Chapter 12 DNA Replication
1/58
Earn XP
Description and Tags
• Semi-conservative • Origin of replication • Leading and lagging strands • DNA polymerases • Telomerase
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
59 Terms
What are the three models of DNA replication originally proposed?
Conservative, dispersive, and semi-conservative.
Characteristics of Conservative Replication?
- Original DNA strands reform original double helix
- Two new DNA strands form a new double helix
Characteristics of Dispersive Replication?
- Original DNA strands break down into pieces
- New DNA reassembled as a mixture of old and new pieces of DNA
Characteristics of Semi-conservative Replication?
- Original DNA strand remains intact and combines with a new DNA strand
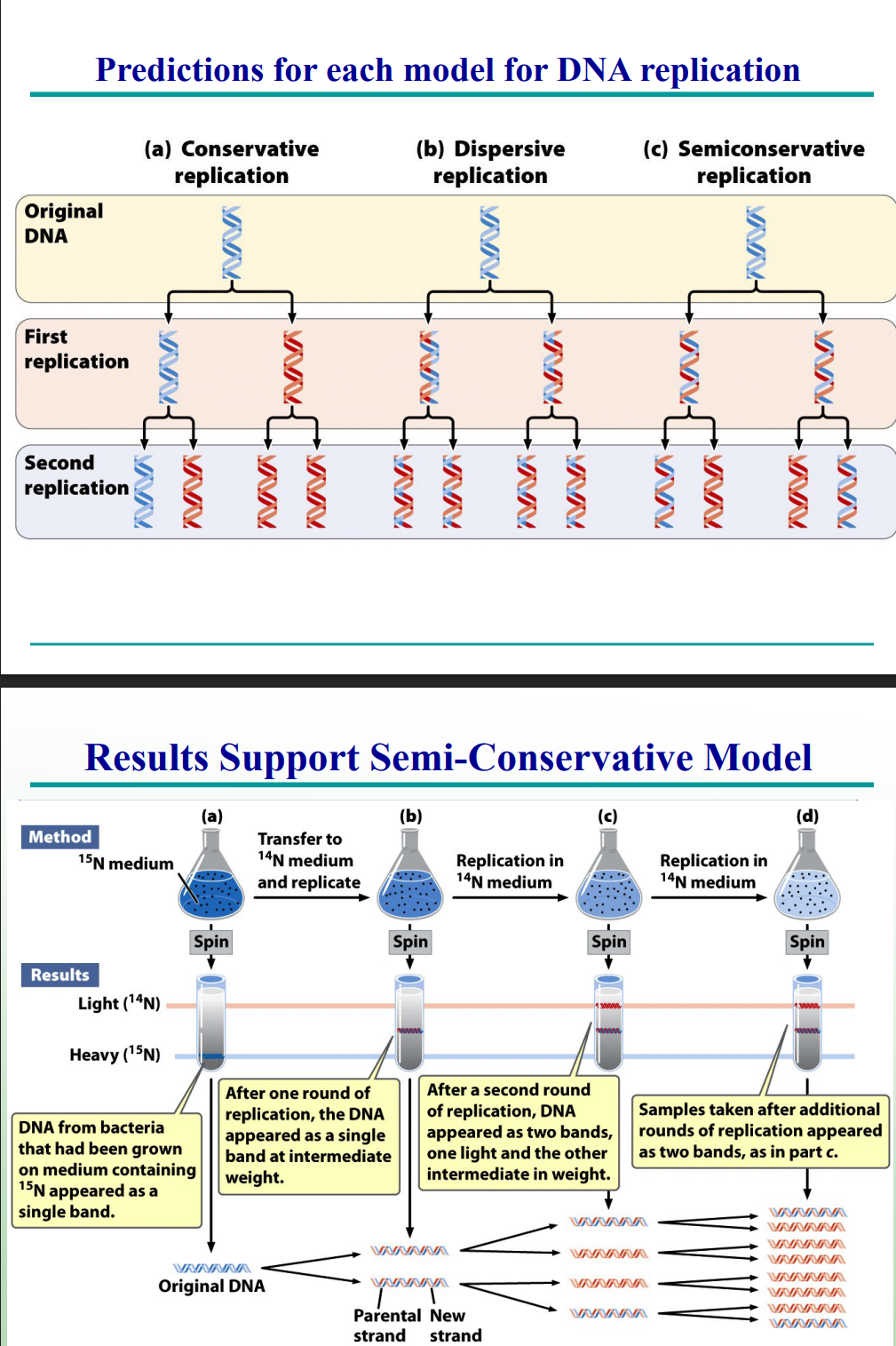
Which experiment confirmed the semi-conservative model?
The Meselson–Stahl experiment using ¹⁵N and ¹⁴N isotopes in E. coli.
How did Meselson and Stahl separate DNA of different densities?
By equilibrium density gradient centrifugation.
Part One Image of Meselson and Stahl Experiment
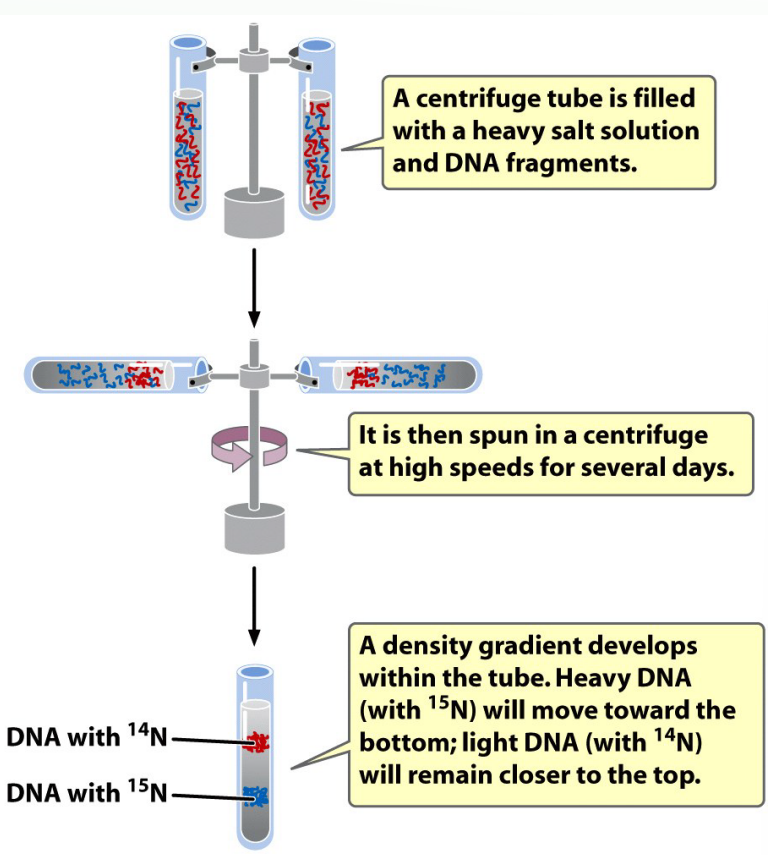
Part 2 Meselson and Stahl Experiment
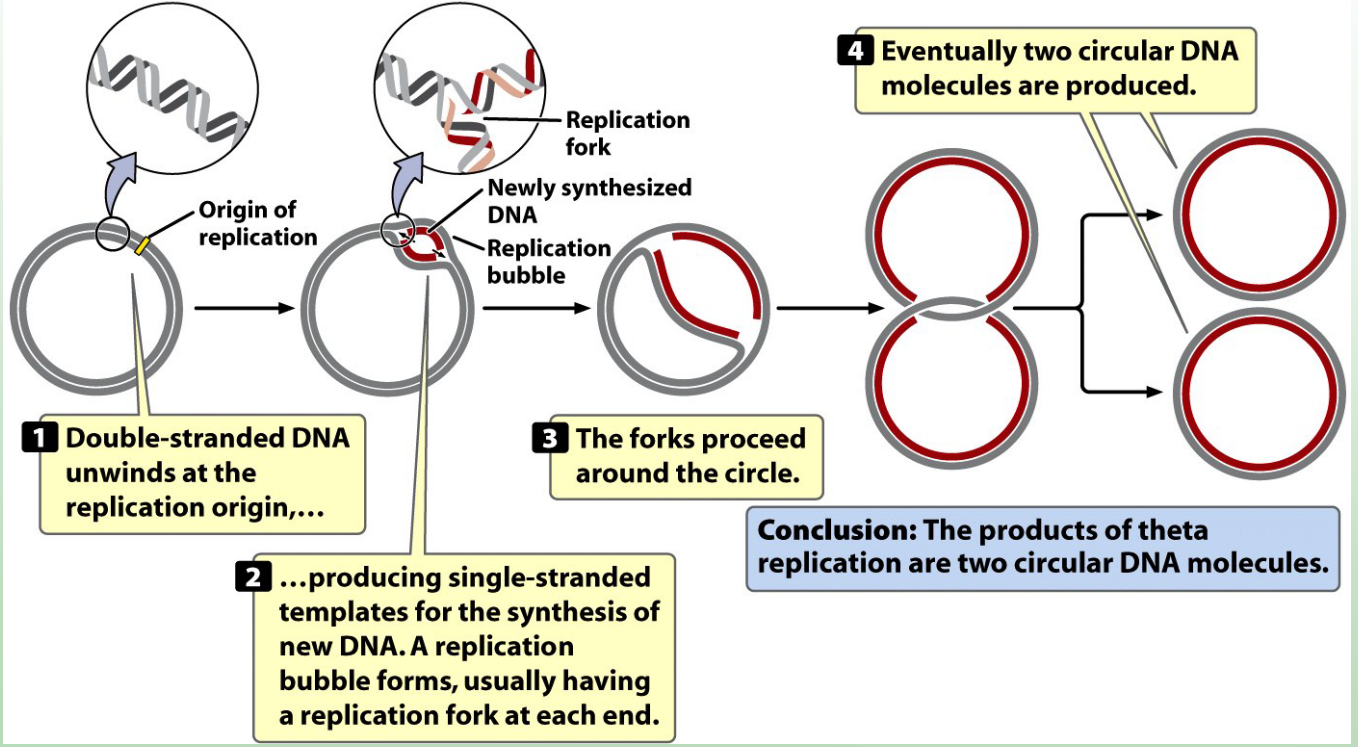
The chromosome of the bacteria is what shape?
Circular
What is the origin of replication (ori)?
A specific DNA sequence where replication begins.
How many origins of replication do bacterial chromosomes have?
One (since bacterial DNA is circular).
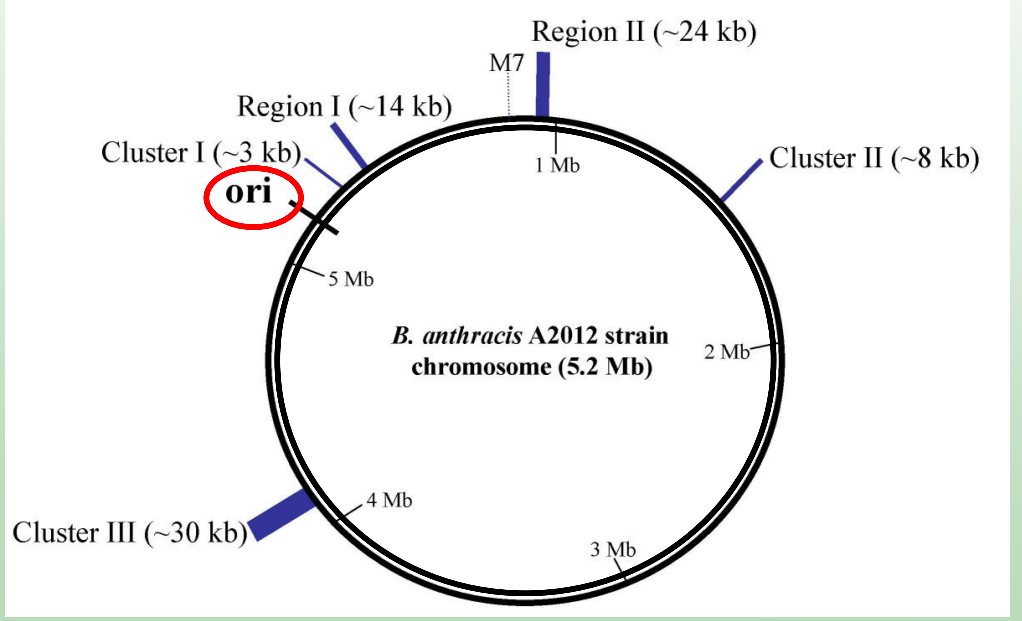
The chromosome of the eukaryote is what shape?
Linear Chromosome
How many origins of replication are there?
Multiple origins of replication
-Ends of linear molecules require special replication
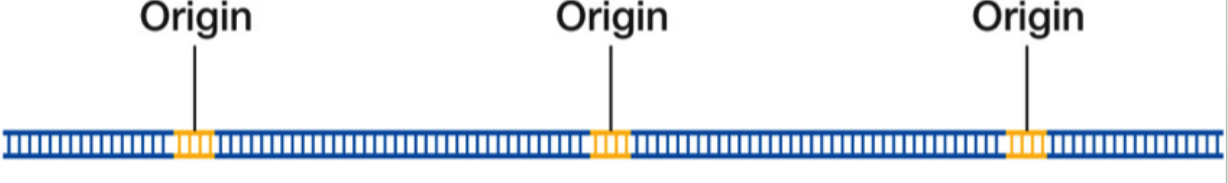
Why do eukaryotic chromosomes have multiple origins of replication?
Their DNA is much longer and linear; multiple origins speed up replication.
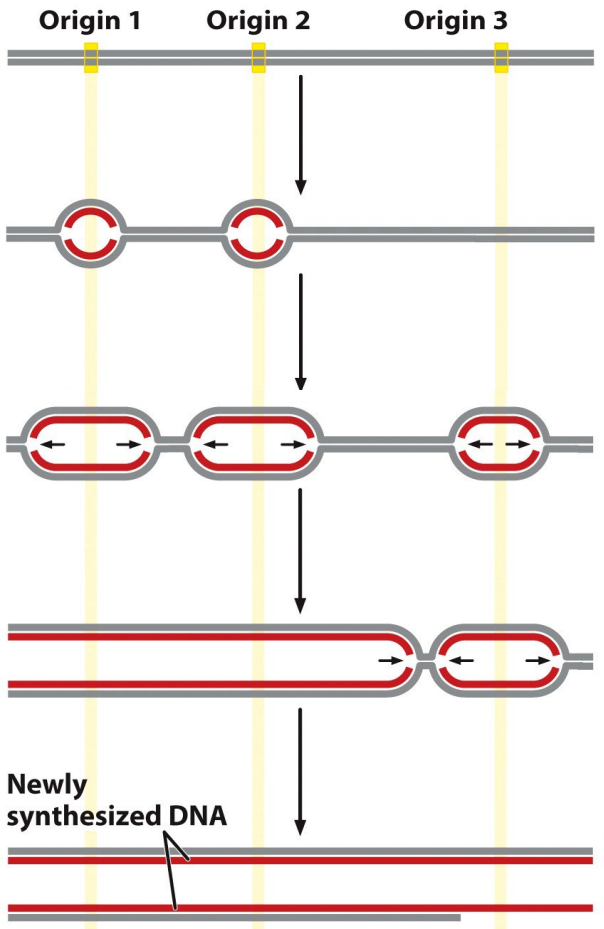
What happens when replication forks from adjacent replicons meet?
They fuse to complete replication, forming two identical DNA molecules
What proteins can bind and destabilize the double helix at ori?
Initiator Proteins
DNA replication is ________, forming a replication bubble and ____ replication forks.
DNA replication is bidirectional, forming a replication bubble and two replication forks
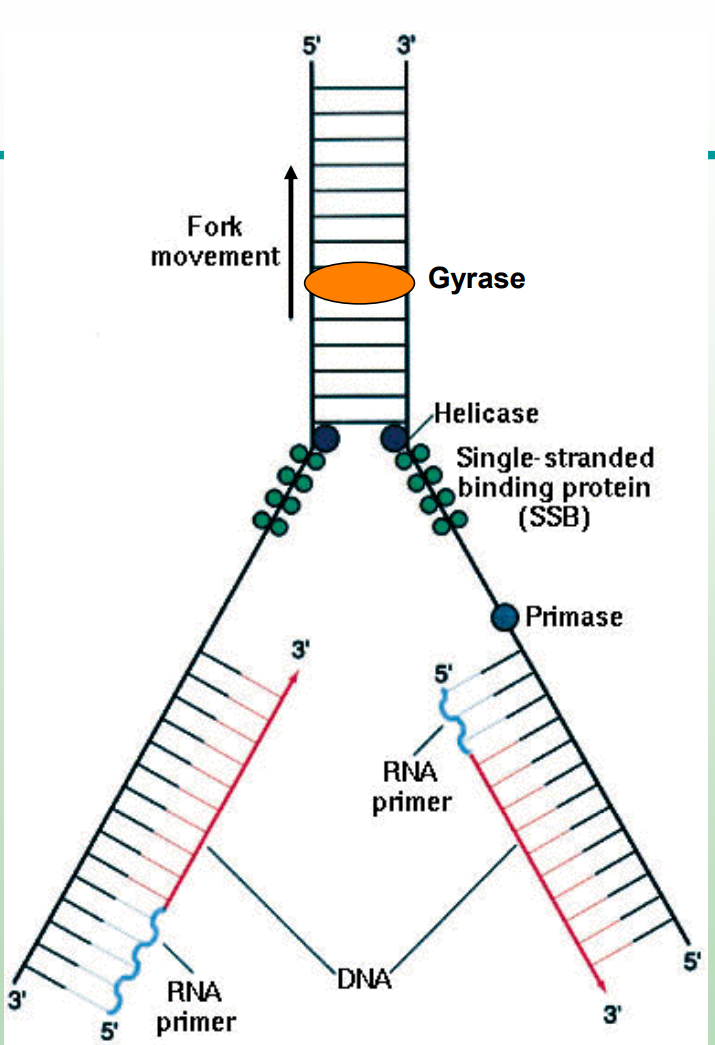
What is the replication fork?
Point at which the two strands of DNA are separated to allow replication of each strand.
Chromosomes are too large to be replicated quickly enough from a _______.
Chromosomes are too large to be replicated quickly enough from a single origin
What are some differences in eukaryotic replication compared to prokaryotic?
The rate of replication is 20x slower than prokaryotes
50-80 origins of replication are activated at one time
What are the 3 requirements for replication?
1) Template of single stranded DNA
2) Substrates (e.g. new dNTP nucleotides)
3) Enzymes to read and assemble nucleotides into new strands
In what direction is DNA synthesized?
5′ → 3′ direction (nucleotides are added to the 3′-OH of the growing strand).
_______ add a nucleotide to the _________ on the growing strand.
DNA polymerase add a nucleotide to the free 3’-OH group on the growing strand
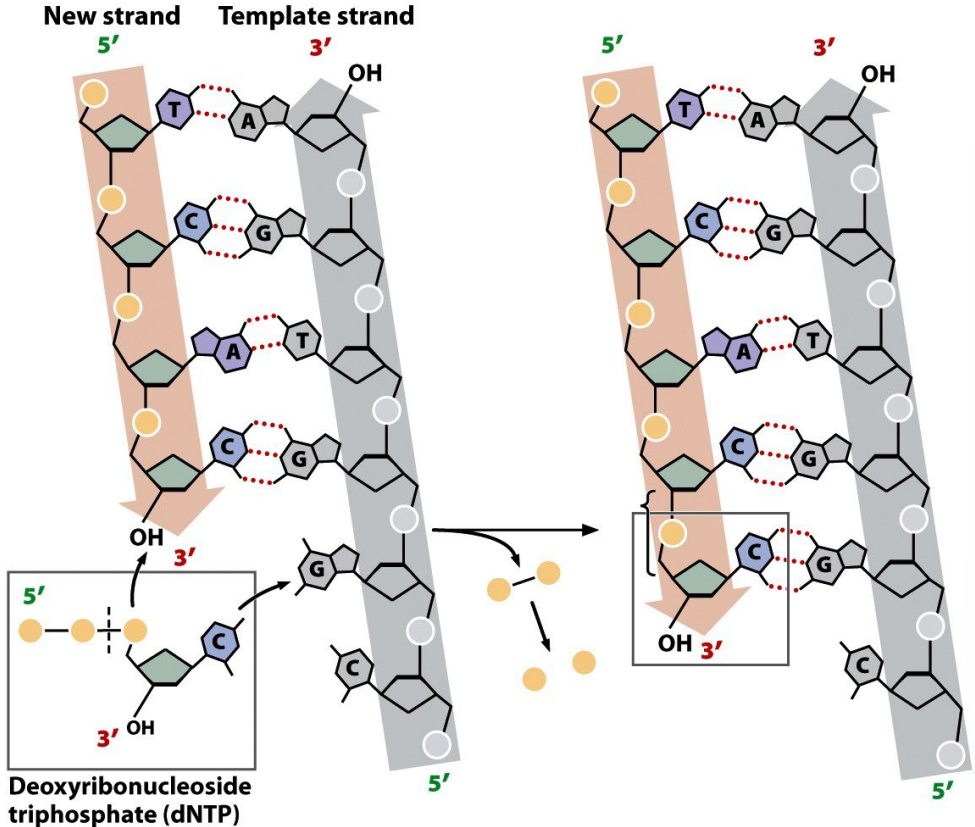
DNA polymerases catalyze the formation of a ___________ between the _________ of the incoming nucleotide and the ________ of the last nucleotide on the growing strand.
DNA polymerases catalyze the formation of a phosphodiester between the 5’-phosphate group of the incoming nucleotide and the 3’-OH group of the last nucleotide on the growing strand.
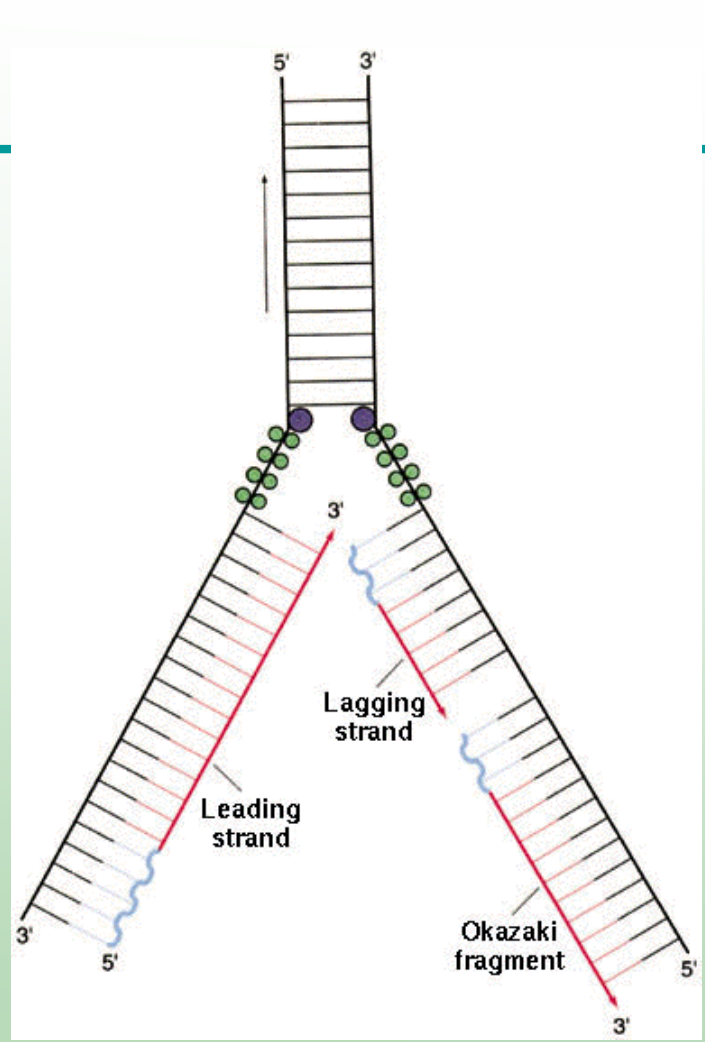
What is the leading strand?
The new DNA strand synthesized continuously in the same direction as the replication fork movement. BOTTOM RED STRAND
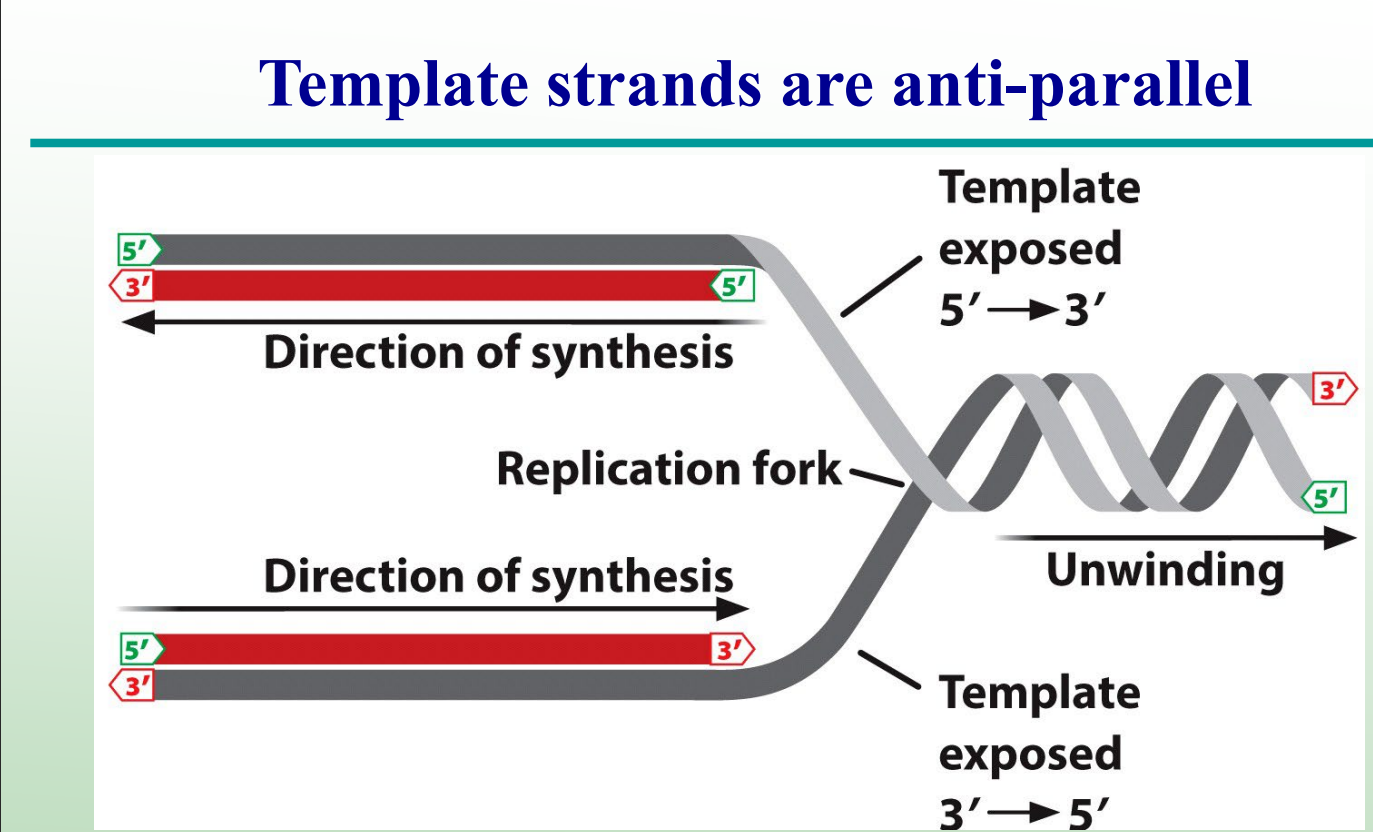
What is the lagging strand?
The new strand is synthesized discontinuously in short Okazaki fragments opposite the fork movement TOPP RED STRAND
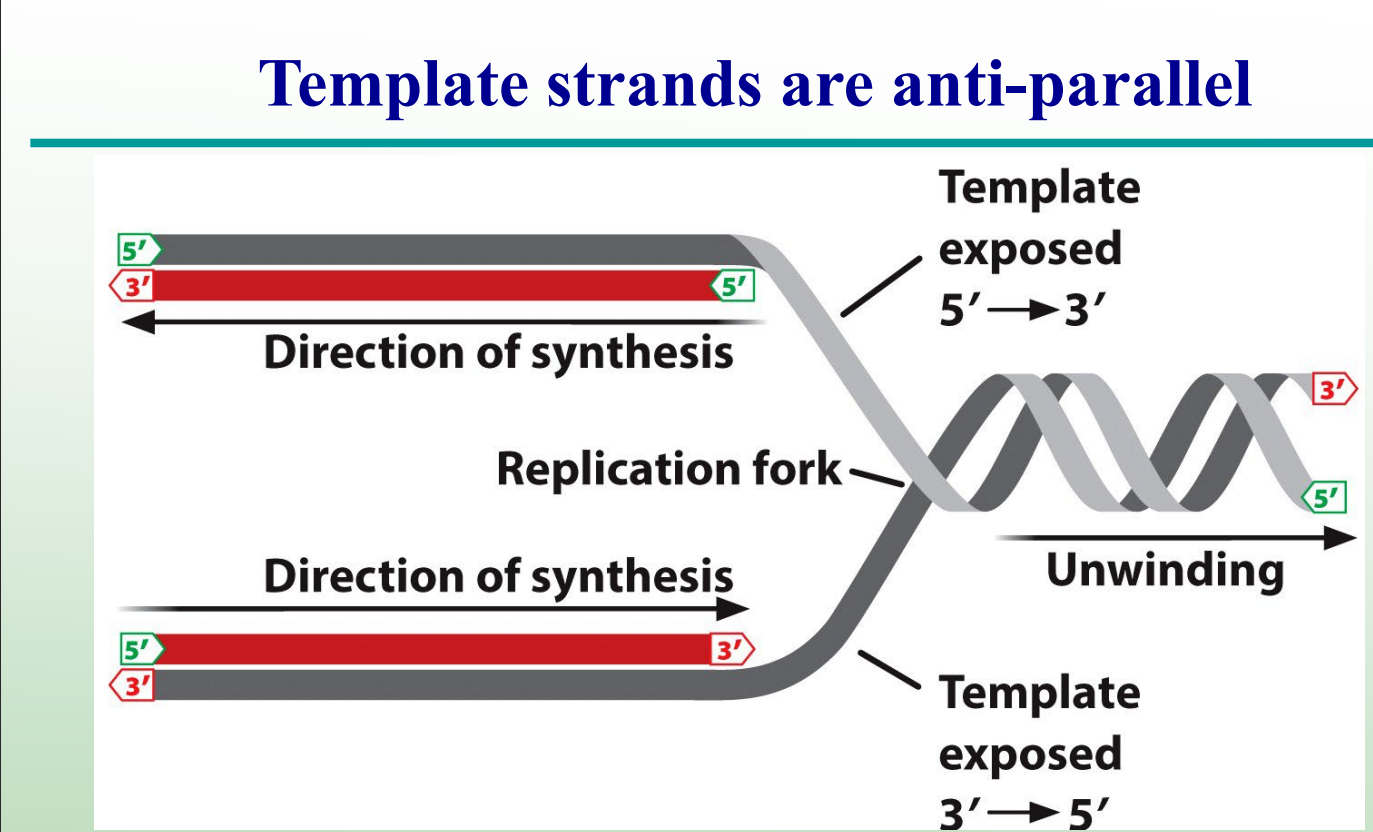
DNA Synthesis is _________ on the lagging strand.
DNA Synthesis is discontinuous on the lagging strand.
Why is the lagging strand synthesized discontinuously?
DNA polymerase can only add nucleotides to the 3′-OH end, so synthesis must restart repeatedly with RNA primers.
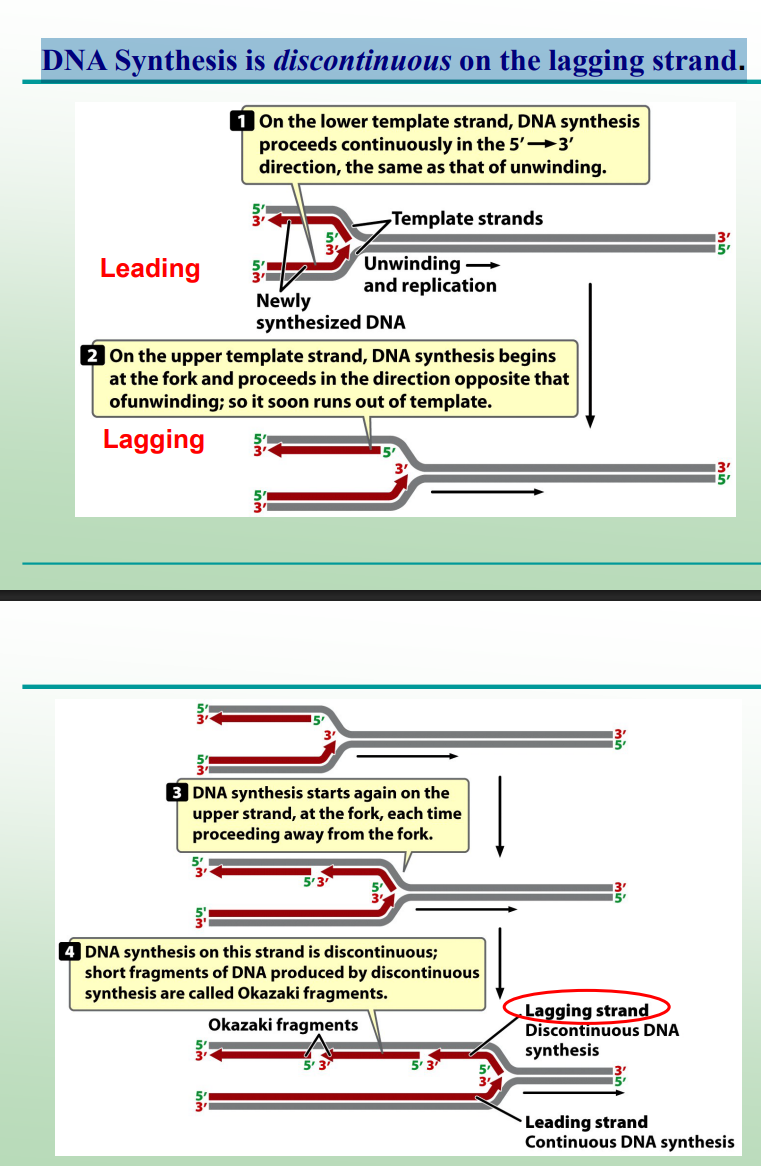
Leading and Lagging strands are synthesized at each ________ simultaneously
Leading and Lagging strands are synthesized at each replication fork simultaneously
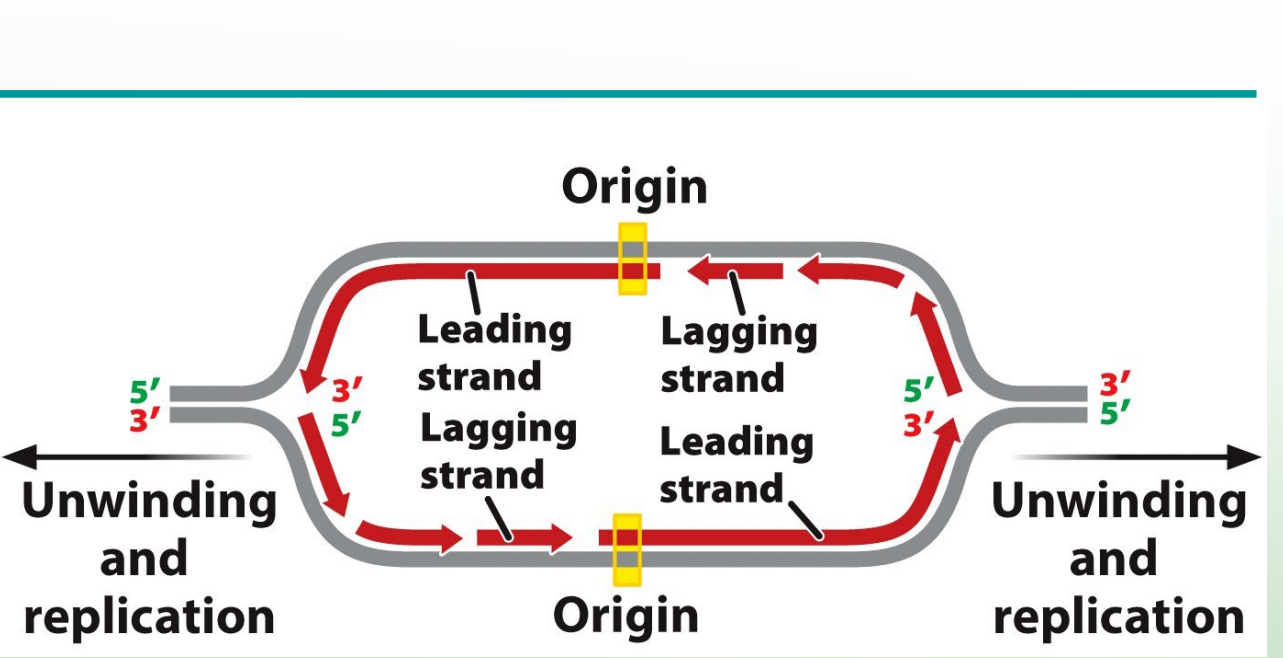
What does helicase do?
Unwinds the DNA helix by breaking hydrogen bonds at the replication fork.
What do single-stranded binding proteins (SSBs) do?
Prevent single-stranded DNA from reannealing or forming secondary structures.
What does DNA gyrase (topoisomerase) do?
Relieves supercoiling tension ahead of the replication fork.
What does primase do?
Synthesizes short RNA primers to provide a 3′-OH group for DNA polymerase III to start.
What does DNA polymerase III do?
Main enzyme that adds nucleotides to the 3′ end during elongation (5′→3′ polymerase activity). ELONGATION
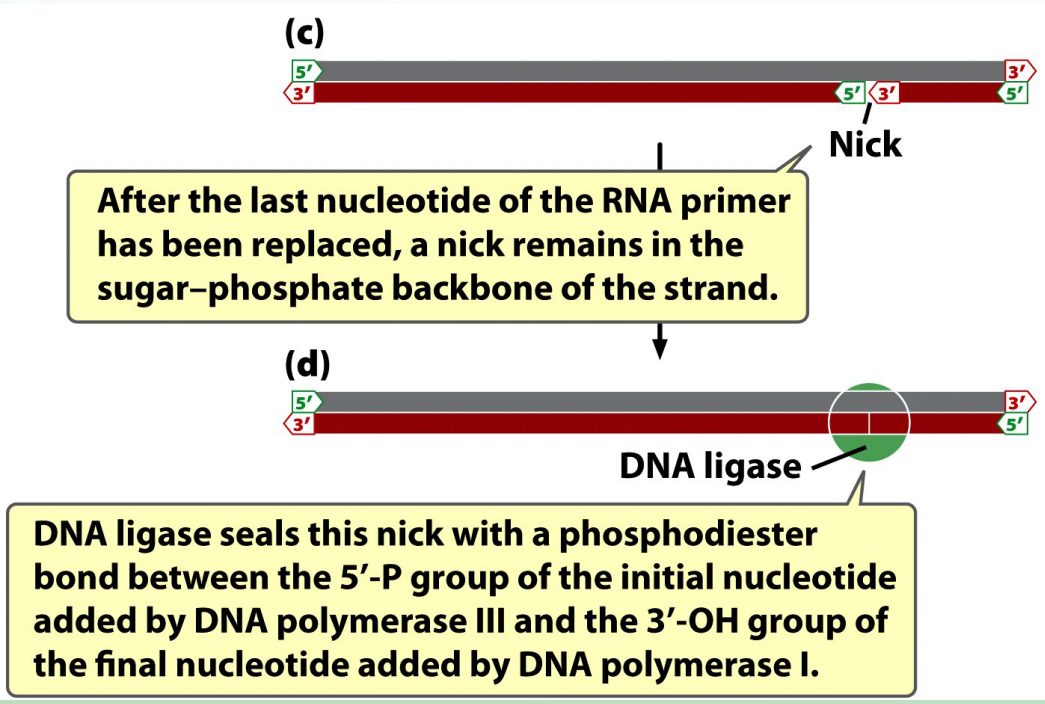
What are the two activities of DNA polymerase I?
5′→3′ exonuclease: removes RNA primers
5′→3′ polymerase: replaces them with DNA nucleotides
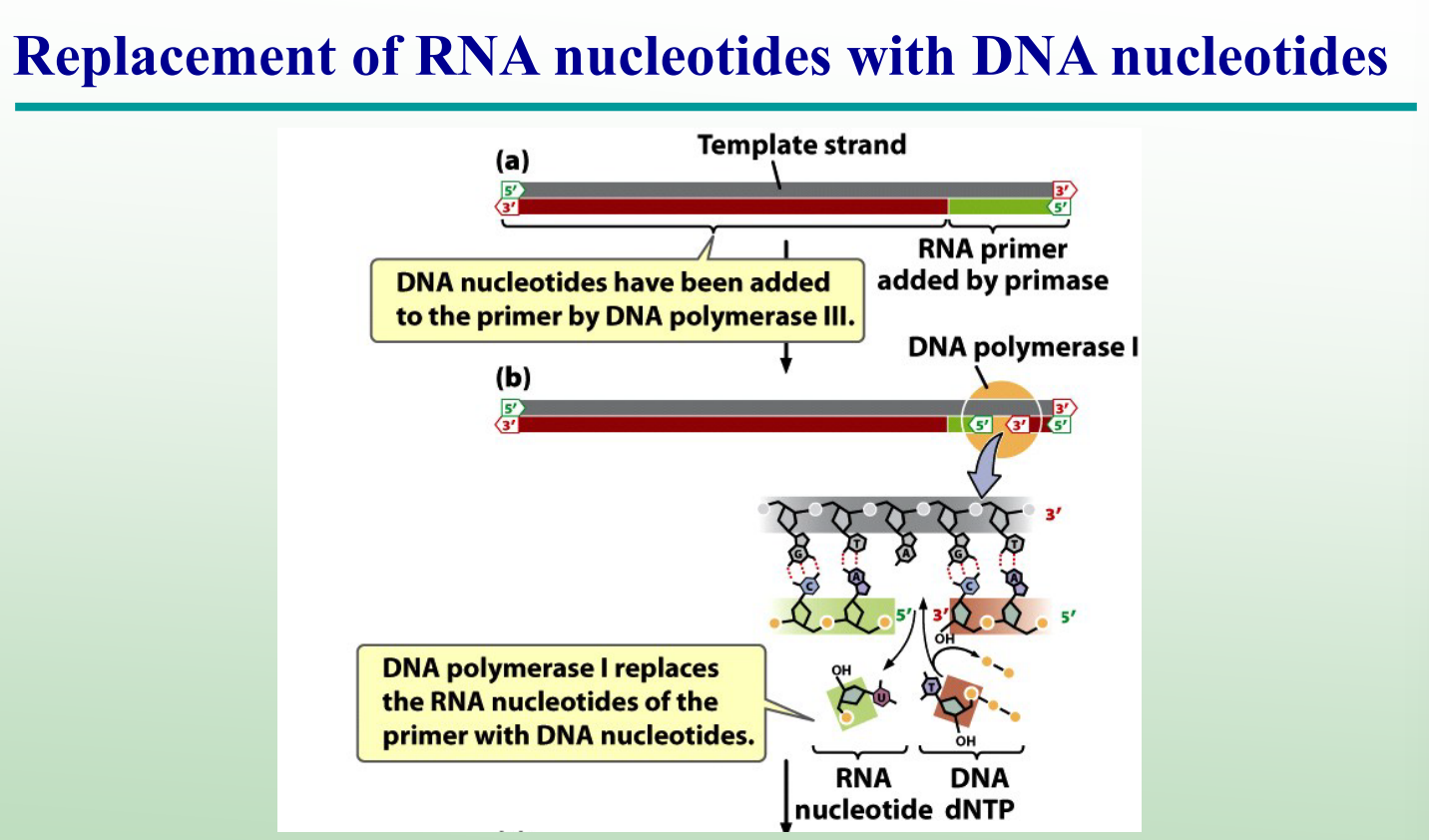
What does DNA ligase do?
Joins Okazaki fragments by forming phosphodiester bonds.
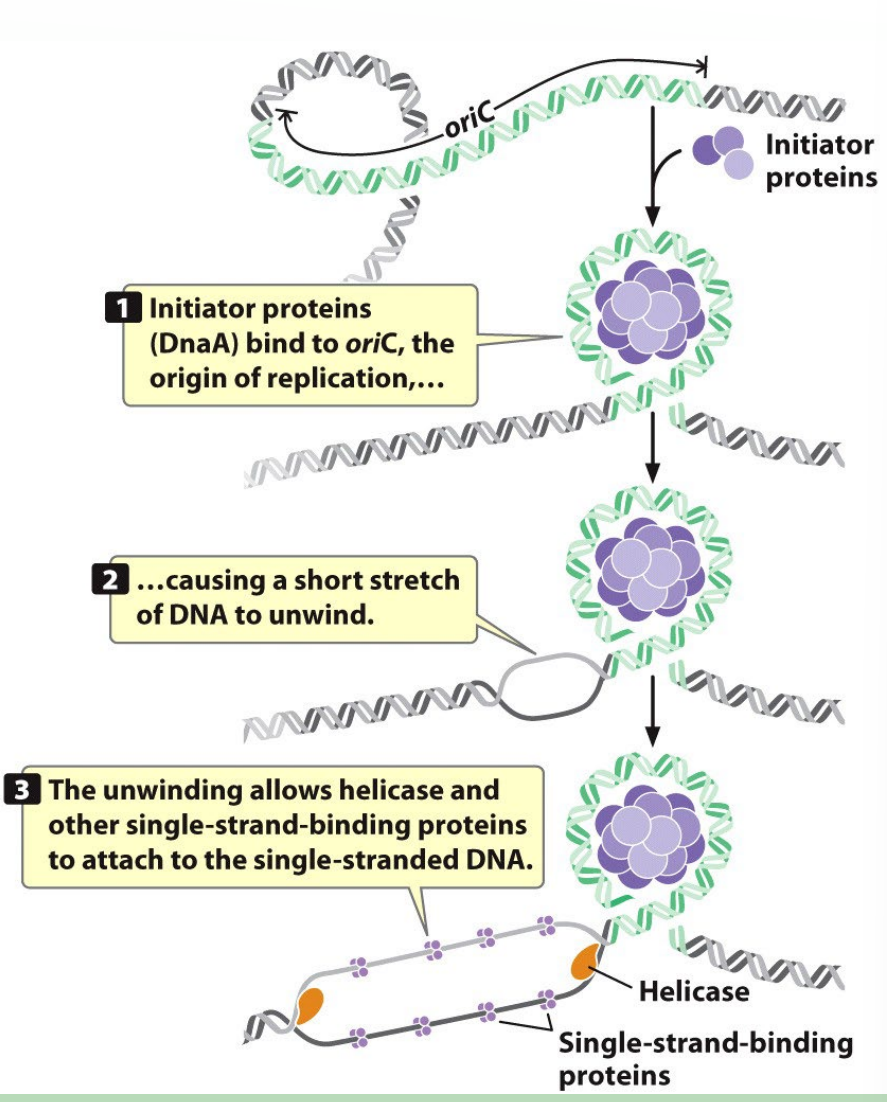
1) Intitiation (Bacterial Replication is the Image on the Flashcard)
- Initiator proteins
- Helicase
- SS Binding proteins
*Ori C is the origin of replication* (WHOLE BUNCH OF A-T that opens up next to the Ori C)
_______________ is similar except for linear nature of the chromosome, multiple origins of replication, and differences in the DNA polymerases
Eukaryotic replication is similar except for linear nature of the chromosome, multiple origins of replication, and differences in the DNA polymerases
2) Unwinding
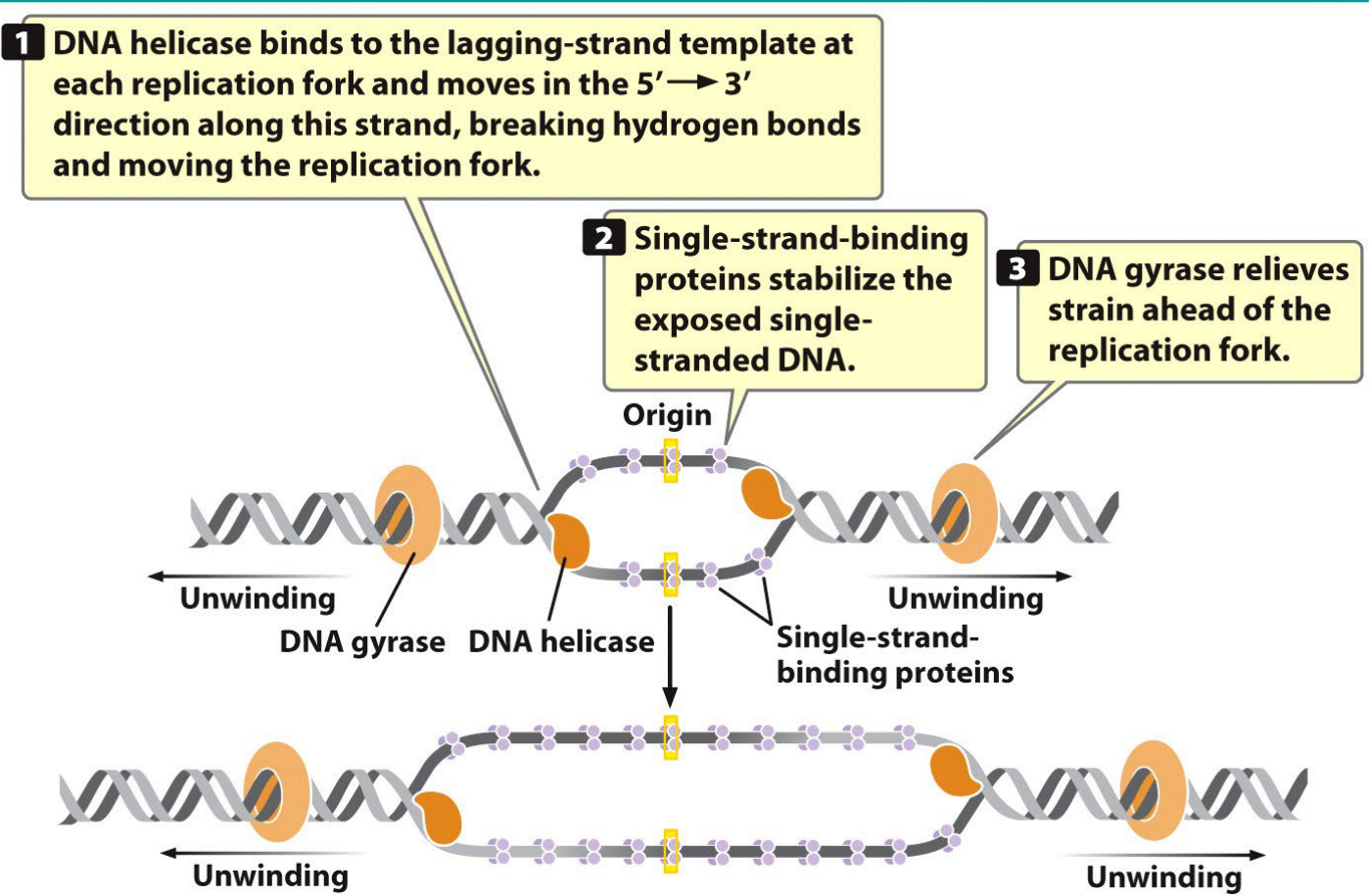
The sequence of enzymes and their functions is what?
(initiation → elongation → termination)
Each Leading strand begins with ______
Each Leading strand begins with 1 primer
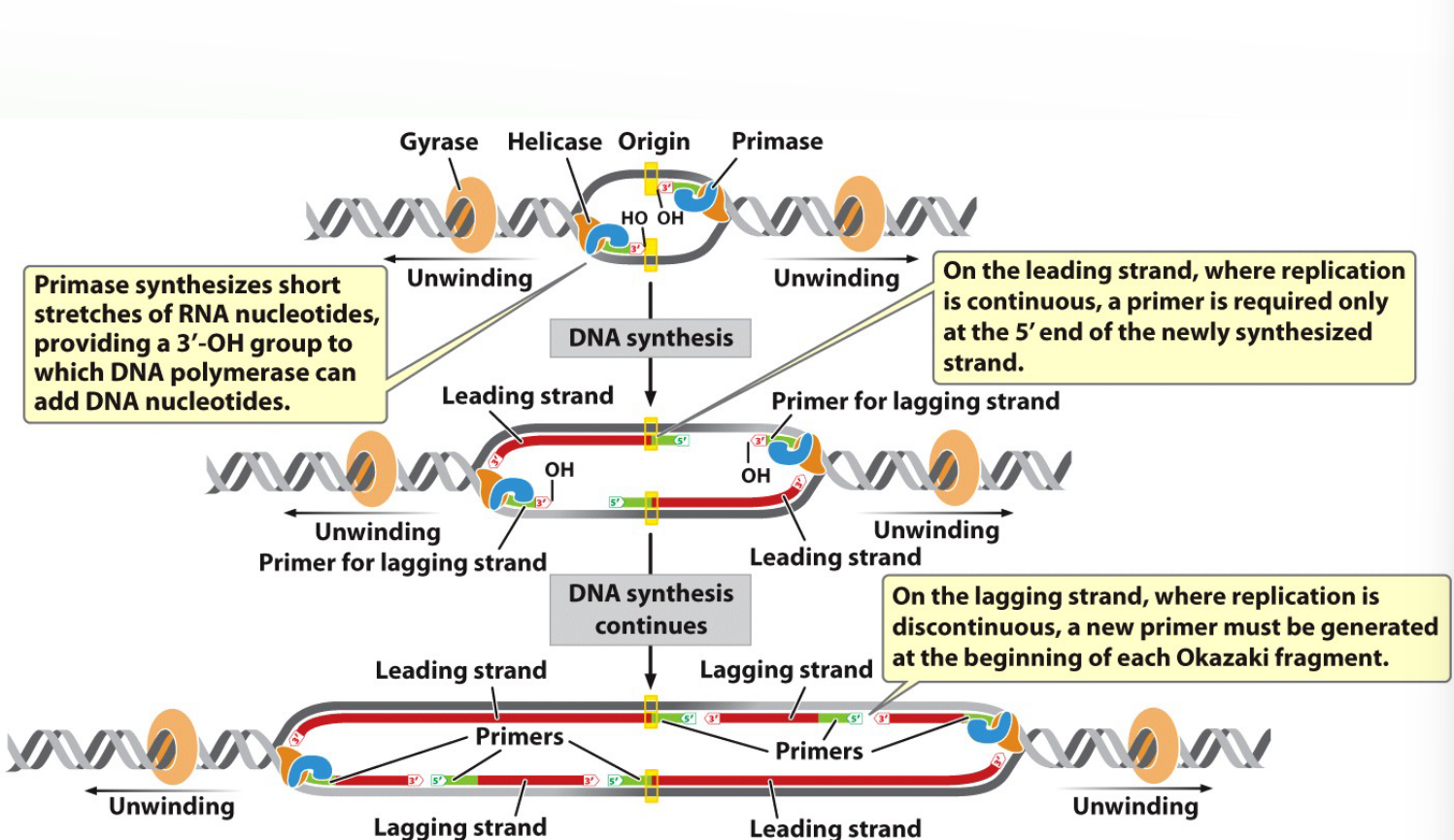
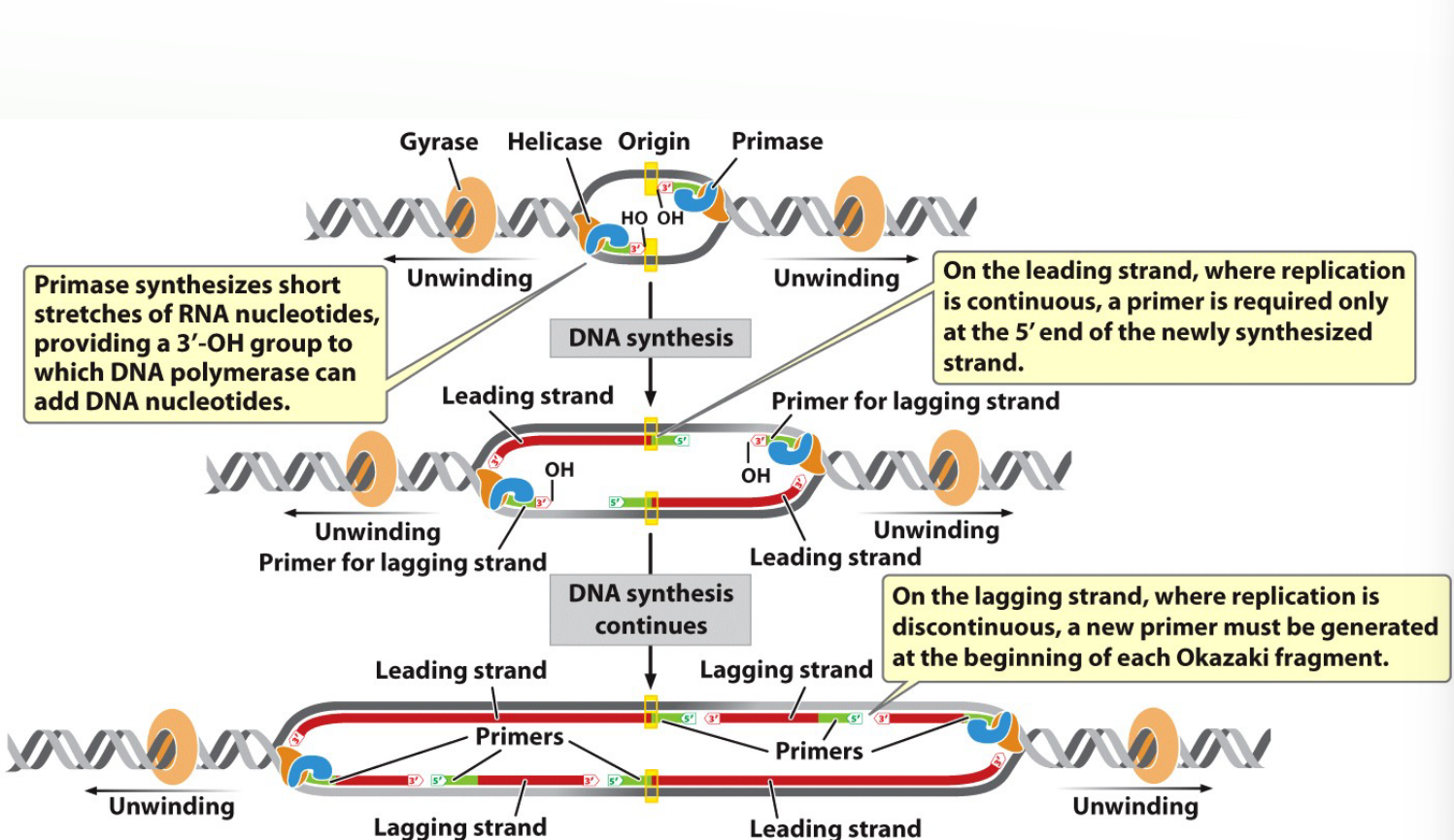
Each Lagging strand _______________________.
Each Lagging strand Okasaki fragment begins with a primer.
Closer view of primers and elongation
A new primer is synthesized for the next lagging strand Okasaki fragment.
Fragments by DNA Ligase
Why can’t DNA polymerase replicate the very end of linear chromosomes?
After the RNA primer is removed from the lagging strand, there’s no 3′-OH for DNA polymerase to add to.
What is the error rate is less than what per BILLION nucleotides?
Error rate is less than 1 mistake per BILLION nucleotides!
What is proofreading?
Any error in DNA polymerase III base-pairing is immediately corrected using its 3’ to 5’ exonuclease activity (backwards removal) and repair
What is the mismatch repair enzyme?
Correct any errors that remain after DNA replication is completed
In bacteria, original DNA is methylated, so repair enzymes know which strand to fix
What happens to chromosomes after multiple rounds of replication without telomerase?
They become progressively shorter, eventually leading to loss of essential genes and cell death.
Why must RNA primers be removed and replaced during DNA replication?
Because primers are made of RNA, not DNA; DNA polymerase I must replace them with DNA nucleotides to complete replication.
Why isn’t primer removal a problem in bacterial (circular) chromosomes?
Circular DNA always has a 3′-OH end available for DNA polymerase to add new nucleotides and fill the gap after primer removal.
Why does linear DNA (eukaryotic chromosomes) face a problem at the ends?
At the end of the lagging strand, once the final RNA primer is removed, there’s no adjacent 3′-OH group for DNA polymerase to extend from — leaving an unreplicated gap.
What happens if the unreplicated gap at the end of linear DNA isn’t fixed?
Each round of replication produces shorter DNA molecules, gradually eroding essential genes.
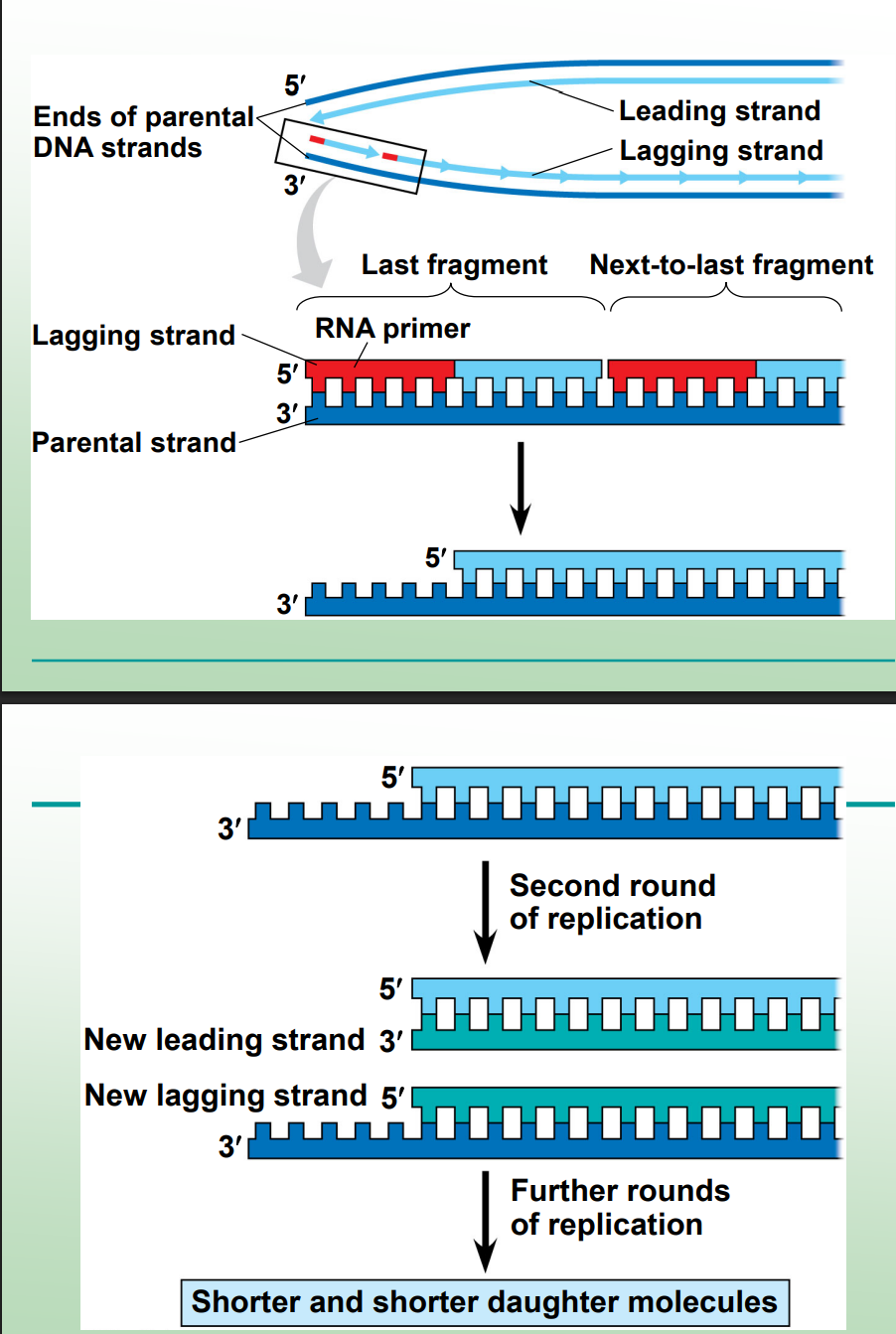
What are telomeres?
Repetitive, heterochromatic DNA sequences at chromosome ends that protect coding DNA.
Postpone the erosion of vital gene sequences
Telomere shortening may contribute to aging. Why is telomerase activity limited in somatic cells?
Most somatic (body) cells have a genetically programmed life span with limits on how many times they can divide before death (apoptosis)
What does telomerase do?
Extends the 3′ end of the parental strand using its RNA template, allowing completion of the lagging strand.
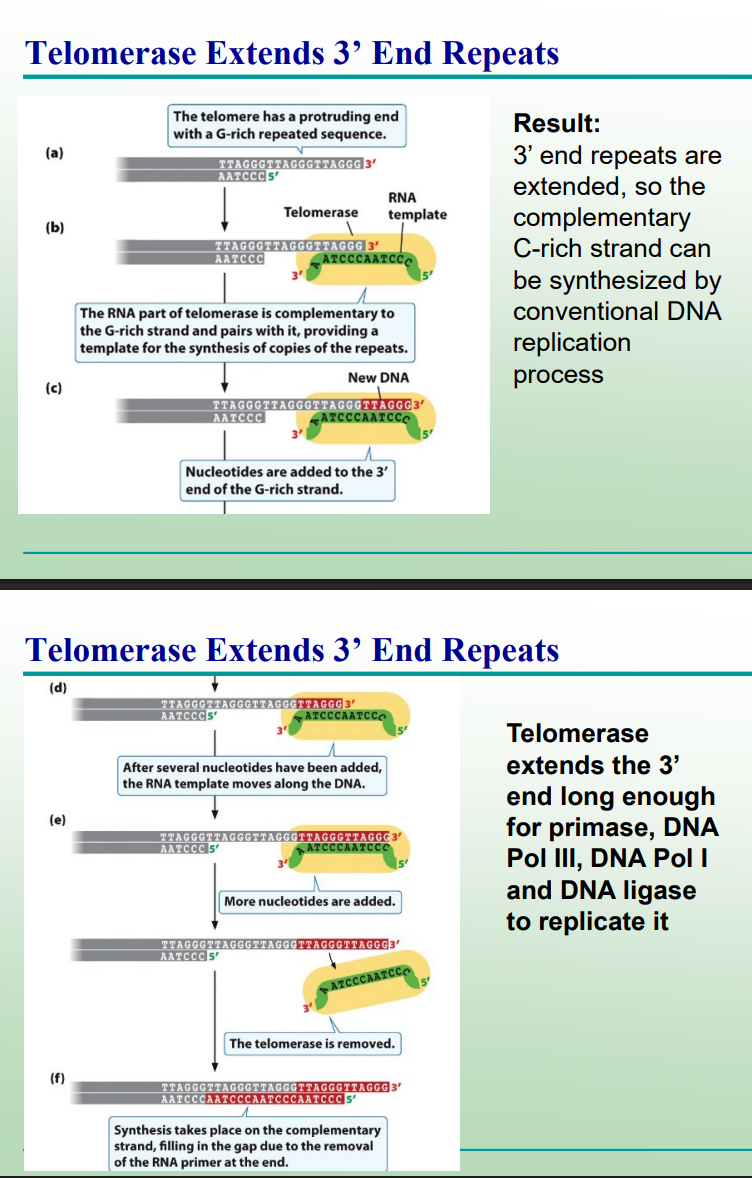
What is the composition of telomerase?
It is a ribonucleoprotein — made of RNA (template) and protein (enzyme). Complementary to a G-rich strand that serves as a template for DNA synthesis
Why do most cancer cells have high telomerase activity?
It allows them to divide indefinitely by maintaining telomere length, contributing to cellular immortality.