lec 22 - precision med (molloy)
1/42
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
43 Terms
what is individualized, personalized, precision medicine?
uses information about genes, proteins and environment to identify patients that most benefit from a particular therapy
uses biomarkers and companion diagnostics to identify genetic variation among subsets of patients
uses identification of responders vs non-responders to target treatment
uses mechanisms to maximize efficacy and safety in pharmacotherapy
goals of precision med
to correlate specific genetic markers to diseases and therapeutic interventions
to produce better predictive and diagnostic molecular tests and drugs
to better select treatments and dosing based on individual need
right patient, right dose, right time
increase efficacy and safety
what is a biomarker?
a characteristic that is objectively measured and evaluated as an indicator of normal biologic processes, pathogenic processes or pharmacologic responses to a therapeutic intervention
identification of biomarkers have led to treatment advancements in oncology, HIV/AIDS, neuroscience, CV and others
why are biomarkers important?
biomarker information is contained in about 60% of FDA approved oncology drug labels between 2015-2019
across all therapeutic areas, the inclusion of pharmacogenomic biomarkers in drug labels has increased, with about 28% of new drugs in 2020
biomarker results play an important role in identifying therapeutic outcomes
biomarkers may categorize responders vs non-responders
biomarkers maximize safety and efficacy
biomarker categories
those that assess susceptibility to a disease (prognostic)
screening for specific strains of HPV to predict disease susceptibility
screening for BRCA gene variants (possible cancer risk)
those that identify disease and outcome (predictive)
predicts the benefit of therapy
avoidance of ADRs associated with unnecessary therapy
example
clopidogrel and 2C19 resistance → some people have genetic variations in CYP2C19 that make clopidogrel less effective
warfarin and CYP2C9/VKORC1 → genetic differences in CYP and VKORC affect how people metabolize warfarin and how sensitive they are to it
predictive biomarker components
efficacy
identifying individuals who are more likely to respond to a particular drug
efficacy pharmacogenomics
safety
identifying individuals who are less likely to have an adverse effect of a particular drug
safety pharmacogenomics
example of predictive biomarker
patients from at risk populations should be screened for presence of HLA-B*1502 allele
carbamazepine may bind to HLA-B15 → leading to activation of cytotoxic T cells that attack skin cells and cause SJS/TEN
what are companion diagnostics?
products and assays used in conjunction with a therapeutic product
inform treatment selection, initiation, dose customization or avoidance
co-development with partner drug
post-marketing experience
FDA guidance
use in clinical trials
landscape of oncology personalized medicine
traditional diagnostic tests in oncology
CT
MRI
PET scans
new research has broadened use of diagnostics
evolving discovery of biomarkers leading world of oncology towards personalized medicine
personalized medicine to have significant impact on development and commercialization of oncology drugs
biomarker methods in cancer
immunohistochemistry (IHC)
application → protein-based assay for detection of expression
advantages
cheap, rapid, widely available
direct visualization of protein expression
limitations
antibody availability
subjective interpretation/quantification
fluorescence in situ hybridization (FISH)
application → hybridization using fluorescent-labeled probes to detect gene copy-number changes or gene rearrangements/fusions
advantages
relatively simple assay design
direct visualization of signals within cells of interest
limitations
probe availability
restricted to specific locus/gene tested
PCR
application → detection of targeted gene mutations, fusions, copy-number alterations, DNA methylation
advantages
high sensitivity and specificity
relative simply assay design
relatively low cost
limitations
limited throughput
restricted to targeted genes and regions of interest interrogated
next-generation sequencing (NGS)
application → massively parallel sequencing of multiple genes for detecting mutations, fusions, copy-number alterations
advantages
high throughput
high sensitivity and specificity
comprehensive coverage
site/tumor-specific applications
limitations
high complexity
bioinformatics requirements
longer turnaround time
gene expression profiling (GEP)
application → differential gene expression between tumor/normal or pre/post treated tumor
advantages
high throughput
limitations
bioinformatics requirement
restricted to targeted genes
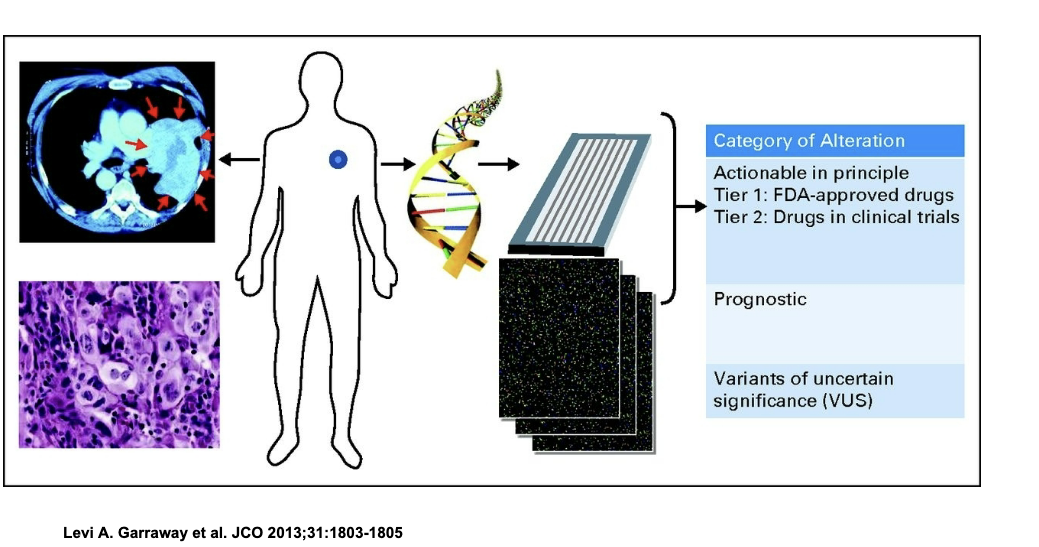
schematic for genomics-driven cancer medicine
pt with tumor is IDed and biopsy is taken
tumor DNA is extracted and sequenced to look for mutations
detected mutations are sorted into 3 categories
actionable
tier 1 → mutations with available FDA approved drugs
tier 2 → mutations with drugs currently in clinical trials
prognostic
mutations that tell us about how aggressive the cancer might be or the patient’s likely outcome
variants of uncertain significance
mutations we’ve found but do NOT know what they mean clinically
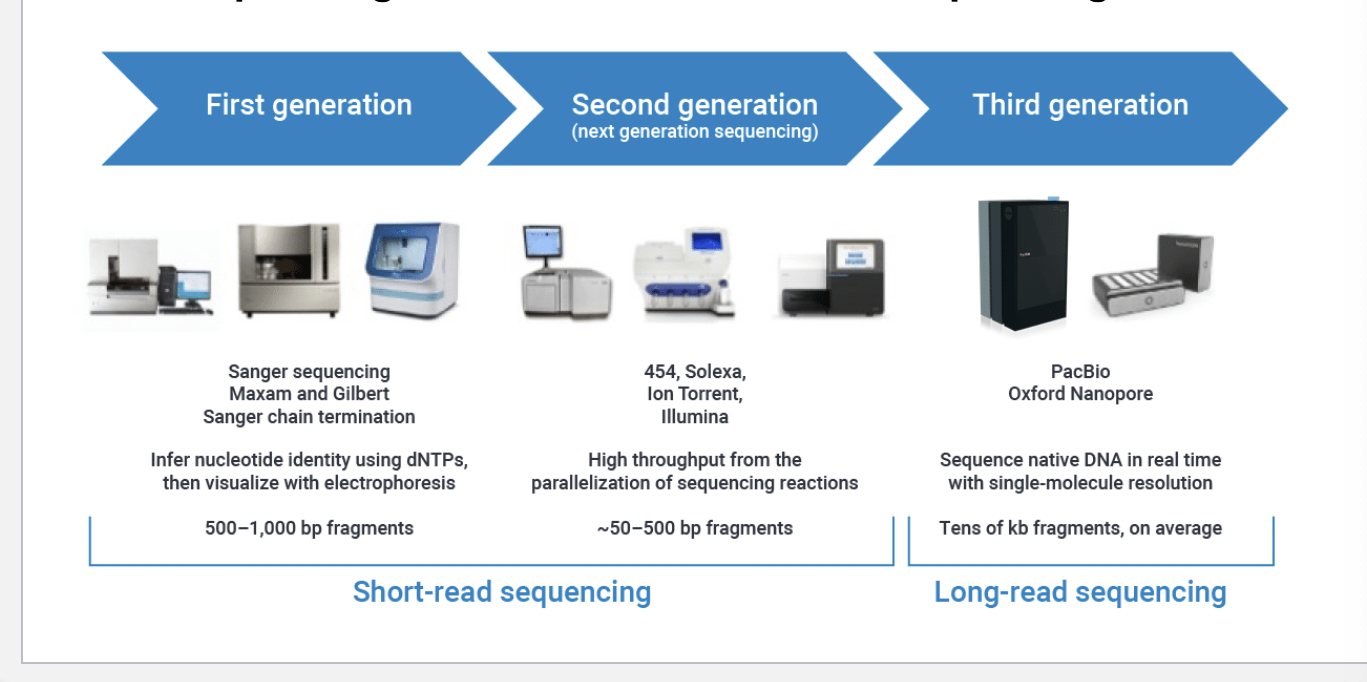
evolution of DNA sequencing tools
first generation (short read sequencing)
sanger sequencing, maxam and gilbert, sanger chain termination
infer nucleotide identity using dNTPs, then visualize with electrophoresis
500-1000 bp fragments
second generation (short read sequencing)
454, solexa, ion torrent, illumnia
high throughput from the parallelization of sequencing rxns
~50-500 bp fragments
third generation (long read sequencing)
pacbio, oxford nanopore
sequence native DNA in real time with single-molecule resolutions
tens of kb fragments on average
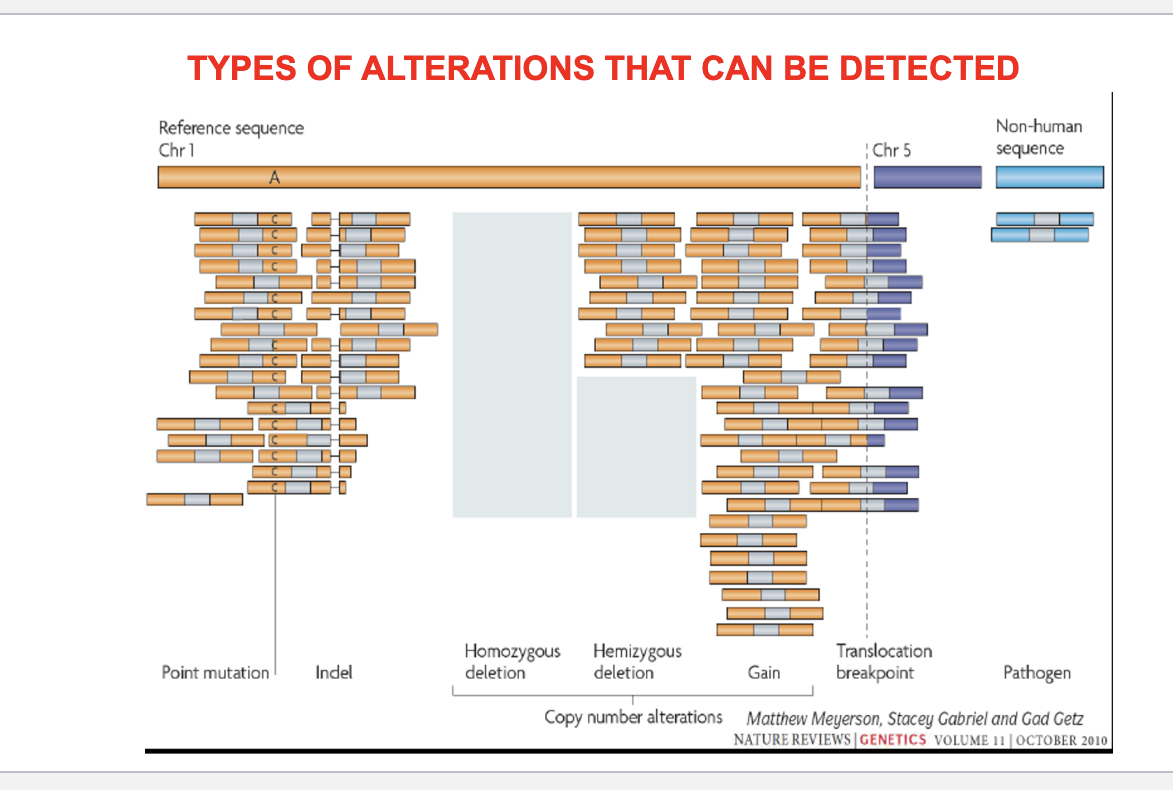
types of alterations that can be detected
point mutation
indel = small insertions or deletions of DNA bases
copy number alterations
homozygous deletion = both copies of a gene are deleted → complete loss of gene
hemizygous deletion = one copy of a gene is deleted → partial loss
gain = extra copies of a gene or region → may lead to overexpression
structural variants
translocation breakpoint = parts of 2 different chromosomes break and reattach to each other → can create fusion genes
pathogen integrated into genome
potential of tissue based analysis
genomic and transcriptomic architecture of 2000 breast tumors reveals novel subgroups
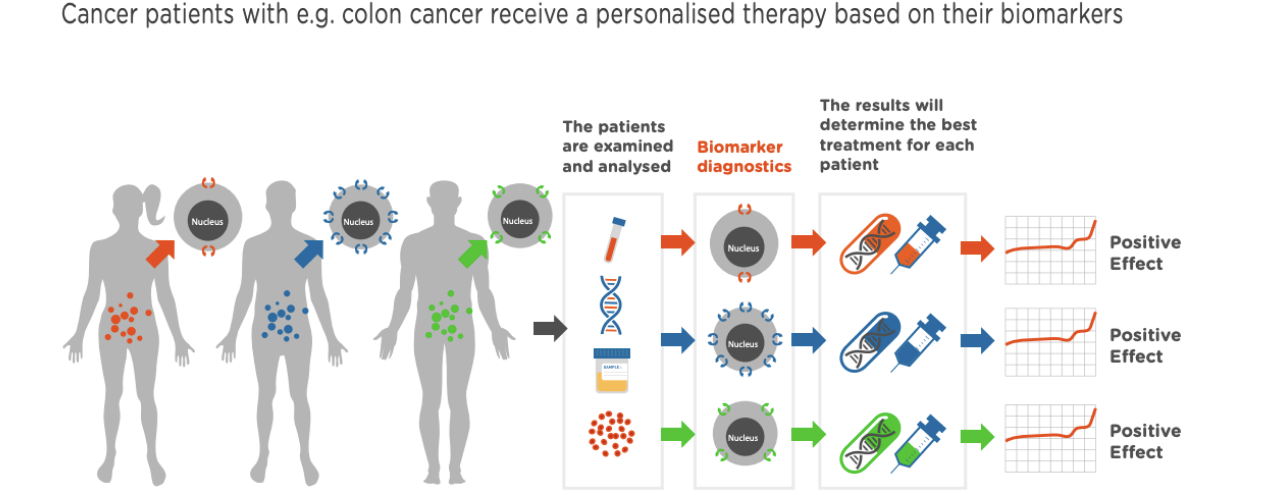
innovative medicine: personalized medicine
analyze patients → biomarker diagnostics → results will determine best treatment for each pt
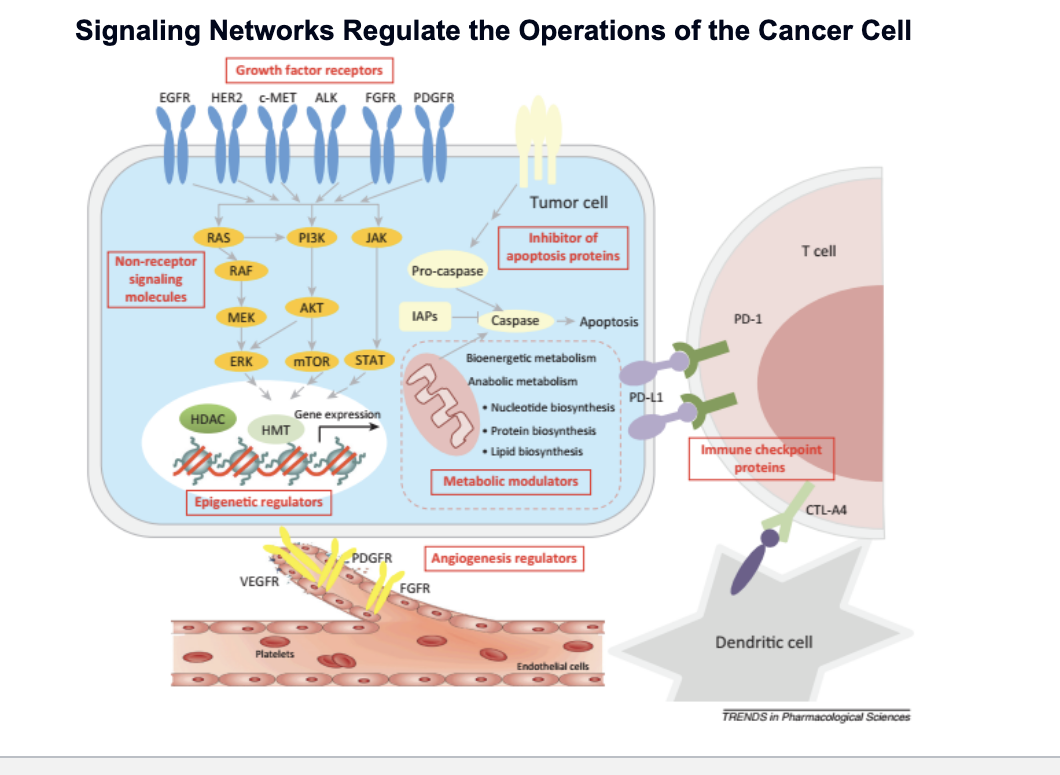
signaling networks regulate the operations of the cancer cell
growth factor receptors
proteins on the surface of the tumor cell
EGFR, HER2, c-MET, ALK, FGFR, PDGFR
normally, detect signals that tell cell to grow or divide; in cancer, often overactive, sending constant “grow” signals
non-receptor signaling molecules
RAS → RAF → MEK → ERK → cell growth and division
PI3K → AKT → mTOR → cell survival, metabolism, growth
JAK → STAT → gene transcription
epigenetic regulators
controls how DNA is packaged and which genes are turned on or off
cancer cells may alter gene expression WITHOUT changing the DNA itself
inhibitor of apoptosis proteins
metabolic modulators
cancer cells change their metabolism to support rapid growth:
nucleotide synthesis
protein & lipid biosynthesis
angiogenesis
cancer cells secrete signals (VEGF, PDGFR) to promote blood vessel growth
immune checkpoint proteins
tumor cells express PD-L1 which binds to PD-1 on T cells which inhibits T cells
CTL-A4 inactivates dendritic cells
reliable biomarkers in clinical use - oncology
HER2 in breast cancer
HER2 amplification reliably predicts response to trastuzumab (herceptin) with standardized IHC/FISH testing protocols
PD-L1 expression
used for checkpoint inhibitors but reliability varies by cancer type (e.g. more predictive in NSCLC than others)
MSI-H/dMMR (high micro-satellite instability/deficient mismatch repair)
highly predictive of immunotherapy responses across multiple cancers
EGFR mutations
strong predictor of EGFR inhibitor efficacy in NSCLC
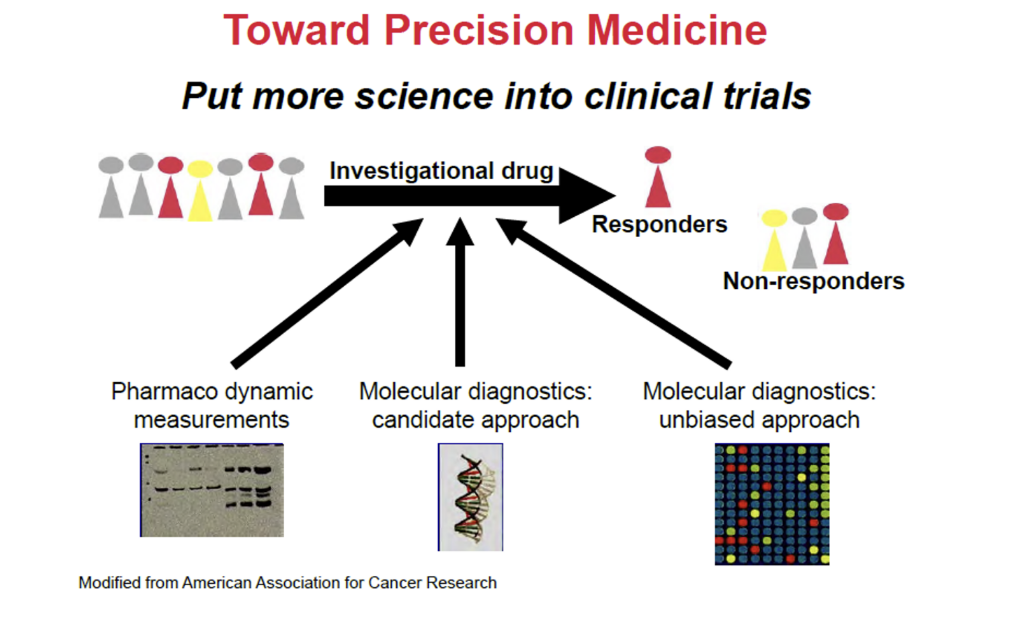
toward precision medicine
put more science into clinical trials
use these methods in investigational drug
PD measurements
molecular diagnostics; candidate approach
molecule diagnostics; unbiased approach
cancer is a disease of the genome
therefore, if we precisely define the cancer genome, we will understand and cure cancer
why we must be cautious about such statements
founder mutations = first genomic mutations
these are often lesions that lead to genomic/chromosomal instability and are often NOT fully transforming
driver mutations = mutations that are required for expression of fully transformed phenotype
driver mutations = mutations that we would like to target and inhibit their function
passenger mutations = these mutations are “collateral damage” resulting from genomic instability and are NOT required for maintaining the transformed phenotype; are “noise” in the system
since most cancers are rapidly evolving biologic entities, major task to sort out drivers from passengers and these may change over time
therapeutic implications
the things not circles are the hallmarks of cancer and the things in shapes are drugs that fight that hallmark
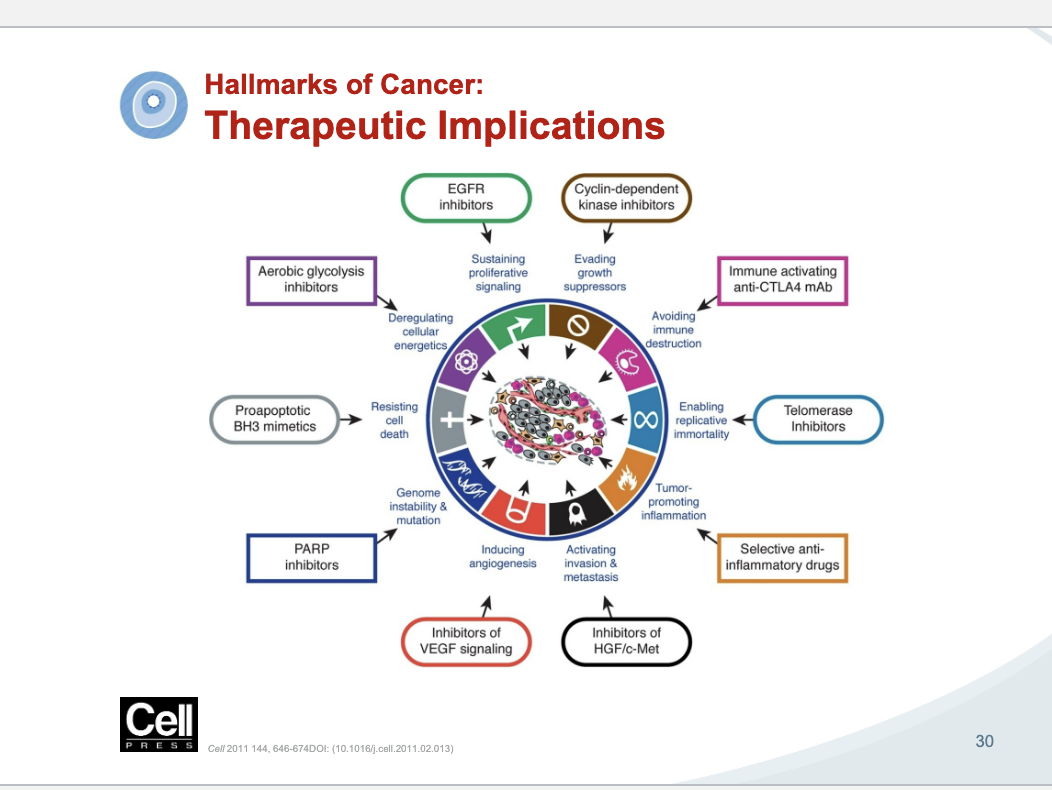
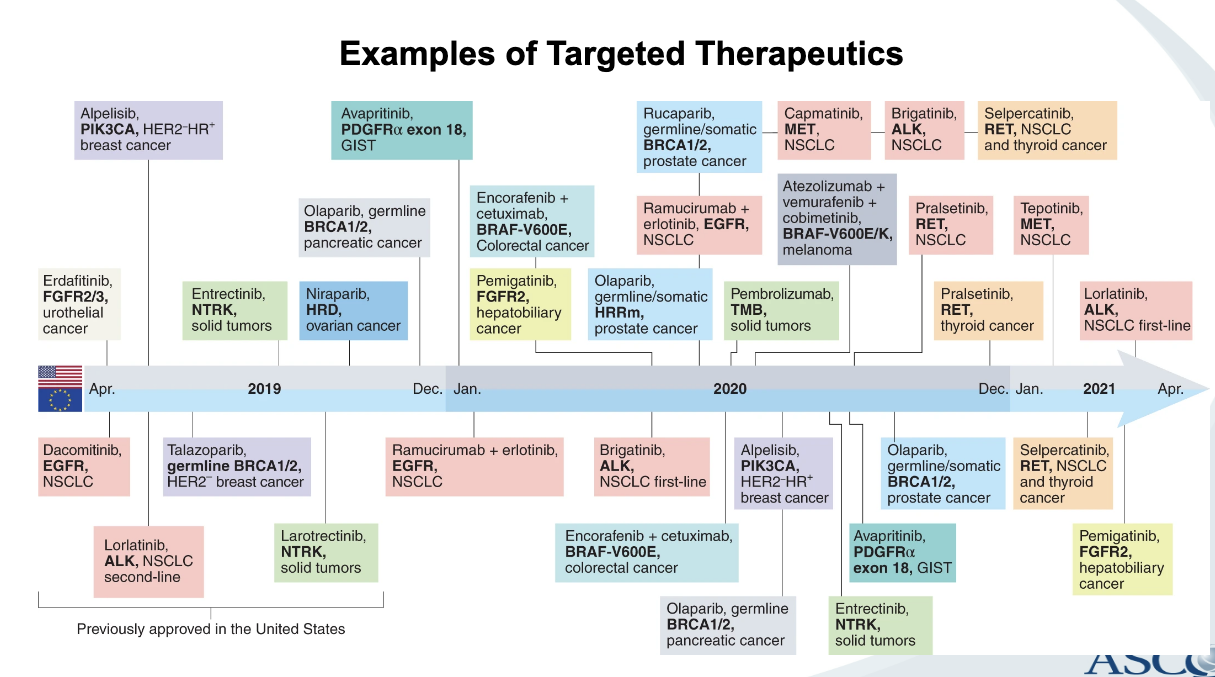
examples of targeted therapeutics
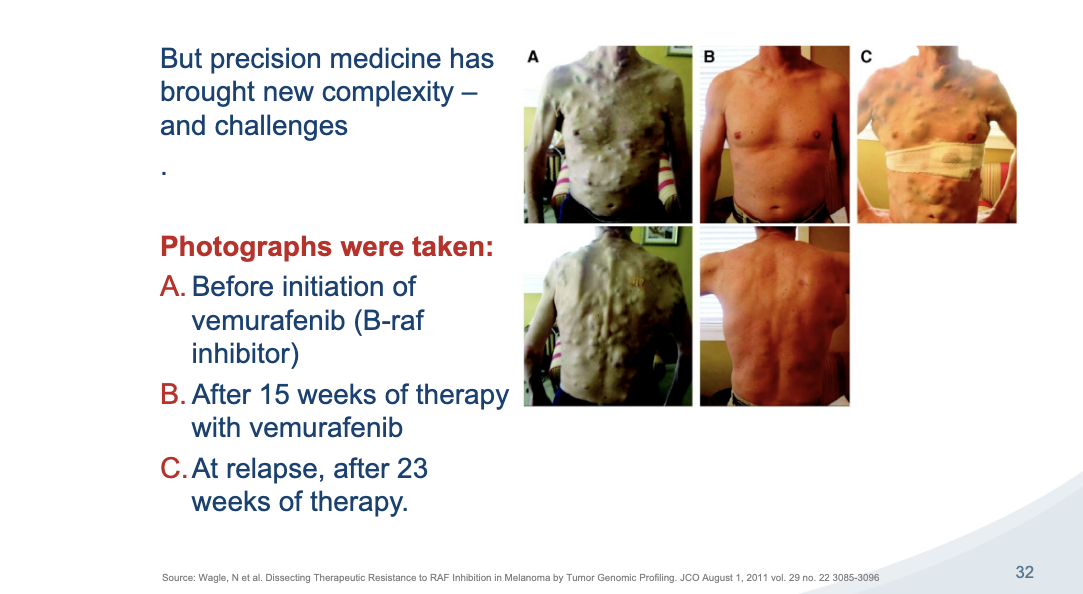
precision med challenges
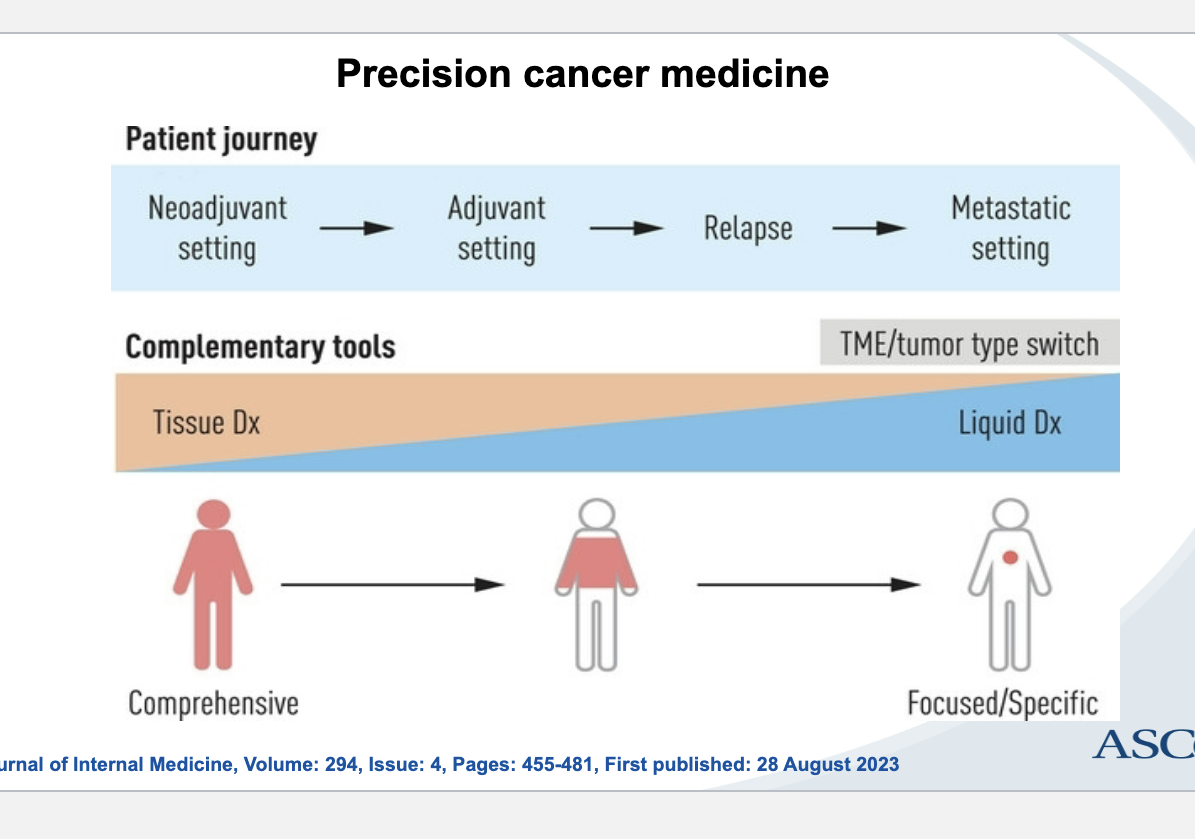
precision med pt journey
neoadjuvant setting (before therapy) → adjuvant therapy (after surgery/therapy) → relapse → metastatic setting
complementary tools
tissue diagnosis → liquid diagnosis
tissue = comprehensive
liquid = focused specific
oncologic biomarker case study: erbitux (cetuximab) for colorectal cancer
indications and usage
as a single agent, for treatment of EGFR-expressing metastatic colorectal cancer after failure of both irinotecan- and oxaliplatin-based regiments or in pts who are intolerant to irinotecan-based regimens
in combination with irinotecan, for treatment of EGFR-expressing metastatic colorectal carcinoma in patients who are refractor to irinotecan-based chemotherapy
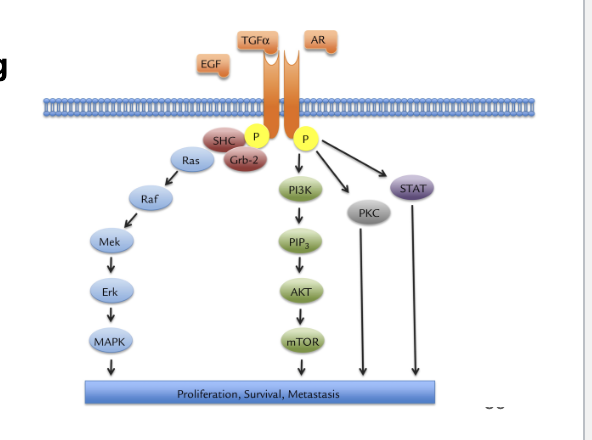
EGFR pathway
transmembrane growth factor receptor belonging to family of HER-related proteins
ligand binding triggers phosphorylation thru tyrosine kinase activity
phosphorylation triggers downstream signaling RAS/RAF/MEK/MAPK and PI3K/AKT pathways
induction of cancer-cell proliferation
blockade of apoptosis
activation of invasion and metastasis
stimulation of neovascularization
search for a predictive biomarker
erbitux initially granted approval only for EGFR-expressing patients with mCRC
60-80% mCRC tumors express EGFR proteins (as measured by IHC)
clinical analyses: NO correlation between extent of EGFR expression and response to anti-EGFR mABs
findings prompted intense research for predictive biomarkers for anti-EGFR mAb
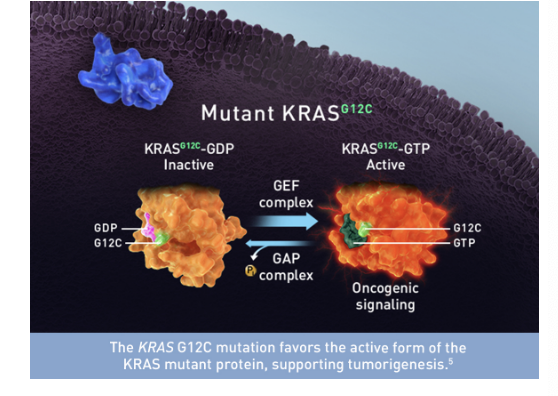
KRAS: the facts
KRAS = gene that makes KRAS protein which plays a key role in cell signaling
KRAS mutation occurs in about 40% of CRC patients
single nucleotide point mutations mainly in codons 12 and 13 of exon 2
mutation maintained throughout CRC development, progression and metastasis
KRAS mutation tested by direct sequencing or PCR
early clinical data on KRAS
2006 study
first reported link between KRAS mutation and lank of response to anti-EGFR mAb
multiple phase 2 studies of KRAS mutation status
cumulative data from 9 studies on 536 patients
KRAS mutation detected in 36% of pts
response rate (RR), tumor to tumor progression (TTP), progression-free survival (PFS), overall survival (OS) significantly better in WT (wild type) patients
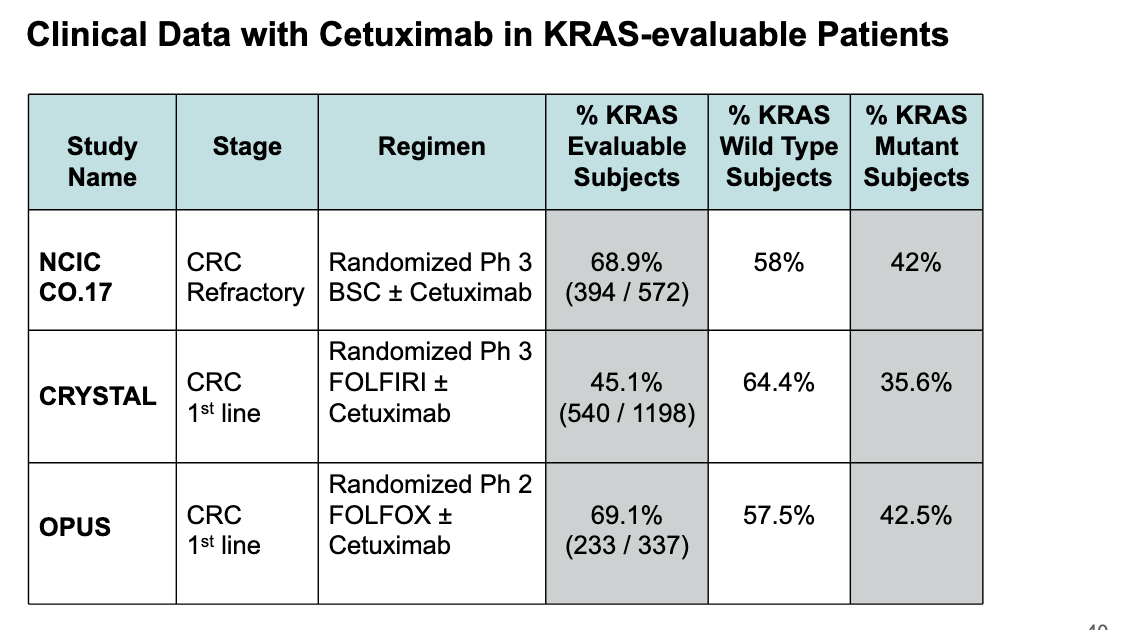
clinical data with cetuximab in KRAS-evaluable patients
across all 3 studies about 35%-42% of pts had KRAS mutations
data reinforces the importance of testing for KRAS mutations before using cetuximab → if pt has KRAS mutation, cetuximab is unlikely to help
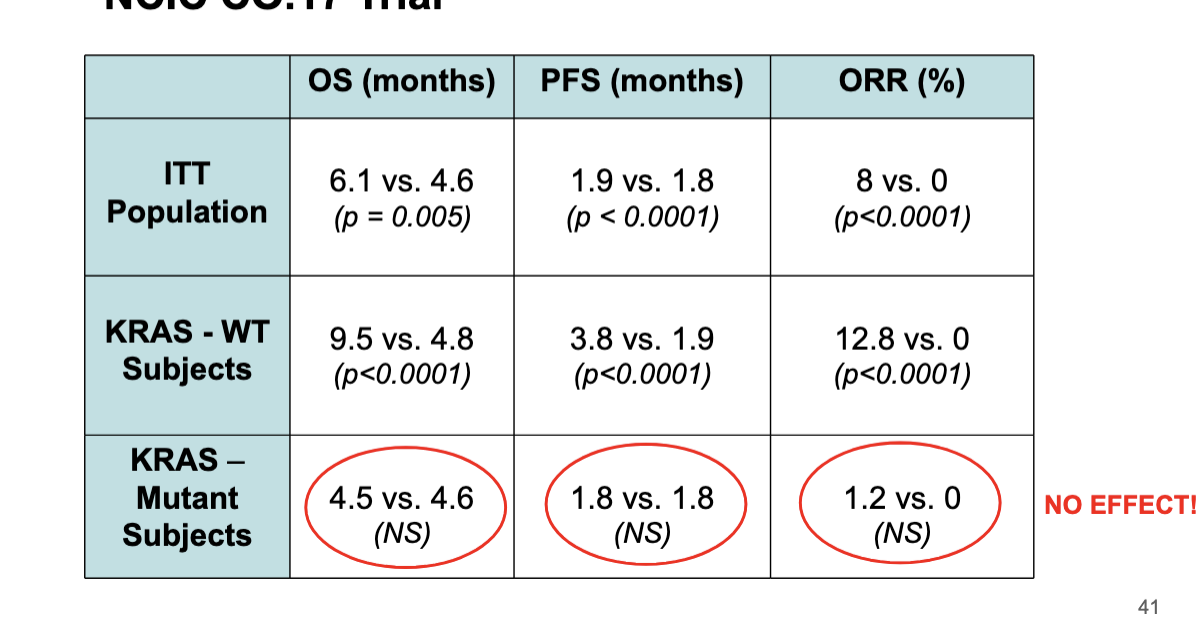
NCIC Co.18 trial
in KRAS wild type pts, cetuximab greatly improved OS, PFS, ORR%
in KRAS mutation pts → NO significant difference whether they got cetuximab or not
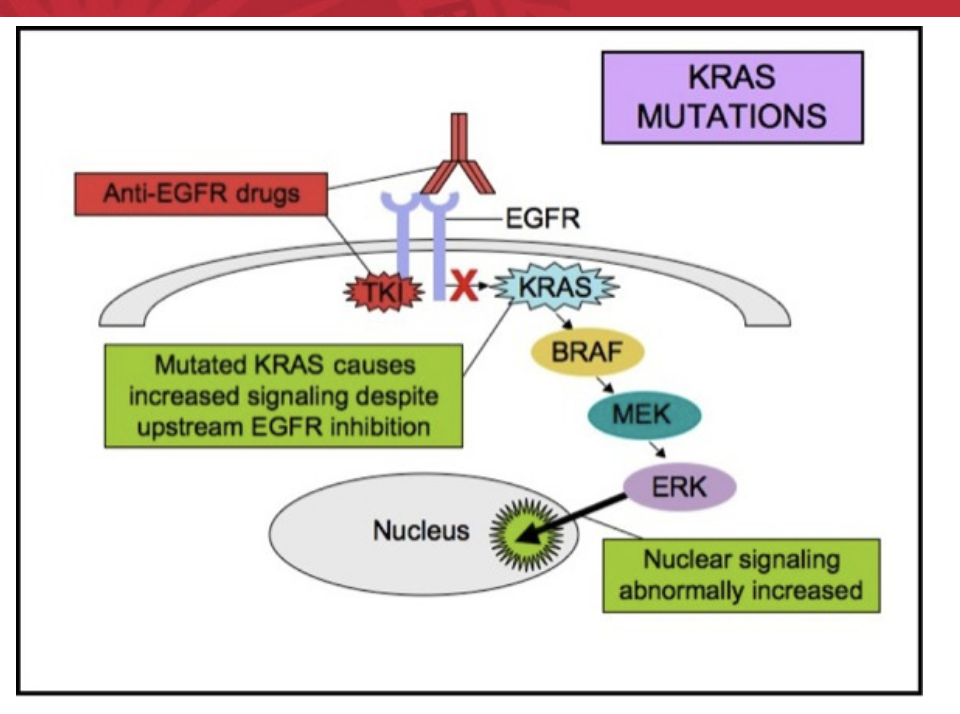
KRAS mutation mechanism
mutated KRAS causes increased signaling despite upstream EGFR inhibition by anti-EGFR drugs hence why drugs like cetuximab do NOT work
tumor with mutant/activated KRAS
if a tumor has a mutant/activated KRAS, it will grow in the presence of GF inhibitors
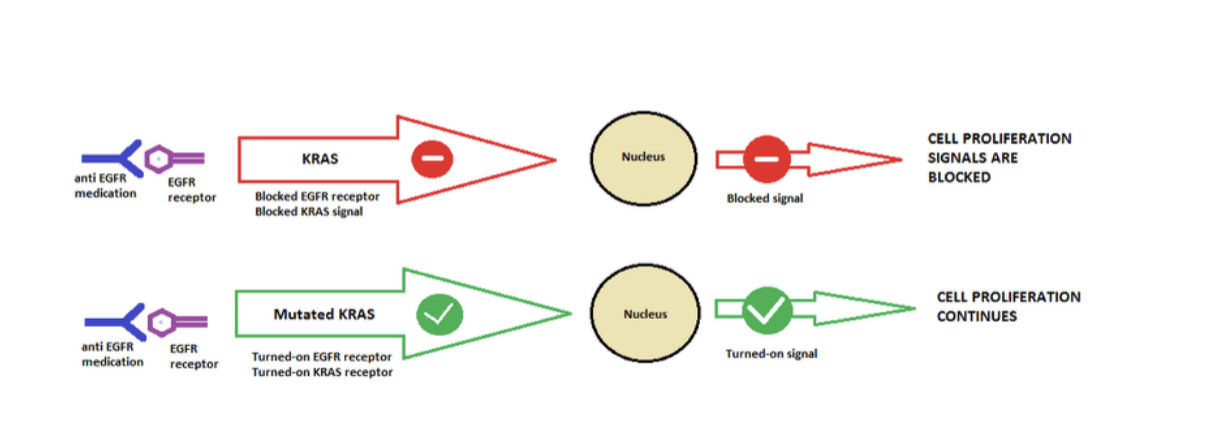
conclusions about KRAS
KRAS = example of how predictive biomarkers can spare patients of using drug WITHOUT potential benefit and with unwanted toxicity and cost
KRAS example likely to be observed by other therapies
identification of KRAS as a predictive biomarker = important step toward fulfilling promise of individualized treatment for mCRC
KRAS drug discovery
after 40 years, “druggable” after all
2021 → FDA approval of first KRAS-targeted therapy → sotorasib
2022 → adagrasib approved
both for mutant lung cancers

conclusions
ability to obtain full genomic data on a given tumor will allow us to make rational choices for therapy
functional genomics may provide help in choosing combination therapy
combinations will NOT be easy due to enhanced toxicities
cancer as a chronic disease is NOT a bad thing as long as we recognize rapid development of resistance and clonal evolution
outstanding questions: targeted cancer therapy
how can we best select ‘biomarker sets’ and properly apply them in clinical treatments of patients to identify optimal target patient subsets, to predict a patient’s response, resistance, and toxicity and to rapidly distinguish between responders and non-responders
is it possible to screen biomarkers using non-invasive approaches such as circulating tumor cells, circulating DNA, cytokines and chemokines? if not, how can we make technical breakthroughs to fully interpret the information of very limited patients’ biopsies?
is biomarker-based combinational therapy, that is, a ‘cocktail’ of highly-specific targeted drugs customized to individual patients according to their genetic aberrations, sufficient to largely overcome the resistance of targeted therapy?
basically saying: if we design a unique combo of targeted drugs for each pt based on their tumor’s genetic mutations, can we prevent the cancer from becoming resistant and make the treatment work better
how can innovative biomarker-based clnical design, that is, stratification of patients, assignment of specific drug therapy and adaptive trial designs, increase the translation of targeted drugs from bench to bedside?
given that the tumor microenvironment has an enormous impact on tumor development, how can we develop models that accurately reflect the tumor microenvironment, in particular the human immune syste, for drug discovery?
cancer characteristics
emerging hallmarks
deregulating celular energetics
avoiding immune destruction
enabling characteristics
genome instability and mutation
tumor-promoting inflammation
immunosuppression and canccer
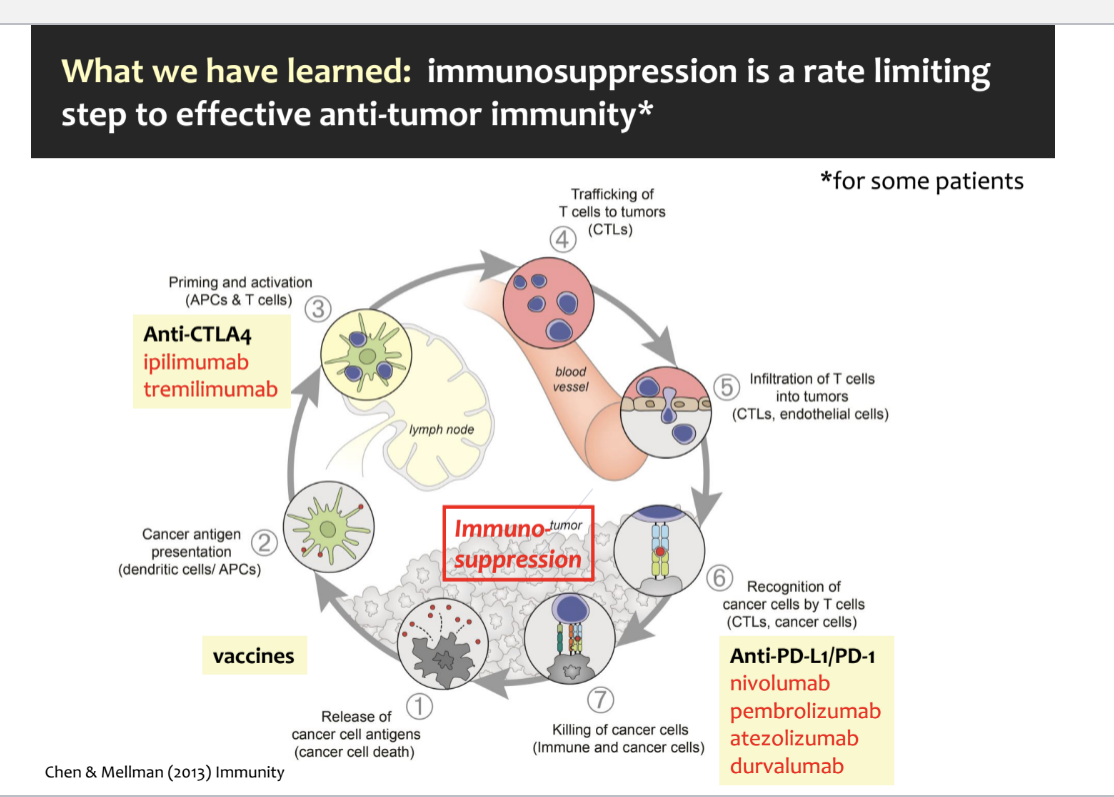
immunosuppression is a rate limiting step to effective anti-tumor immunity (for some patients)
key steps in the cancer-immunity cycle
release of cancer cell antigens → tumor cells die and release antigens
cancer antigen presentation → dendritic cells or APCs capture these antigens and present them to the immune system
priming and activation → APCs activate native T-cells in lymph nodes; anti-CTLA-4 drugs work here by preventing the “off” signals that would normally suppress T cell activation
trafficking of T cells to tumors → activated cytotoxic T cells (CTLs) travel thru the blood stream toward the tumor
infiltration of T cells into the tumor → cytotoxic T cells move into the tumor tissue
recognition of cancer cells by T cells → CTLs recognize cancer cells by binding to antigens presented on their surface; but many cancer cells express PD-L1 which binds to PD-1 on T cells, turning them off; anti-PD1/PD-L1 drugs block this signal off so T cells can stay active
killing of cancer cells → CTLs kill the cancer cells
where immunosuppression happens
tumor-induced immunosuppression can interfere at multiple steps:
prevent antigen presentation
stop T-cell activation
prevent T cells from reaching tumors
supress T cell killing ability
rise of immunotherapy
new therapeutics
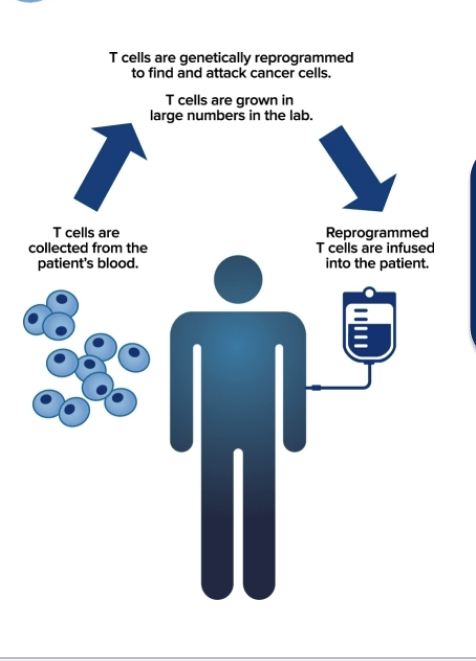
CART cell therapy
customized vaccines
process of immunotherapy
T cells are collected from the patient’s blood
T cells are genetically reprogrammed to find and attack cancer cells; T cells are grown in large numbers in lab
reprogrammed T cells are infused into the patient
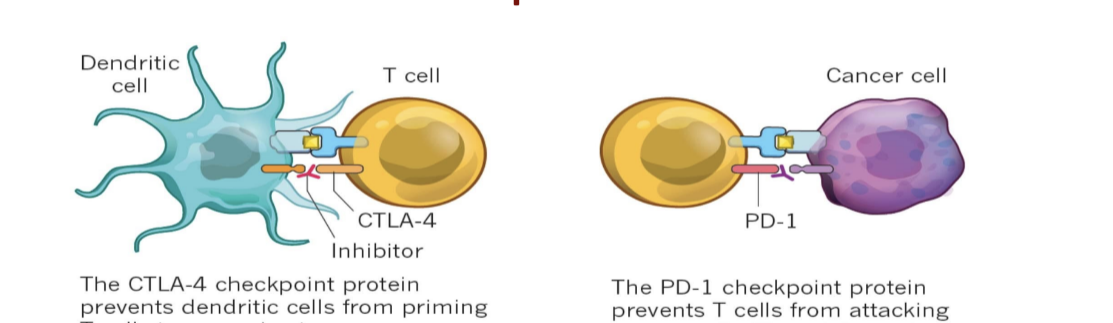
immune checkpoint inhibitors
CTLA-4 blockage (e.g. ipillimumab)
CTLA-4 checkpoint protein prevents dendritic cells from priming T cells to recognize tumors → inhibitor drug blocks the checkpoint
PD-1 blockage (e.g. nivolumab, pembrolizumab, atezolizumab)
PD-1 checkpoint protein prevents T cells from attacking cancer cells → inhibitor drug allows T cells to act
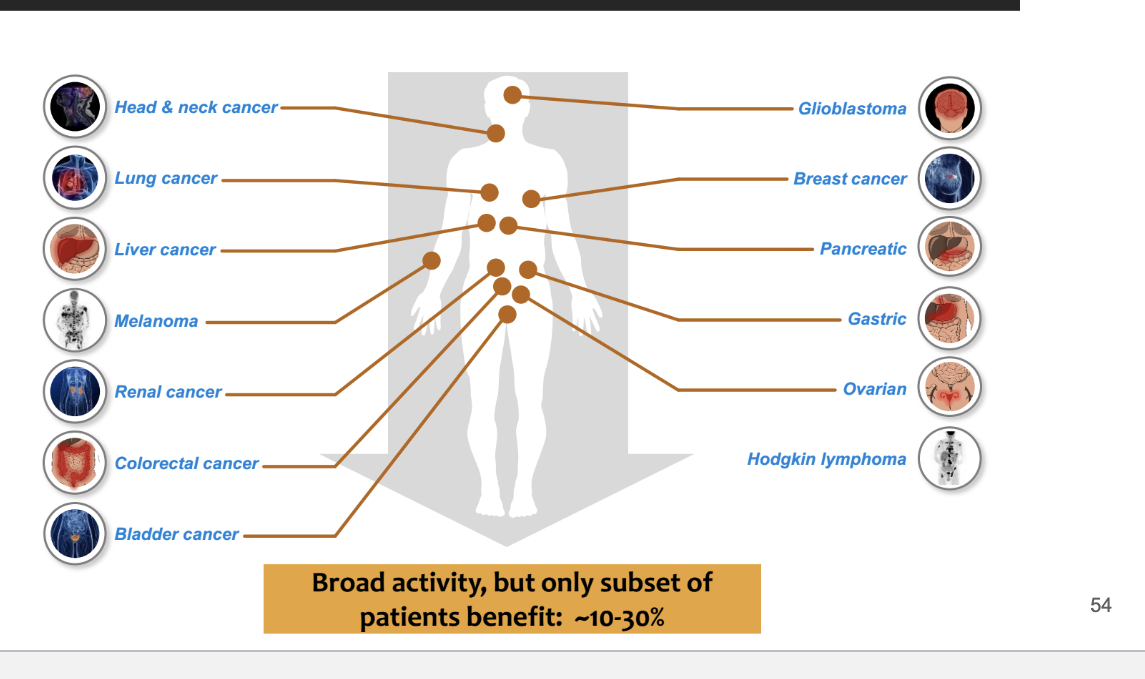
broad activity for anti-PD-L1/PD-1 in human cancer
basically affects a bunch of cancer like
head and neck cancer
lung cancer
gastric
broad activity but only subset of patients benefit → ~10-30%
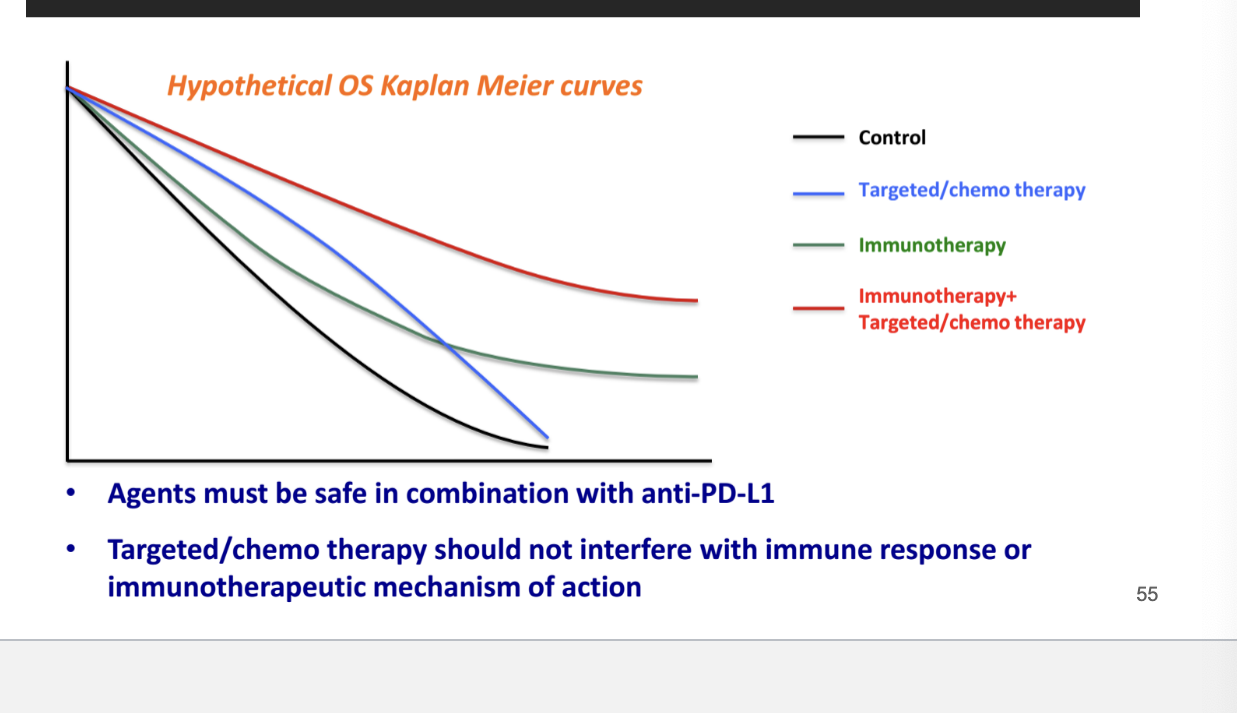
combo of immunotherapeutics or immunotherapeutics with SOC/targeted therapies
shows that best survival outcomes = immunotherapy + targeted/chemotherapy
agents must be safe in combination with anti-PD-L1 (immunotherapy)
targeted/chemotherapy should NOT interfere with immune response or immunotherapeutic MOA
conclusions
personalized medicine can be regraded as the 21st century’s answer to the rational use of drugs → right drug for the right pt
key drivers in personalized medicine:
molecular diagnostics
academic groups
patient advocacy groups
authorities and health insurance companies
implementing personalized medicine into clinical practice will be a challenge that needs to address various other issues such as regulatory requirements, reimbursement, education and logistics