Lecture 2: Intracellular Signaling II
1/24
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
25 Terms
What are the 3 types of PKCs?
Conventional PKC: requires coincidental binding of DAG/Ca2+
Novel PKC: only requires DAG binding
Atypical PKC: requires phosphorylation by enzyme PDK1 (all PKCs need)
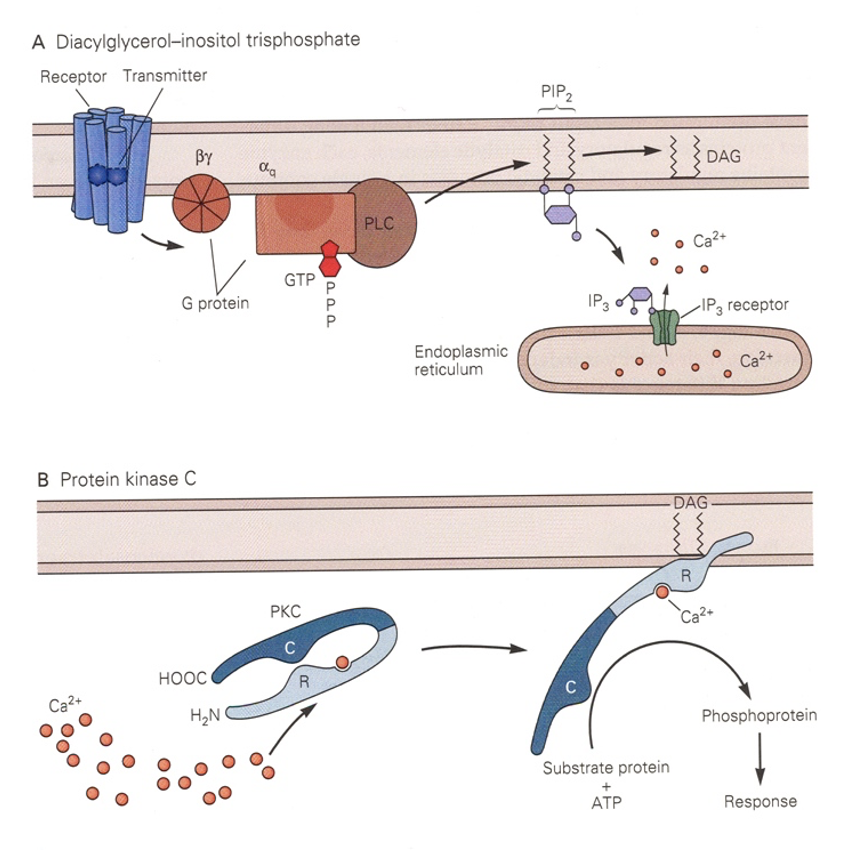
What is special about the DAG binding site?
Phorbol esters bind at DAG binding sites. They produce malignant tumors because they mimic DAG + spur unregulated growth of cells by activating various PKCs.
Why is both DAG and Ca2+ needed by conventional PKC?
Ca2+ binds R and separates it from C → R is free to bind DAG and leaves C free to phosphorylate substrate proteins.
Turns PKC from cytoplasmic protein to membrane-associated one.
What’s the difference between PKCz and PKMz?
PKCz = full-length atypical PKC with R+C activated by phosphorylation via PDK1
PKMz = unregulated, truncated form of PKCz w/ only C
Produced by independent transcription/translatio
Always ON
How do you control PKMz?
Control translation of its mRNA
PKMz also needs PDK1
Once synthesized, PKMz also promotes translation of its own mRNA
Structure of CaMKII?
Assembly of 12 kinase molecules held together (diagram there would be another group of 6 above or below)
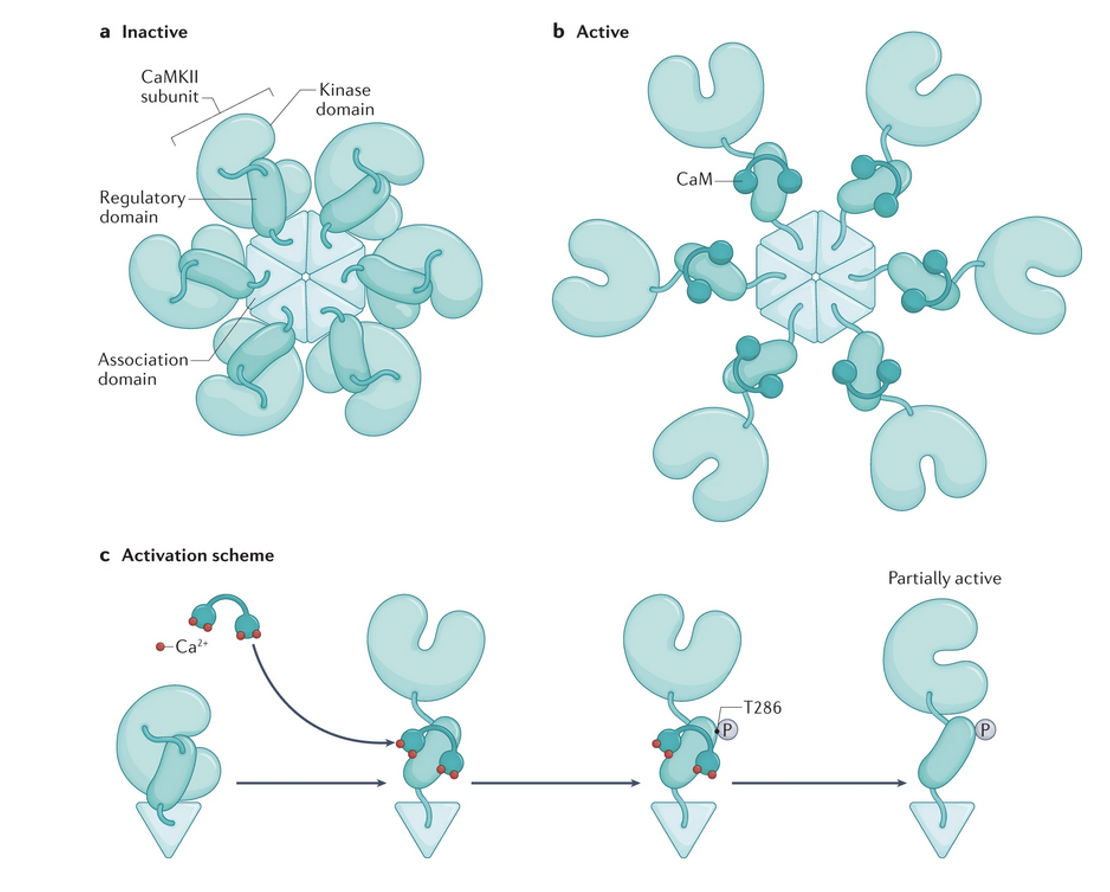
How does CaMKII work?
Ca2+ influx and its binding to CaM → CaM binds R → R dissociates → neighboring enzymes both bind CaM → each phosphorylates 286th threonine residue (T286).
Phosphorylation of T286 leaves C partially active (60%) even when CaM dissociates.
Binding to NMDA-Rs also keeps C open → can prolong kinase activity for 20 minutes after CaM detaches
CaMKII is protected from inactivation at postsynaptic density of a glutamate synapse
What is autophosphorylation?
When activated CaMKII subunit can phosphorylate its activated neighbor.
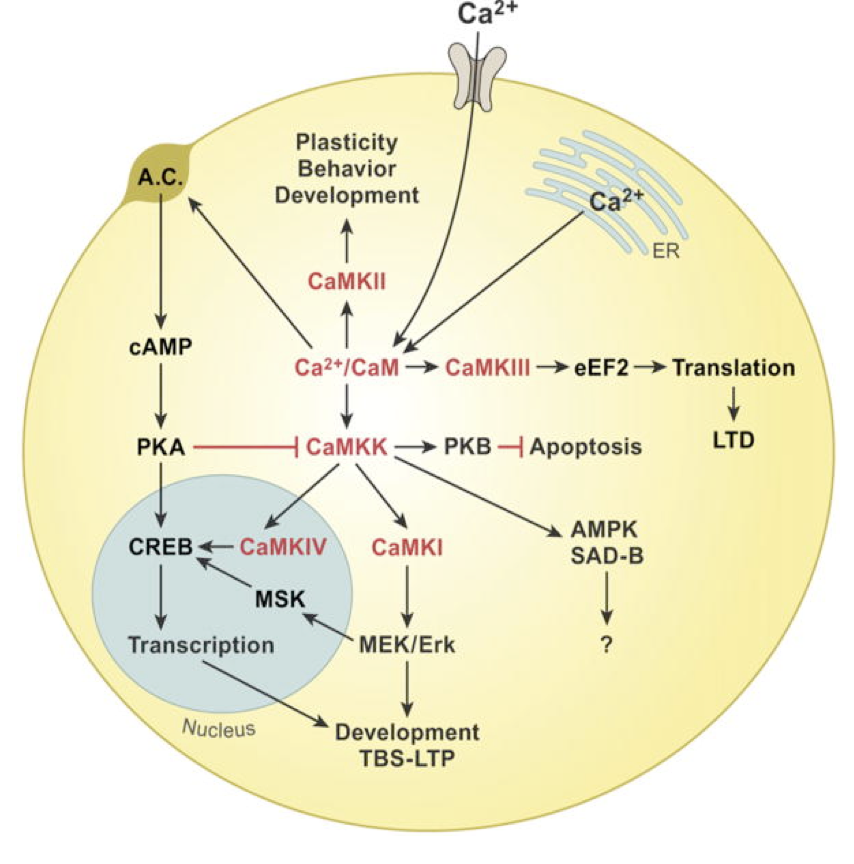
Describe CAMKK pathways
Phosphorylation of CAMKK by PKA is inhibitory
CAMKK → PKB (Akt) → apoptosis
CAMKK → CAMKIV → CREB
CAMKK → CAMKI → MEK/Erk → MSK → CREB
What are the most common phosphotases?
Mammalian CNS has 6 types of protein phosphatases w/ multiple subtypes.
PP-1, PP-2A, PP2B (calcineurin; the only Ca2+-activated phosphotase)
How is PP-2A specific?
PP-2A is a trimer with A (scaffold), B (regulatory), and C (catalytic) subunits
There are many different B subunits that determine what gets phosphorylated and where.

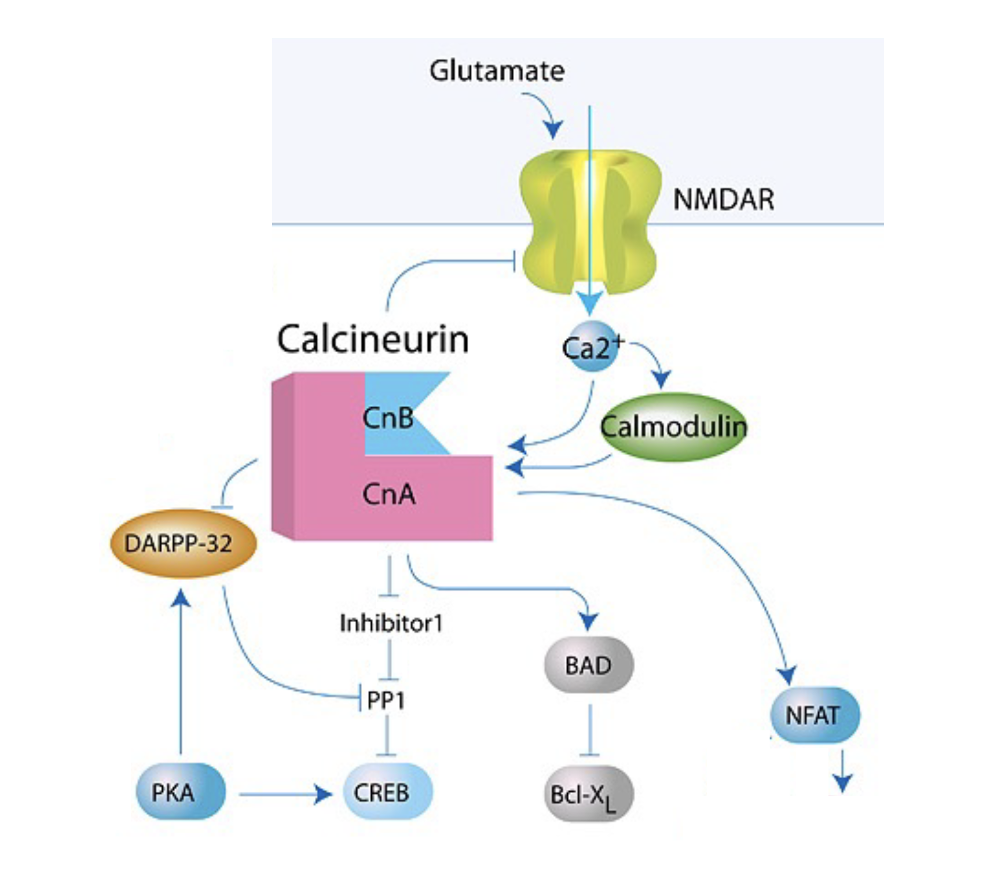
What is the structure of calcineurin?
Has 2 subunits: catalytic CnA + regulatory CnB
CnA binds Ca2+/CaM w/ high affinity; CnB binds free Ca2+ (C inactivates at high concentrations)
What kind of effects can calcineurin exert?
Synaptic depression (LTD)
Activate PP1 → dephosphorylate CREB
Dephosphorylate NFAT (T cell activatoin)
Dephosphorylate BAD (pro-apoptosis)
What are tyrosine kinases?
They catalyze phosphorylation of tyrosine residues (90 genes in humans)
2/3 of them are receptor tyrosine kinases (RTKs) (enzyme-linked receptors)
½ are NRPTKs (e.g. Jak, Src, Abl)
How do RTKs work?
They need to form a dimer and each member phosphorylates the tyrosine residue on other.
Phosphotyrosine sites on RTKs act as docking sites where adaptor proteins or enzymes bind.
What do phosphorylated tyrosine residues on RTKs do?
Provide binding site for proteins with SH2 domains:
Ex: PI 3-kinase (T740, T751), PLC (T1009, T1021), GAPs (T771), GEFs, adapter proteins
Multiple SH2-containing proteins can bind at smae time to cross-phosphorylate RTK → simultaneous activation of multiple mathways
What are the 3 major pathways of RTKs?
Regulate cell growth, differentiation, and survival with 3 major pathways:
MAP kinase pathway: GEF → ras → kinaseds → MAPK → neurite growth + differentiation
PI-3 kinase pathway: adapter proteins → PI 3 kinase → Akt kinase → cell survival
PLC pathway: PLC → IP3 + DAG → neurite outgrowth + differentiation
How does the MAPK pathway work?
RTKs activate Ras by recruiting an adapter protein (Grb2; has SH2 domain) + GEF (SOS) that turns on MAPK pathway
Ras-GTPase → Raf → MEK
How does the PI 3-kinase pathway mediate survival?
RTKs activate PI3K → PI3K phosphorylates PIP2 to form PIP3 → PIP3 binds Akt + PDK1→ Akt blocks apoptosis by phosphorylating BAD → cell survives.
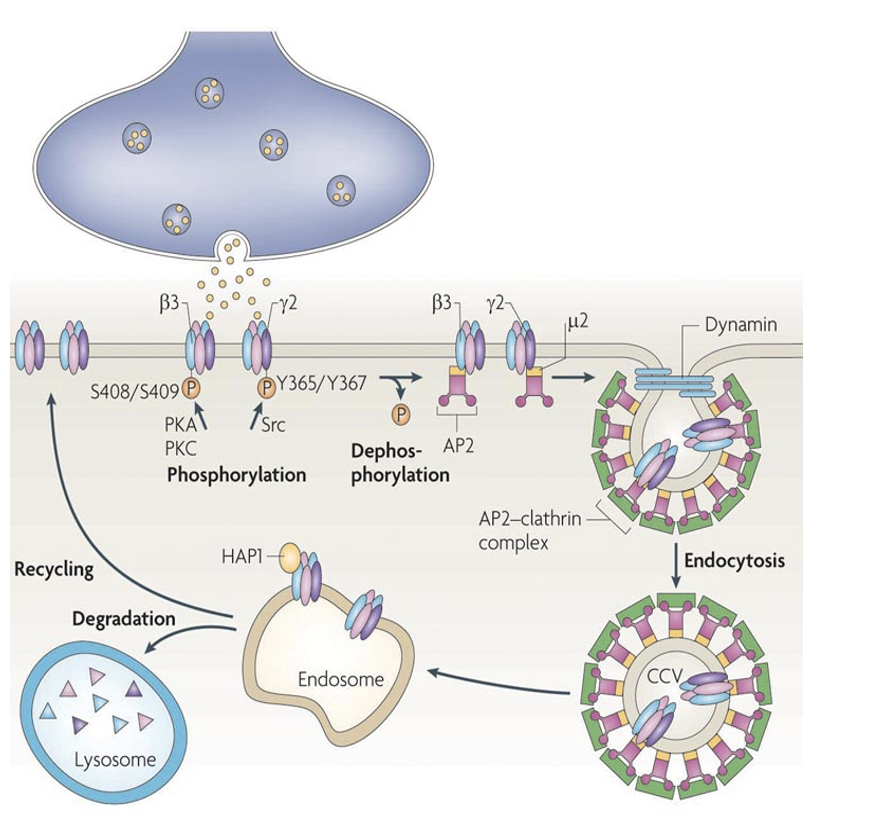
Why do we internalize RTKs and how?
To control transcription:
RTKs removed through clathrin-mediated endocytosis.
Internalized RTKs can be recycled back to plasma membrane or routed to lysosome/proteolytic pathway for degradation (inactivating RTKs)
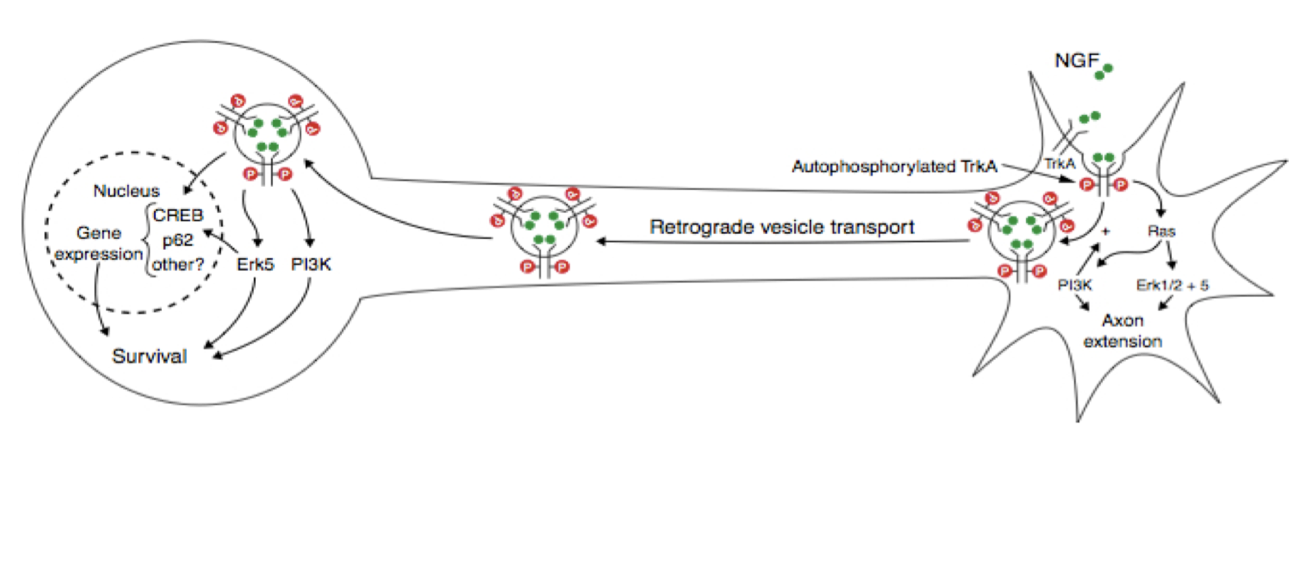
Why + how do we retrogradely transport RTKs
NGF binds RTK at axon terminals, and you need to transport it back to the nucleus to control gene expression.
RTKs are first internalized, then retrogradely transported to cell body via signaling endosomes
Binding of trophic factor keep receptor catalytically active.
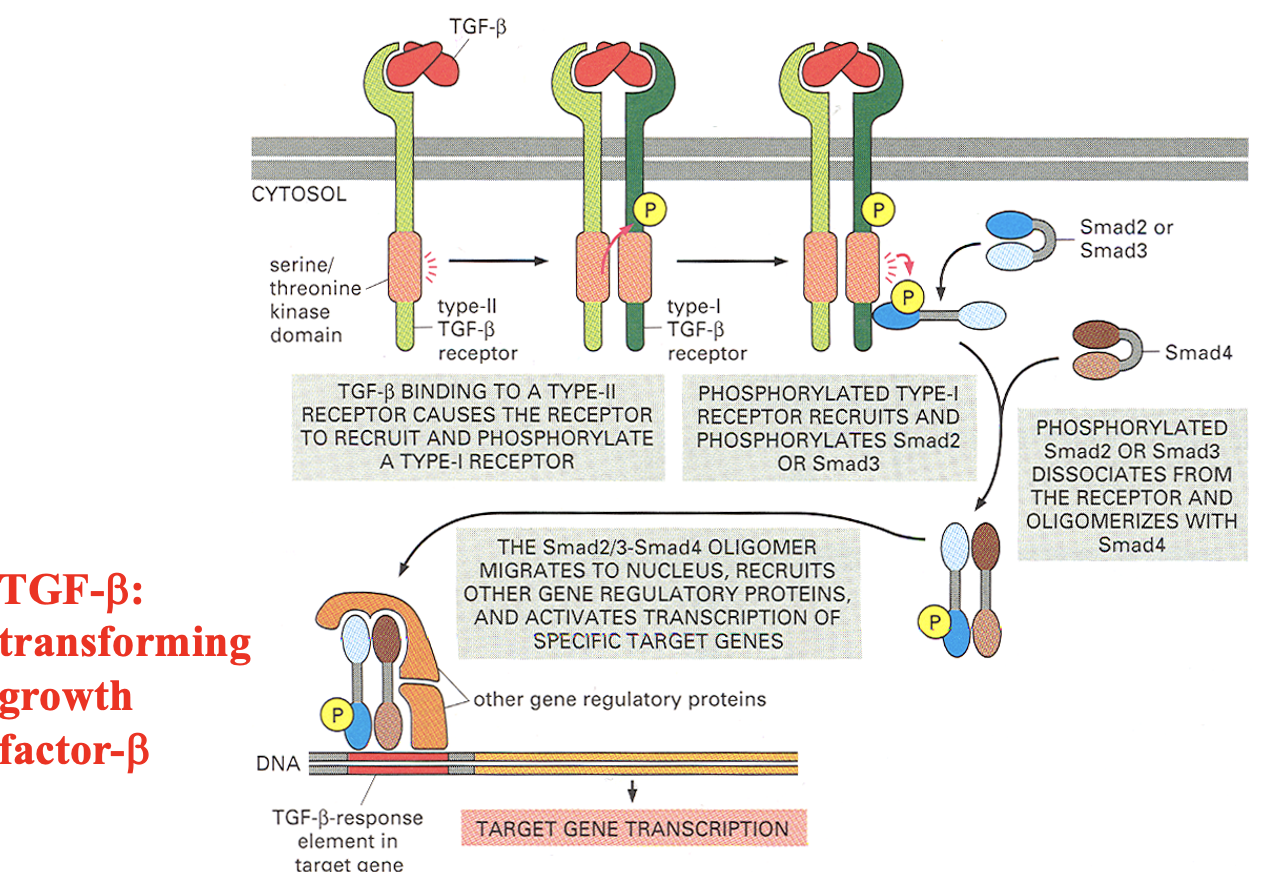
What are TGF-b receptors?
Receptor serine/threonine kinase
Has 2 receptors:
Type II TGF-β receptor (always active kinase)
Type I TGF-β receptor (gets activated by Type II)
How do TGF-b receptors work?
TGF-β binds a Type II ser/thr kinase receptor, which activates a Type I receptor that phosphorylates Smad2 or Smad3 → Smad2/3 then complexes with Smad4 (dissociates from receptor) and enters the nucleus to regulate gene transcription.
What are NRPTKs?
Intracellular enzymes that use regulatory domains (like SH2) to bind activated receptors and propagate signaling downstream.
How do NRPTKs produce inhibition?
Src-mediated phosphorylation recruits and stabilizethes GABAA-Rs to synaptic membrane.
Loss of phosphorylation → receptor removal → weaker inhibition
This is a mechanism for inhibitory synaptic plasticity