11. Generation of Diversity
1/23
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
24 Terms
Could ancient pathogens from permafrost harm humans?
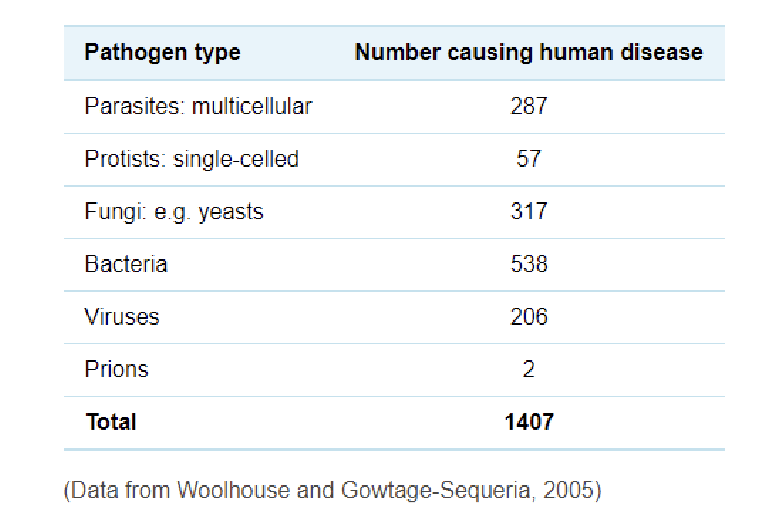
Over 10,000 bacterial species, ~300,000 viruses exist
Only ~1,400 pathogens cause disease in humans
Adaptive immune system can likely recognize any human pathogen
Most microbes (~99.9%) are harmless
How many environmental antigens can the adaptive immune system respond to?
Estimated 10¹² (1 trillion) possible antigens
Adaptive immune system can generate specific responses to all
How does each B cell recognize an antigen?
Each B cell expresses one antibody sequence
Multiple copies per cell
Clones: B cells with identical antibody sequences → arise via cell division
How does the Lego analogy explain antibody diversity?
Few parts → many combinations → lots of diversity
Example: 8 heads × 4 bodies × 2 legs = 64 Lego people with only 14 parts
To make all these people simultaneously you need 64 × 3 = 192 parts
Antibodies: small gene segments combine to make huge diversity without needing 1 trillion genes

What was the Germline Theory (Leroy Hood)?
Proposal: One gene → one antibody
Human genome too small (~19,000 genes, 3 billion nucleotides)
Couldn’t explain 10¹² specificities
Plot twist: We didn’t know genome could change
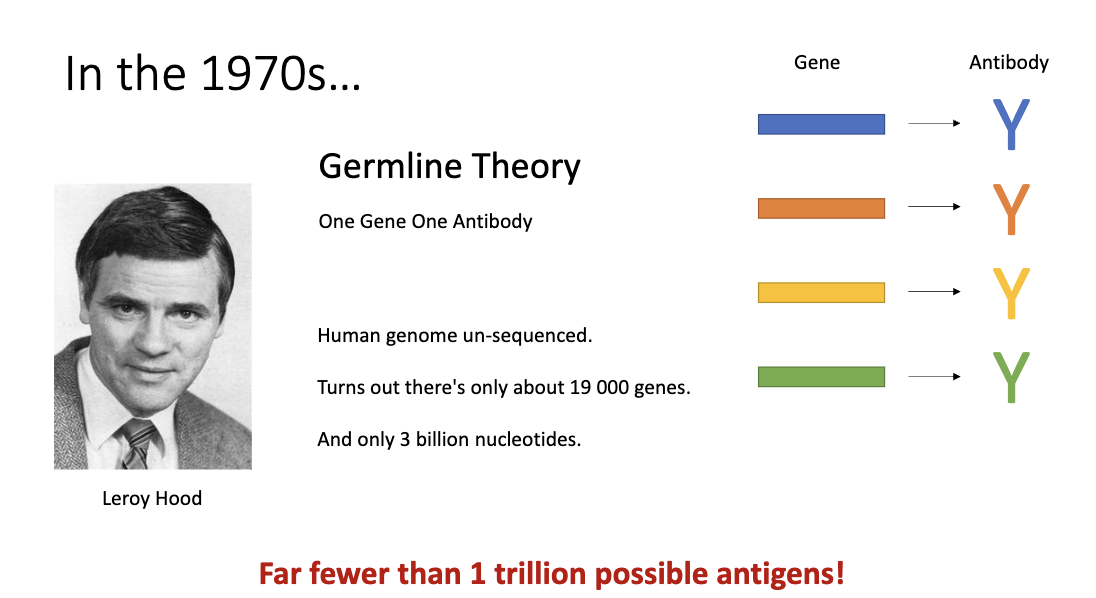
How is huge diversity achieved with a small genome (Melvin Cohn, Susumu Tonegawa)?
Somatic diversification theory: combine smaller gene segments (V, D, J)
Explains enormous antibody diversity with a small genome
However, back then they didn’t think it was possible for cells to rearrange their genes until Tonegawa proved it
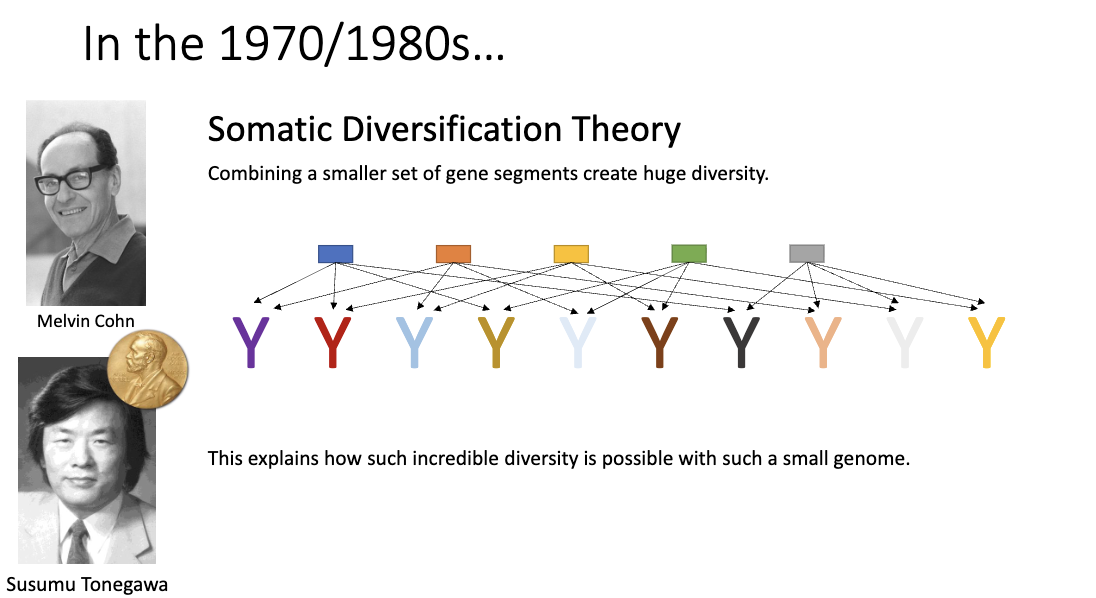
What receptors bind antigens in adaptive immunity and which parts of the receptors bind the antigen?
B cells: B-cell receptor / surface immunoglobulin (has transmembrane region), antibody (circulating)
T cells: T-cell receptor (TCR)
Most of receptor: constant region (defines effector function)
Small variable region: tips are hyper-variable determines antigen specificity
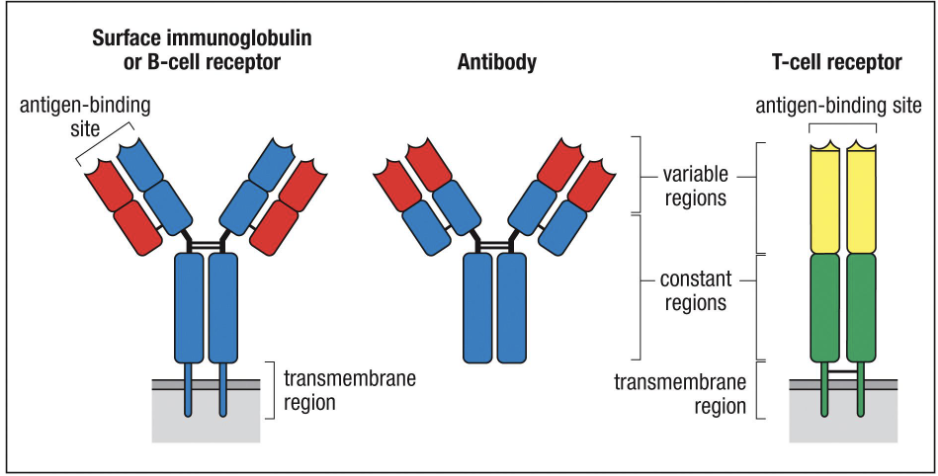
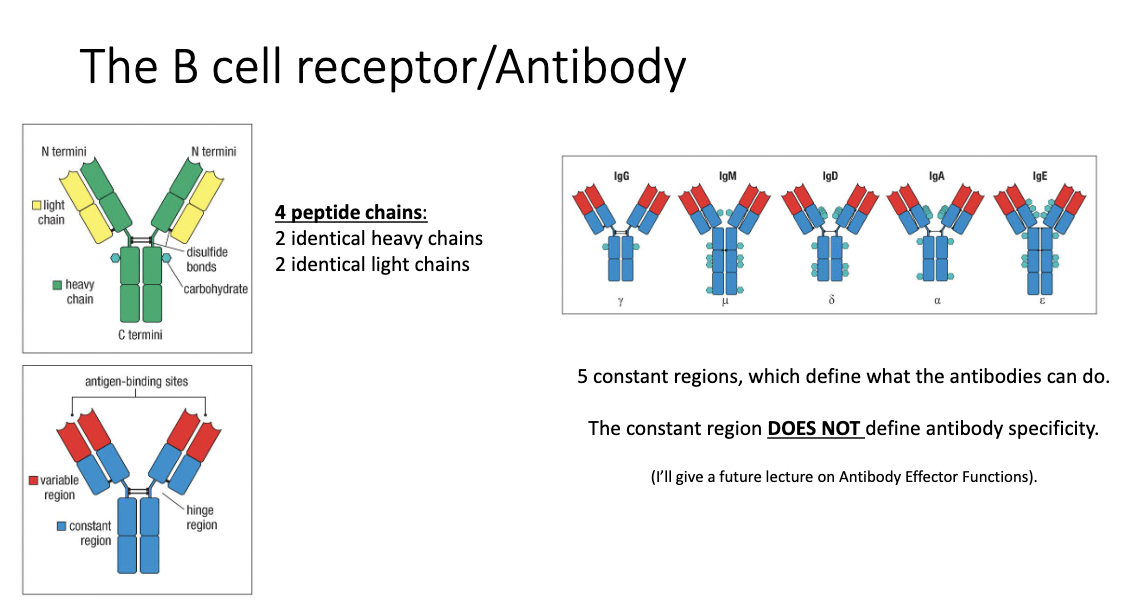
What is the structure of an antibody?
4 peptide chains: 2 identical heavy chains + 2 identical light chains (held by disulphide bonds)
5 constant regions:
Control effector functions (what the antibody can do)
Have different glycan numbers, affecting antibody function
Do NOT determine antigen specificity
Variable region:
Determines antigen specificity (what the antibody binds)
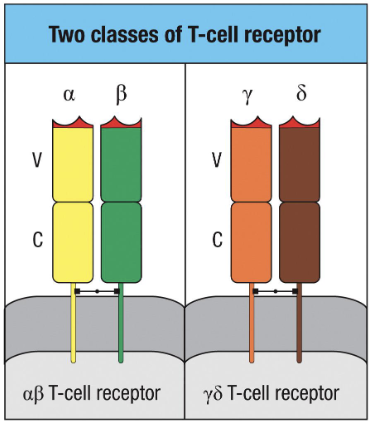
How is a TCR structured and what are the two classes of TCR?
Two chains: αβ or γδ (not identical)
Each chain has variable (V) + constant (C) regions (held by disulphide bonds)
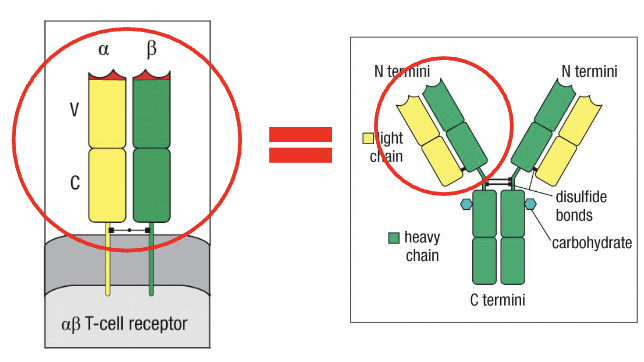
Why focus on variable regions?
B-cell receptors (antibodies) and T-cell receptors (TCRs) are both immunoglobulin (Ig)-like receptors
Antibody vs TCR: Similar overall structure, but antibody has a Y-shape
Variable region:
Binds the antigen
Diversity in this region → determines specificity of antigen recognition
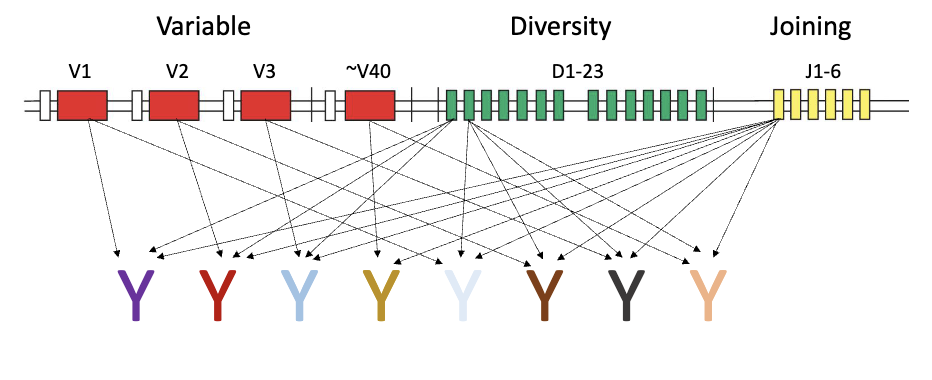
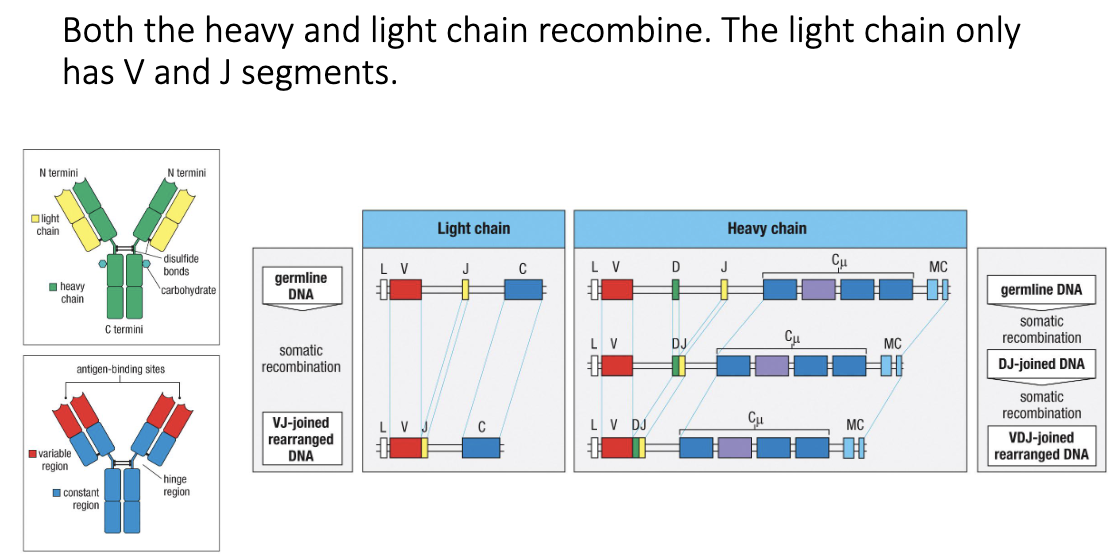
How is receptor diversity generated at the gene level?
Variable region made of 3 segments: V, D, J
Variable (up to 40), Diversity (up to 23), Joining (up to 6)
Heavy chain: V + D + J
Light chain: V + J only
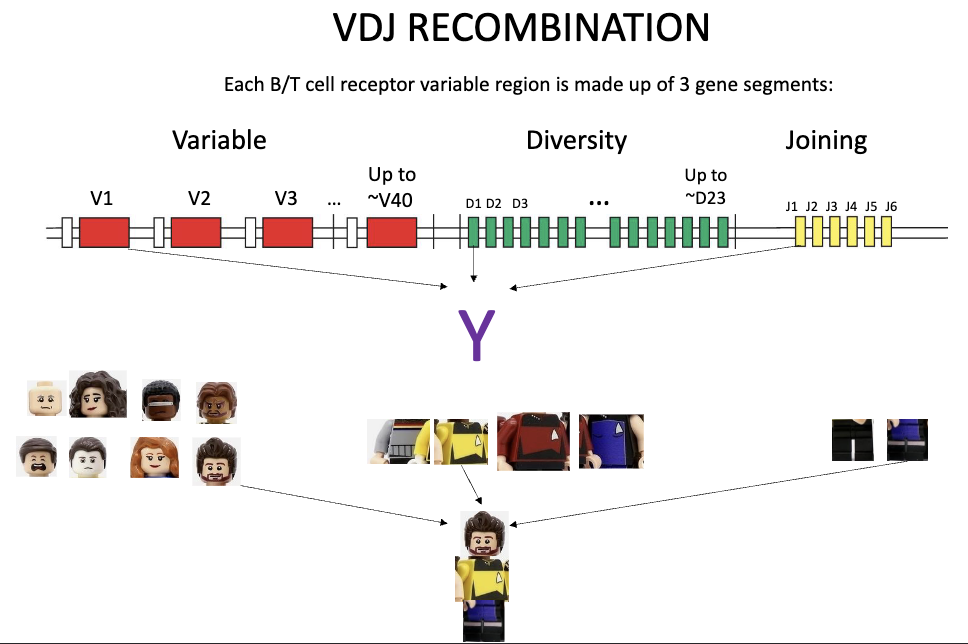
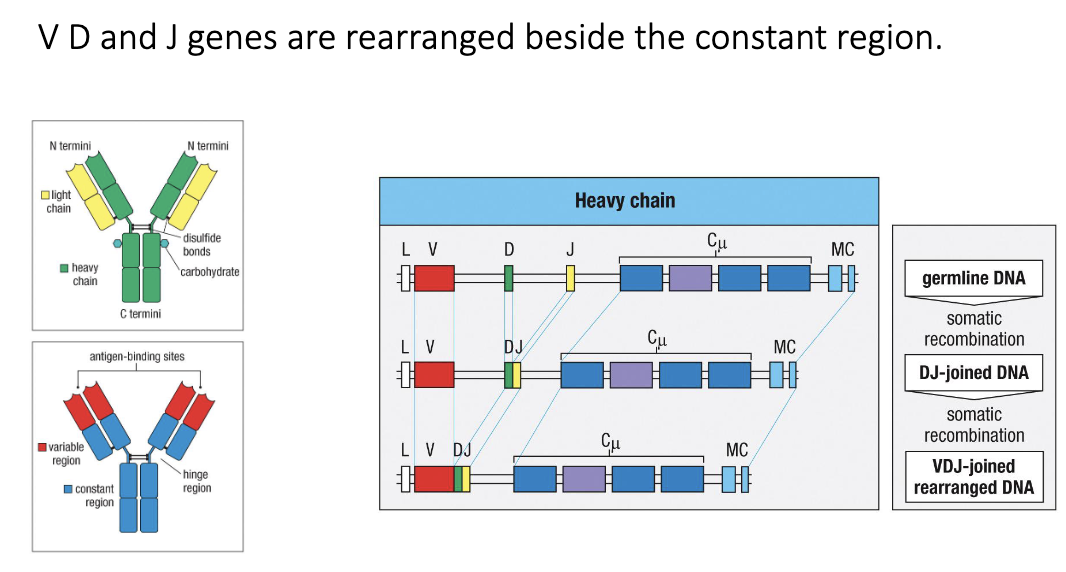
How does recombination occur?
Heavy chain: germline DNA → D+J joined → V+DJ joined → VDJ rearranged DNA sits beside constant region
Generates unique antibody genes
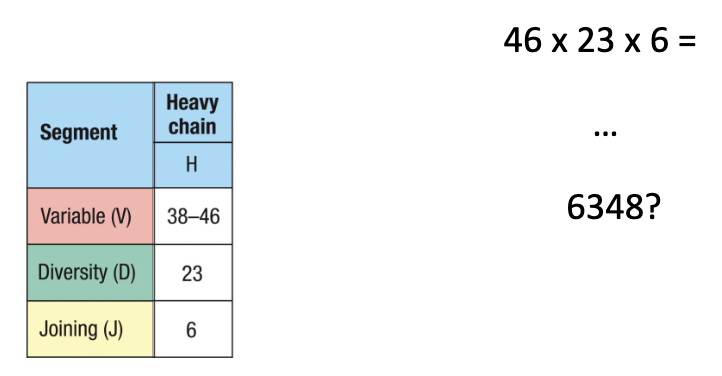
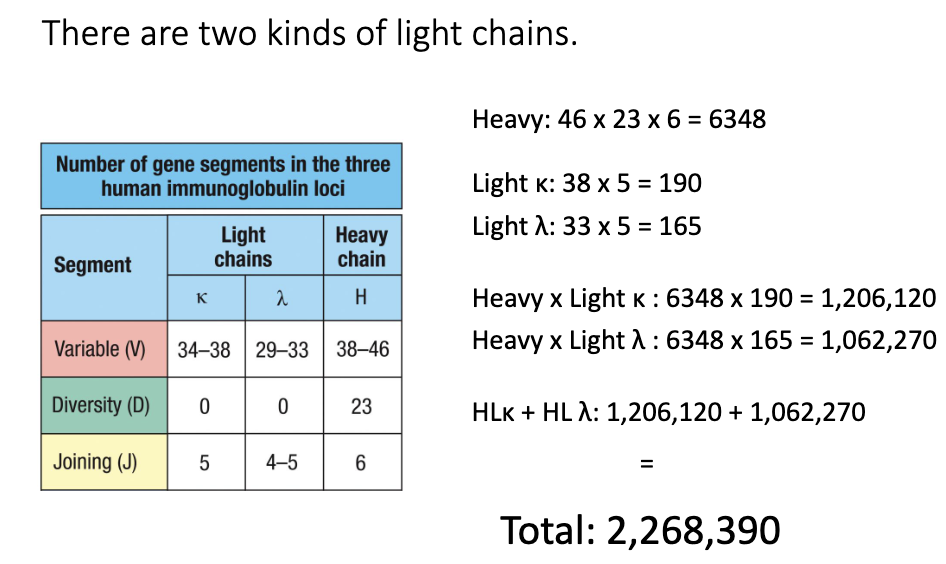
How many heavy chain combinations?
V: 38–46 × D: 23 × J: 6
Total combinations: 38×23×6 = 6,348
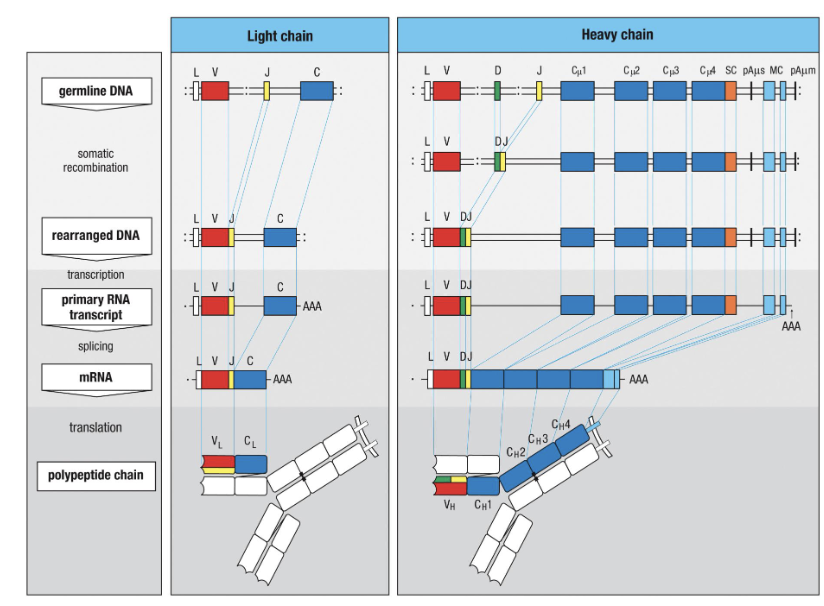
How do light chains contribute?
Light chains only use V + J segments
Two types: κ (kappa) and λ (lambda) light chains
Combine with heavy chains to form the complete antibody
Formation process:
Germline DNA → somatic recombination → rearranged DNA → transcription → primary RNA transcript → splicing → mRNA → translation → polypeptide chainLight blue = transmembrane domain which is removed to release the antibody
Total possible antibody sequences?
Heavy × Light κ = 1,206,120
Heavy × Light λ = 1,062,270
Total: 2,268,390 combinations (without junctional diversity)
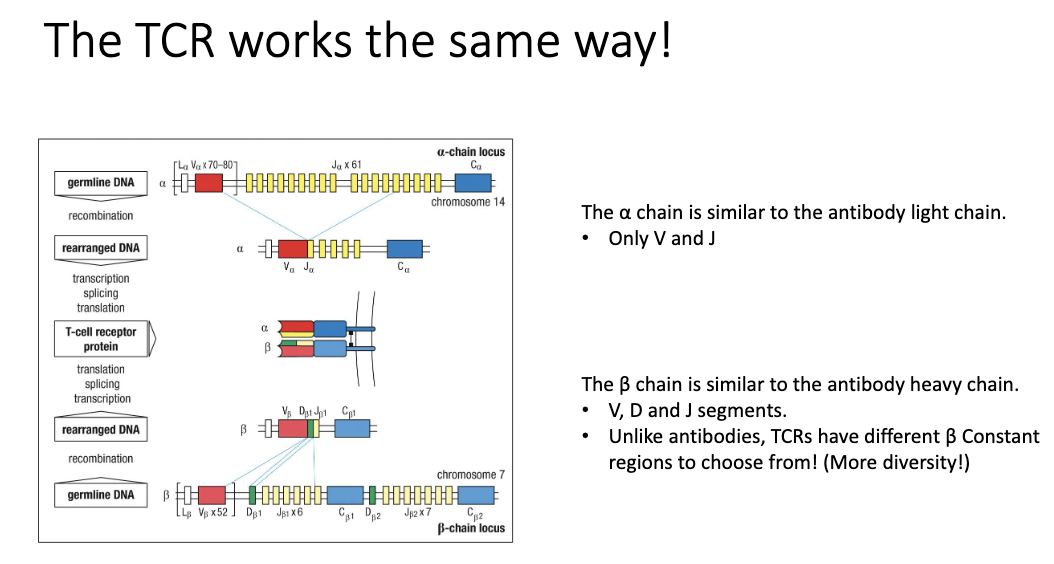
How do TCRs generate diversity?
α chain: like light chain (V+J)
β chain: like heavy chain (V+D+J)
Unlike antibodies, TCRs have multiple β constant regions → even more diversity
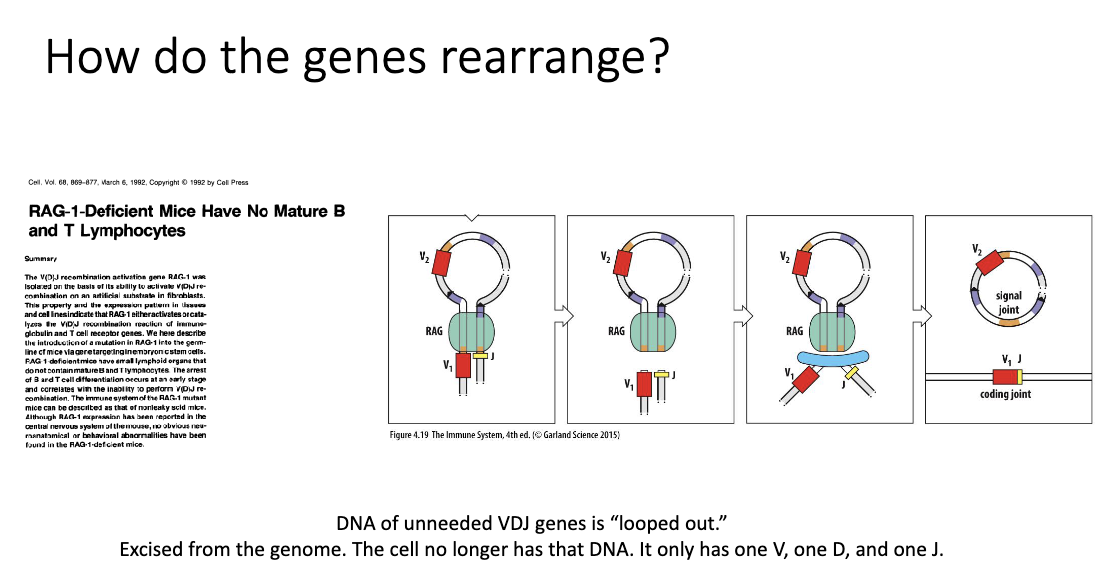
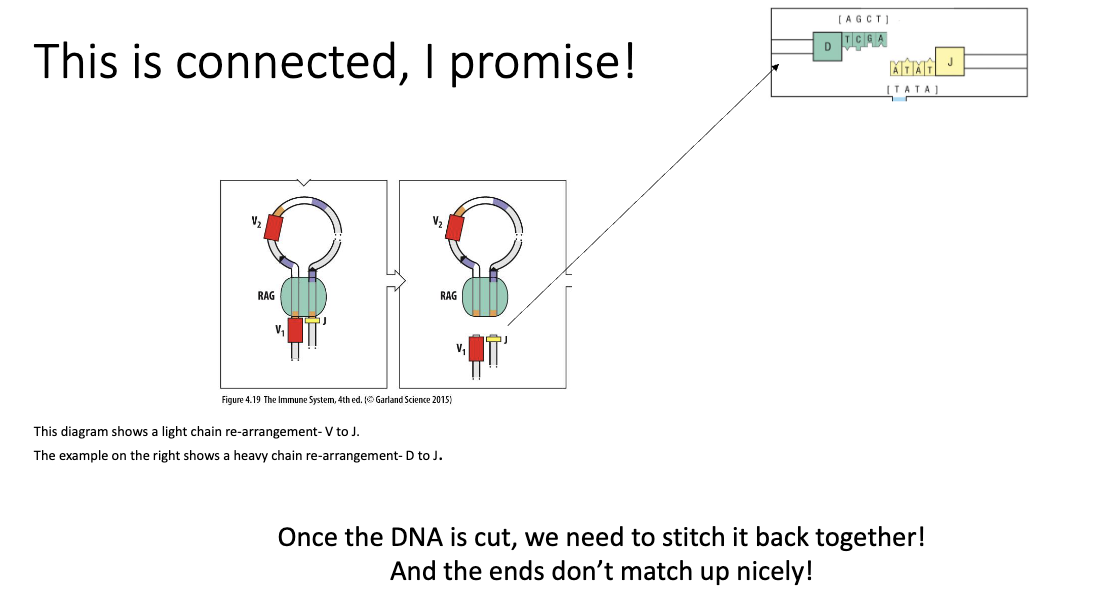
How are unneeded VDJ segments removed and gene rearrangement occur?
RAG-1 enzymes cut DNA → form loops to bring selected V, D, and J segments together
Unneeded DNA is looped out and permanently excised from the genome
During stitching (joining) of V, D, and J → extra diversity added (junctional diversity)
Result: Only one V, D, and J combination remains → creates a unique receptor for each cell
What happens after DNA is cut in VDJ recombination?
DNA ends need to be stitched back together
Random junctions introduce additional diversity
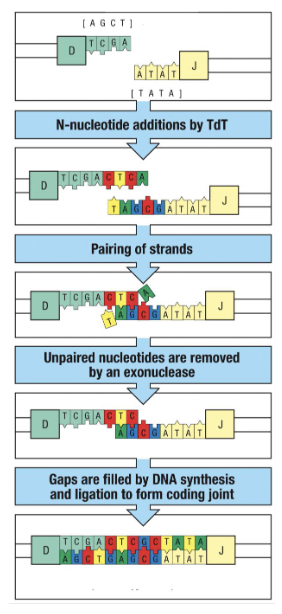
How does junctional diversity increase receptor diversity?
Junctional diversity = extra diversity created during joining of V, D, and J segments
TdT enzyme adds a random number (2–20) of random nucleotides at the junctions
DNA strands anneal, and any unpaired bases are removed by exonuclease
DNA synthesis + ligation fill gaps → form a coding joint
Results:
Added nucleotides → extra amino acids in the variable region
Variable loop lengths (diverse V regions), allowing receptors to reach different antigen shapes
Antibodies with long loops for tight spaces
Short loops to avoid steric hindrance
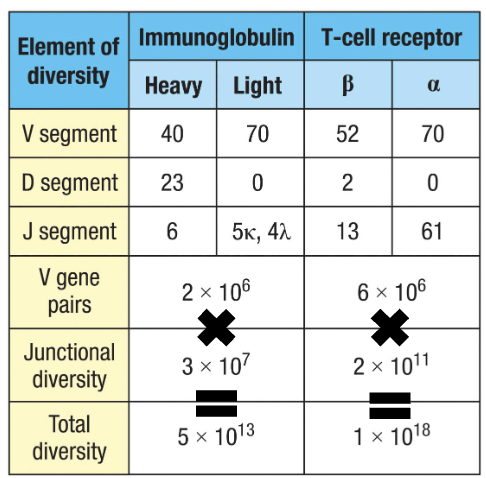
How diverse are B and T cell receptors (with junctional diversity)?
Junctional diversity takes up vast majority of diversity
Heavy chain has junctional diversity twice (D+J) and (V+DJ)
Immunoglobulin (B cell): ~5 × 10¹³
T-cell receptor: ~1 × 10¹⁸
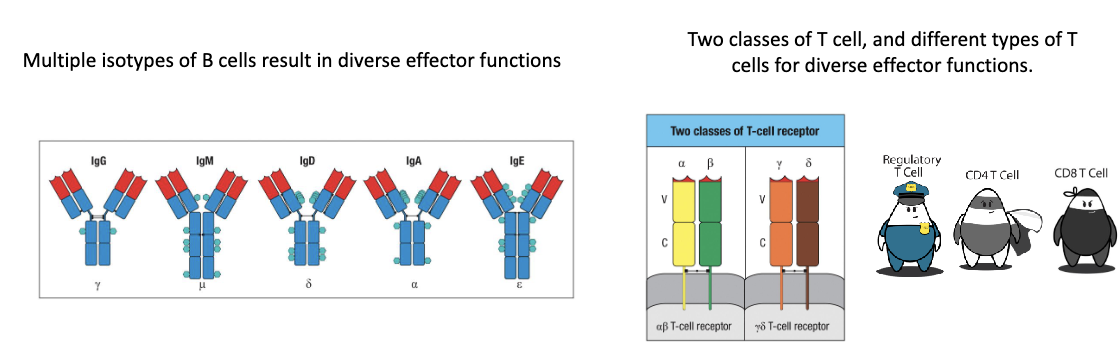
How does diversity continue beyond gene rearrangement (antibody receptor)?
Diversity in response:
Multiple B cell isotypes → different effector functions
Different T cell types → diverse effector functions
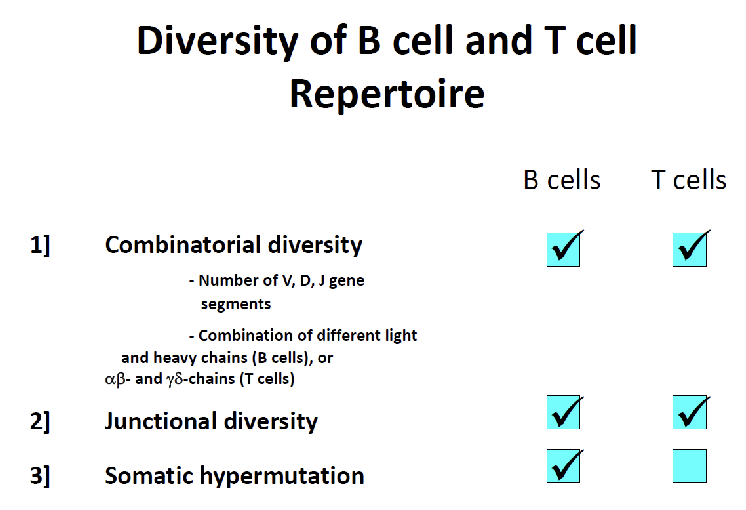
What mechanisms generate receptor diversity?
Combinatorial diversity: different V, D, J segments + chain combinations
Junctional diversity: random nucleotides when joining
Somatic hypermutation: B cells only, after antigen exposure
Do all antibodies bind antigen equally?
High affinity: tight binding, may never dissociate
Moderate/low affinity: loose binding, may fall off
Repulsion: antibody actively pushed off (ex: due to charge repulsion)
Multiple antibodies can recognize the same epitope, but each has a different binding affinity for it.

What is somatic hypermutation and why is it important?
Occurs only in B cells after activation.
The B cell mutates its antibody (BCR) gene DNA to slightly change the variable region.
Some mutations increase affinity for the antigen — those B cells survive and proliferate.
Lower-affinity mutants die off → this creates a Darwinian “survival of the fittest” process inside the body.
Over time, antibodies become higher affinity for the same antigen.
→ VDJ recombination creates initial diversity, but somatic hypermutation refines it for stronger antigen binding.
