cas nuclease typically consists of one protein (most common is Cas9)
* only 1 RNA = crRNA
* Cas nuclease typically many protein subunits
type 2:
* 2 RNAs: tracrRNA + crRNA = guide RNA
* Cas nuclease typically consists of one protein (most common is Cas9)
60
New cards
in developing the CRISPR-Cas9 system
bacterial vs simplified
bacterial vs simplified
bacterial type 2 CRISPR:
* 3 components: 2x sRNAs + 1 protein (Cas9)
* bacterial defense system against infection
simplified type 2 CRISPR:
* 1 sRNA (single guide RNA)
* 1 protein Cas9
* humans use bc its simple, easy system and very programmable
* 3 components: 2x sRNAs + 1 protein (Cas9)
* bacterial defense system against infection
simplified type 2 CRISPR:
* 1 sRNA (single guide RNA)
* 1 protein Cas9
* humans use bc its simple, easy system and very programmable
61
New cards
uses of CRISPR-Cas9 system
inserts, deletions/ knockouts, change the gene sequence,
gene is disrupted
gene has new sequence
increase gene expression: attached to an activator protein which activates gene expression = visualization
gene is disrupted
gene has new sequence
increase gene expression: attached to an activator protein which activates gene expression = visualization
62
New cards
why CRISPR good?
precision and control
63
New cards
CRISPR history, nobel
identified in 1987, 2007 first experimental evidence,
2020 Nobel Prize in Chemistry Emmanuelle Charpentier and Jennifer Doudna
2020 Nobel Prize in Chemistry Emmanuelle Charpentier and Jennifer Doudna
64
New cards
restriction modification systems and CRISPR-Cas used for?
restriction modification systems: bacterial innate immunity
CRISPR-Cas: bacterial adaptive immunity
CRISPR-Cas: bacterial adaptive immunity
65
New cards
disinfectant, antiseptic, antibiotic
antimicrobial compounds applied to inanimate objects and surfaces
\
antimicrobial compounds applied to the skin/outside the body
\
antimicrobial compounds that can be taken inside the body that either inhibits or kills microorganisms
\
antimicrobial compounds applied to the skin/outside the body
\
antimicrobial compounds that can be taken inside the body that either inhibits or kills microorganisms
66
New cards
disinfectants types and mode of killing 7
alcohols: denature proteins
alkylating agents: form epoxide bridges that inactive proteins
halides: oxidizing agents
heavy metals: bind SH denature proteins
phenols: denature proteins
QACs: disrupt cell membranes
UV radiation: blocks DNA replication
alkylating agents: form epoxide bridges that inactive proteins
halides: oxidizing agents
heavy metals: bind SH denature proteins
phenols: denature proteins
QACs: disrupt cell membranes
UV radiation: blocks DNA replication
67
New cards
some features of antimicrobial drugs 5
selective toxicity
antimicrobial action
spectrum of activity
pharmacokinetic properties
adverse effects
antimicrobial action
spectrum of activity
pharmacokinetic properties
adverse effects
68
New cards
selective toxicity
how to select - 2
how to select - 2
= efficacy vs toxicity - how good is it at killing vs huch will it harm human host
cause greater harm to microorganisms than to host
how can we select for this?
1. antibiotic target is ONLY found in the bacteria
2. antibiotic target in the bacteria is sufficiently different from homologue in humans
cause greater harm to microorganisms than to host
how can we select for this?
1. antibiotic target is ONLY found in the bacteria
2. antibiotic target in the bacteria is sufficiently different from homologue in humans
69
New cards
antimicrobial action
bactericidal: kill bacteria
bacteriostatic: slow down or inhibit growth of bacteria
bacteriostatic: slow down or inhibit growth of bacteria
70
New cards
spectrum of activity
antibiotics vary with respect to the range of bacteria they kill or inhibit
broad-spectrum antibiotic: active against a wide range of bacteria, both Gram-positive __*and*__ Gram-negative
narrow-spectrum antibiotic: limited range, may only be effective against either Gram-positive __*or*__ Gram-negative bacteria
broad-spectrum antibiotic: active against a wide range of bacteria, both Gram-positive __*and*__ Gram-negative
narrow-spectrum antibiotic: limited range, may only be effective against either Gram-positive __*or*__ Gram-negative bacteria
71
New cards
pharmacokinetic properties
pharmacokinetics: examines the fate of a drug from the moment that it is administered up to the point at which it is completely eliminated from the body
\
\
important parameters are:
* absorption - The process of a substance entering the blood circulation
* distribution - The dispersion or dissemination of substances throughout the fluids and tissues of the body
* metabolism - The drug is recognized by the body and broken down
* excretion - The removal of the substances from the body
\
bioavailability – How much of the administered dose gets into the system
\
\
important parameters are:
* absorption - The process of a substance entering the blood circulation
* distribution - The dispersion or dissemination of substances throughout the fluids and tissues of the body
* metabolism - The drug is recognized by the body and broken down
* excretion - The removal of the substances from the body
\
bioavailability – How much of the administered dose gets into the system
72
New cards
adverse effects
allergic reactions: some people develop hypersensitives to antimicrobials
suppression of normal microbiota: when normal microbiota are killed other pathogens may be able to grow to high numbers (cdiff)
suppression of normal microbiota: when normal microbiota are killed other pathogens may be able to grow to high numbers (cdiff)
73
New cards
antibiotic development timeline
antibiotic drug discovery
types of AB drugs
antibiotic drug discovery
types of AB drugs
10+ years, $50 million dollars
less antibiotics being created bc money
natural products, semisynthetic compounds, synthetic compounds
less antibiotics being created bc money
natural products, semisynthetic compounds, synthetic compounds
74
New cards
natural products
by definition antibiotics are compounds __produced by microbes__ that kill or inhibit growth of other microbes
most antibiotics come from soil microbes (bacteria)
antibiotics are compounds produced by microorganisms typically as **secondary metabolites** or as a response to stress = antibiotics
most antibiotics come from soil microbes (bacteria)
antibiotics are compounds produced by microorganisms typically as **secondary metabolites** or as a response to stress = antibiotics
75
New cards
Why do microorganisms produce compounds that kill other microorganisms?
must compete with other microbes for habitat
76
New cards
soil microbes?
less than 1% can be grown in lab
scientists grew unculturable bacteria in agar in the soil
scientists grew unculturable bacteria in agar in the soil
77
New cards
semisynthetic compounds
chemical modification of naturally occurring antibiotics leads to semisynthetic antibiotics
penicillin G and V = natural
ampicillin, amoxicillin, methicillin = semisynthetic
penicillin G and V = natural
ampicillin, amoxicillin, methicillin = semisynthetic
78
New cards
synthetic compounds
synthetic antibiotics are compounds that are __**not found in nature**__
they are completely synthesized in a lab
how many chemical structures are possible?
two major pathways in synthetic drug design:
1. combinatorial chemistry
2. rational drug design
they are completely synthesized in a lab
how many chemical structures are possible?
two major pathways in synthetic drug design:
1. combinatorial chemistry
2. rational drug design
79
New cards
combinatorial chemistry
begin with a core compound
synthesize thousands of derivatives by adding groups at specific positions (e.g. R1 and R2)
combine different compounds together in groups (1,000’s of compounds in each group)
pool 1,000’s of compounds together
test pooled compounds for antibacterial activity
when activity is detected determine which compound is responsible by __**deconvolution**__
breaking large group into smaller groups and re-testing
HTS – High Throughput Screening
(creating sub-libraries to search for activity)
synthesize thousands of derivatives by adding groups at specific positions (e.g. R1 and R2)
combine different compounds together in groups (1,000’s of compounds in each group)
pool 1,000’s of compounds together
test pooled compounds for antibacterial activity
when activity is detected determine which compound is responsible by __**deconvolution**__
breaking large group into smaller groups and re-testing
HTS – High Throughput Screening
(creating sub-libraries to search for activity)
80
New cards
rational drug design
rational drug design is a targeted approach to antibiotic development
select a specific bacterial target (typically one that is required for survival/infection)
using a variety of approaches design a molecule that will inhibit that target
depends heavily on computer based modeling of target:inhibitor
HTS also used to optimize compounds
(doesn’t work as well as we hoped)
select a specific bacterial target (typically one that is required for survival/infection)
using a variety of approaches design a molecule that will inhibit that target
depends heavily on computer based modeling of target:inhibitor
HTS also used to optimize compounds
(doesn’t work as well as we hoped)
81
New cards
11 major classes of antibiotics
β-lactams
Glycopeptides
Aminoglycosides
Tetracyclines
Macrolides/lincosamides
Streptogramins
Fluoroquinolones
Rifampin
Trimethoprim/sulfonamides
Metronidazole
Oxazolidinones
Glycopeptides
Aminoglycosides
Tetracyclines
Macrolides/lincosamides
Streptogramins
Fluoroquinolones
Rifampin
Trimethoprim/sulfonamides
Metronidazole
Oxazolidinones
82
New cards
antibiotic targets 4
cell wall
protein synthesis
DNA/RNA
tetrahydrofolic acid biosynthesis
protein synthesis
DNA/RNA
tetrahydrofolic acid biosynthesis
83
New cards
cell wall synthesis inhibitors
1 AB: 4 subgroups
1 AB: 4 subgroups
β-lactam antibiotics
* penicillins
* cephalosporins
* carbapenems
* monobactams
kill bacteria by inhibiting the last step in peptidoglycan biosynthesis
transpeptidation: crosslinking peptide side chains
carried out by penicillin binding proteins = transpeptidase
PBP’s inhibited by beta-lactam antibiotics
(PBP’s crosslink peptidoglycan)
* penicillins
* cephalosporins
* carbapenems
* monobactams
kill bacteria by inhibiting the last step in peptidoglycan biosynthesis
transpeptidation: crosslinking peptide side chains
carried out by penicillin binding proteins = transpeptidase
PBP’s inhibited by beta-lactam antibiotics
(PBP’s crosslink peptidoglycan)
84
New cards
protein synthesis inhibitors 5
euk ribosome v pro
euk ribosome v pro
ribosomes in pro are very different that euk
\
eukaryotes – __80S Ribosome__
40S -Subunit
60S –Subunit
\
prokaryotes – __70S Ribosome__
30S -Subunit
50S -Subunit
\
pro ribosome is sufficiently different than eukaryotic, compounds that inhibit prokaryotic ribosome do not inhibit eukaryotic (or do so poorly)
makes prokaryotic ribosome an attractive target
\
types:
* aminoglycosides - block 30
* tetracycline - block 30
* macrolides - block 50
* azithromycin
* lincosimides - block 50
\
eukaryotes – __80S Ribosome__
40S -Subunit
60S –Subunit
\
prokaryotes – __70S Ribosome__
30S -Subunit
50S -Subunit
\
pro ribosome is sufficiently different than eukaryotic, compounds that inhibit prokaryotic ribosome do not inhibit eukaryotic (or do so poorly)
makes prokaryotic ribosome an attractive target
\
types:
* aminoglycosides - block 30
* tetracycline - block 30
* macrolides - block 50
* azithromycin
* lincosimides - block 50
85
New cards
aminoglycosides
structure
functional against
function
3 examples
structure
functional against
function
3 examples
trisaccharide structure
against many Gram- and some Gram+
bind to specific sites in the 30S subunit
block protein synthesis
* gentamicin – Used for acute, life-threatening Gram- infections
* neomycin – Too toxic for internal use. Used topically for skin infections
* streptomycin – Active against Mycobacterium tuberculosis
against many Gram- and some Gram+
bind to specific sites in the 30S subunit
block protein synthesis
* gentamicin – Used for acute, life-threatening Gram- infections
* neomycin – Too toxic for internal use. Used topically for skin infections
* streptomycin – Active against Mycobacterium tuberculosis
86
New cards
tetracyclines
four fused cyclic ring structure
broad-spectrum
bind to specific sites in the 30S subunit
generally bacteriostatic
low toxicity
Tet binds to Ca in growing bones and teeth and can discolor teeth
should be avoided in children < 8 yrs old
broad-spectrum
bind to specific sites in the 30S subunit
generally bacteriostatic
low toxicity
Tet binds to Ca in growing bones and teeth and can discolor teeth
should be avoided in children < 8 yrs old
87
New cards
DNA/RNA synthesis inhibitors
metronidazole: inhibits NA synthesis by disrupting the DNA of microbial cells, interacts with alcohol = projectile vomiting
\
quinolone: nalidixic acid
fluoroquinolones: norfloxacin, ciprofloxacin
both inhibits DNA replication by binding to and inhibiting DNA gyrase after gyrase has introduced double stranded nicks in DNA
\
rifampin: inhibits RNA polymerase by binding to the beta subunit and preventing transcription
\
quinolone: nalidixic acid
fluoroquinolones: norfloxacin, ciprofloxacin
both inhibits DNA replication by binding to and inhibiting DNA gyrase after gyrase has introduced double stranded nicks in DNA
\
rifampin: inhibits RNA polymerase by binding to the beta subunit and preventing transcription
88
New cards
tetrahydrofolic acid inhibitors
tetrahydrofolic acid is an essential co-factor in bacteria
required for synthesis of nucleic acids and formylmethionine (fMet)
bacteria make their own via the FH4 biosynthesis pathway
humans require FH4 but do not make it
makes it an ideal target for antibiotics
* **Sulfonamides**
* **Trimethoprim**
bacteria can make FH4 but not uptake, humans cannot make and must uptake
required for synthesis of nucleic acids and formylmethionine (fMet)
bacteria make their own via the FH4 biosynthesis pathway
humans require FH4 but do not make it
makes it an ideal target for antibiotics
* **Sulfonamides**
* **Trimethoprim**
bacteria can make FH4 but not uptake, humans cannot make and must uptake
89
New cards
resistant
susceptible
\*get wording right
susceptible
\*get wording right
somewhat arbitrary designation that implies that an antimicrobial **WILL NOT** inhibit bacterial growth at clinically achievable concentrations
\
somewhat arbitrary designation that implies that an antimicrobial **WILL** inhibit bacterial growth at clinically achievable concentrations
\
somewhat arbitrary designation that implies that an antimicrobial **WILL** inhibit bacterial growth at clinically achievable concentrations
90
New cards
MIC
MBC
MBEC
MBC
MBEC
MIC: minimal inhibitory concentration, lowest concentration of antimicrobial that completely inhibits growth of bacteria, common in clinical lab (no growth in tube)
\
MBC: minimal bactericidal concentration, concentration of an antimicrobial that completely kills bacteria, used only in special circumstances (no growth on plate)
\
MBEC: minimum biofilm eradication concentration, concentration of an antimicrobial agent required to kill a bacterial biofilm (typically have highest concentration)
\
MBC: minimal bactericidal concentration, concentration of an antimicrobial that completely kills bacteria, used only in special circumstances (no growth on plate)
\
MBEC: minimum biofilm eradication concentration, concentration of an antimicrobial agent required to kill a bacterial biofilm (typically have highest concentration)
91
New cards
antibiotics vs AB resistance
happening forever
increase in resistance with increase in AB use (humans)
bacteria usually become resistant in 3-5 years loosely
increase in resistance with increase in AB use (humans)
bacteria usually become resistant in 3-5 years loosely
92
New cards
antibiotic misuse
recognized for several decades that up to 50% of antimicrobial use is inappropriate
given when they are not needed/at the wrong dose
continued when they are no longer necessary
discontinued prematurely = resistant bacteria grows
broad spectrum agents are used to treat very susceptible bacteria
agricultural misuse: additional of sub-therapeutic levels of antibiotics in livestock feed results in 10% increase in body mass, unknown why, banned in europe, 70-80% antibiotics sold in US used for animal production
given when they are not needed/at the wrong dose
continued when they are no longer necessary
discontinued prematurely = resistant bacteria grows
broad spectrum agents are used to treat very susceptible bacteria
agricultural misuse: additional of sub-therapeutic levels of antibiotics in livestock feed results in 10% increase in body mass, unknown why, banned in europe, 70-80% antibiotics sold in US used for animal production
93
New cards
antimicrobial action
bactericidal: kill bacteria
bacteriostatic: slow down or inhibit growth of bacteria (immune system then kills bacteria)
bacteriostatic: slow down or inhibit growth of bacteria (immune system then kills bacteria)
94
New cards
mechanisms of antibiotic resistance
intrinsic resistance
acquired resistance
acquired resistance
95
New cards
intrinsic resistance
intrinsic resistance is the innate ability of a bacterial species to resist activity of an antibiotic through its inherent structural or functional characteristics
organisms that are intrinsically resistant have __**never been susceptible**__ to that particular drug
* lack of __affinity__ of the drug for the bacterial target
* inaccessibility of the drug into the bacterial cell
* extrusion of the drug by chromosomally encoded active exporters
* innate production of enzymes that inactivate the drug
organisms that are intrinsically resistant have __**never been susceptible**__ to that particular drug
* lack of __affinity__ of the drug for the bacterial target
* inaccessibility of the drug into the bacterial cell
* extrusion of the drug by chromosomally encoded active exporters
* innate production of enzymes that inactivate the drug
96
New cards
organisms, natural resistance against, mechanism
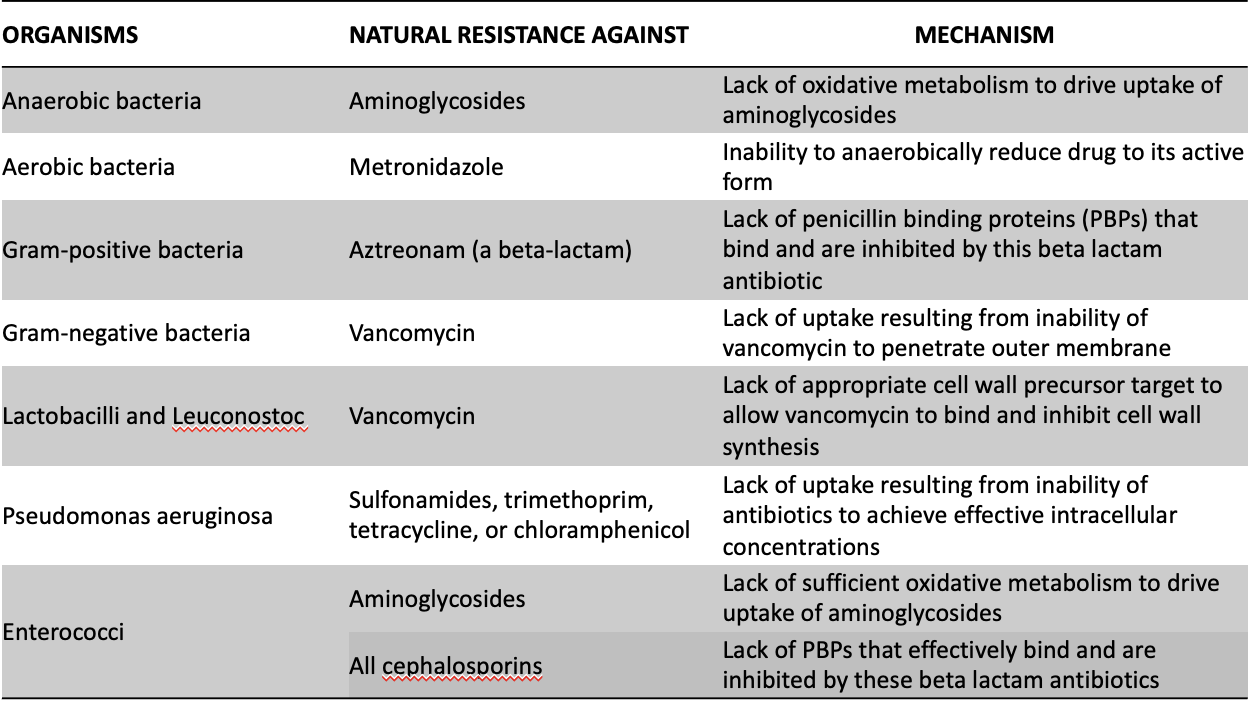
97
New cards
tolerance/ persister cells
persister cells are a small sub-population of dormant bacteria
metabolically inactive
because they are not growing they are able to tolerate antibiotics
not true resistance
not a spore, everything inside cell is shut down
how they survive? AB target protein synthesis/DNA/RNA but cannot affect persister cells bc they are shut down
normally AB would kill persister cells
unknown length of dormancy
always maintain a subset of persister cells, incase a colony gets wiped out the persister cells can regrow when AB leaves
biofilms with persister cells: AB kills all but persister cells, ECM is left behind, persister cells can regrow and regrow biofilm. In the same ECM as usual
metabolically inactive
because they are not growing they are able to tolerate antibiotics
not true resistance
not a spore, everything inside cell is shut down
how they survive? AB target protein synthesis/DNA/RNA but cannot affect persister cells bc they are shut down
normally AB would kill persister cells
unknown length of dormancy
always maintain a subset of persister cells, incase a colony gets wiped out the persister cells can regrow when AB leaves
biofilms with persister cells: AB kills all but persister cells, ECM is left behind, persister cells can regrow and regrow biofilm. In the same ECM as usual
98
New cards
acquired resistance
acquires resistance occurs when a particular microorganism obtains the ability to resist the activity of a particular antimicrobial agent to which it was previously susceptible
this can result from 2 different genetic processes:
1. mutation of genes
2. the acquisition of foreign resistance genes (most common - HGT)
this can result from 2 different genetic processes:
1. mutation of genes
2. the acquisition of foreign resistance genes (most common - HGT)
99
New cards
mutations of genes
random changes in the DNA
100
New cards
acquired resistance
mech of action 3
mech of action 3
acquisition of foreign resistance genes
HGT: resistance bacteria transfer DNA to sensitive bacteria
3 mechanisms of acquired resistance:
1. inactivation of the antibiotic
2. target modification
3. removal of antibiotic
HGT: resistance bacteria transfer DNA to sensitive bacteria
3 mechanisms of acquired resistance:
1. inactivation of the antibiotic
2. target modification
3. removal of antibiotic