MCAT Biochemistry - RNA and the Genetic Code
1/103
Earn XP
Description and Tags
383
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
104 Terms
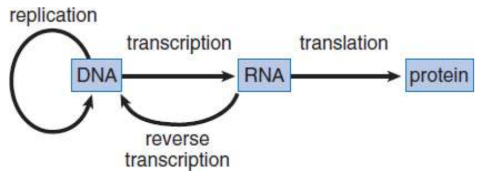
central dogma of molecular biology
major steps involved in the transfer of genetic information
gene
a unit of DNA that encodes a specific protein or RNA molecule; can be expressed through transcription and translation
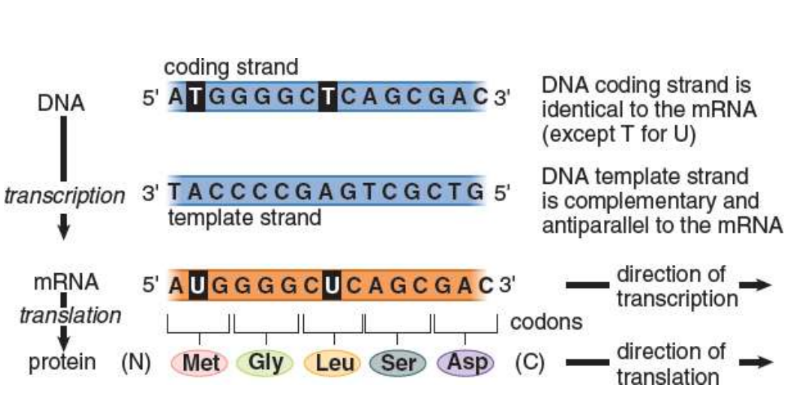
direction of transcription
5’ to 3’
direction of translation
5’ to 3’
Messenger RNA (mRNA)
carries the information specifying the amino acid sequence of the protein to the ribosome; transcribed from template DNA strands by RNA polymerase enzymes in the nucleus; may undergo a host of posttranscriptional modifications
codons
three-nucleotide segments that designate a specific amino acid
monocistronic
each eukaryotic mRNA molecule translates into only one protein product
polycistronic
prokaryotic mRNA may code multiple proteins; starting the process of translation at different locations in the mRNA can result in different proteins
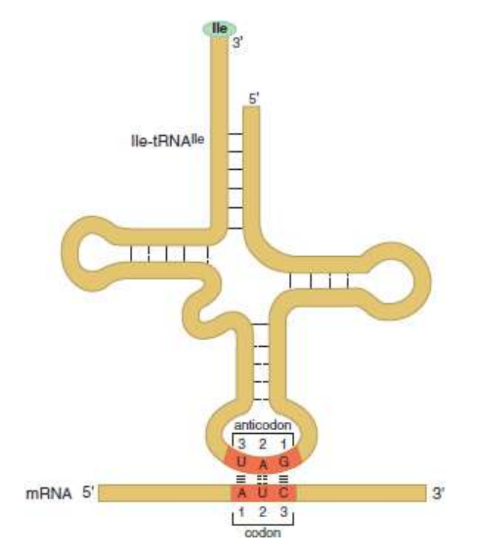
Transfer RNA (tRNA)
responsible for converting the language of nucleic acids to the language of amino acids and peptides; contains a folded strand of RNA that includes a three-nucleotide anticodon that recognizes and pairs with the appropriate codon on an mRNA molecule; found in the cytoplasm

charged/activated tRNA
connected to an amino acid at the 3’ end by aminoacyl-tRNA synthetase
aminoacyl-tRNA synthetase
each amino acid is attached to tRNA at the 3’ end (CCA sequence) with this enzyme; requires two ATP; bond supplies energy for peptide bond
Ribosomal RNA (rRNA)
synthesized in the nucleolus; functions as an integral part of the ribosomal machinery used during protein assembly in the cytoplasm; helps catalyze the formation of peptide bonds; splices out its own introns within the nucleus
ribozymes
enzymes made of RNA molecules instead of peptides
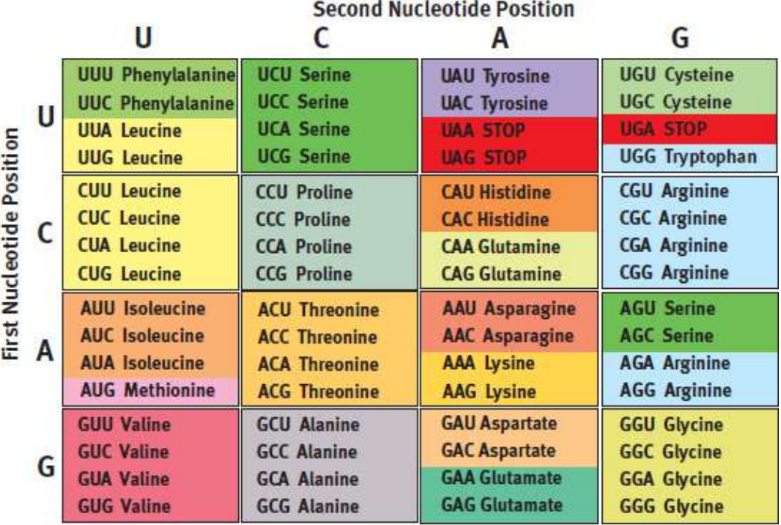
Genetic code tables
easy way to determine the amino acid that is translated from each mRNA codon (4³ or 64 codons); unambiguous - each codon is specific for one and only one amino acid; degenerate - multiple codons code for each of 20 amino acids; universal - same across all species
anticodon
on tRNA; recognises codon on mRNA; antiparallel. orientation
start codon
signals the beginning of translation
methionine (AUG)
stop codons
encode for termination of protein translation; no amino acids on the associated tRNA
UGA, UAA, UAG
degeneracy
more than one codon can specify a single amino acid (except methionine and typtophan)
wobble position
for most amino acids, the first two bases are usually the same, and the third base in the codon is variable; protect against mutation
silent/degenerate mutation
no effect on the expression of the amino acid and therefore no adverse effects on the polypetide sequence

point mutation
affects one nucleotide in a codon
expressed mutations
can affect the primary amino acid sequence of the protein
Missense mutation
one amino acid substitutes for another

Nonsense (truncation) mutation
the codon now encodes for a premature stop codon

reading frame
three nucleotides of a codon
frameshift mutation
some number of nucleotides are added to or deleted from the mRNA sequence, shifting the reading frame, usually resulting in changes in the amino acid sequence or premature truncation of the protein; typically more serious than point mutations

Cystic fibrosis
most commonly caused by a frameshift mutation; results in a defective chloride ion channel that can’t reach cell membrane, leading to blocked passage of salt and water into and out of cells; cells that line the passageways of the lungs, pancreas, and other organs produce an abnormally thick, sticky mucus that traps bacteria, increasing the likelihood of infection in patients
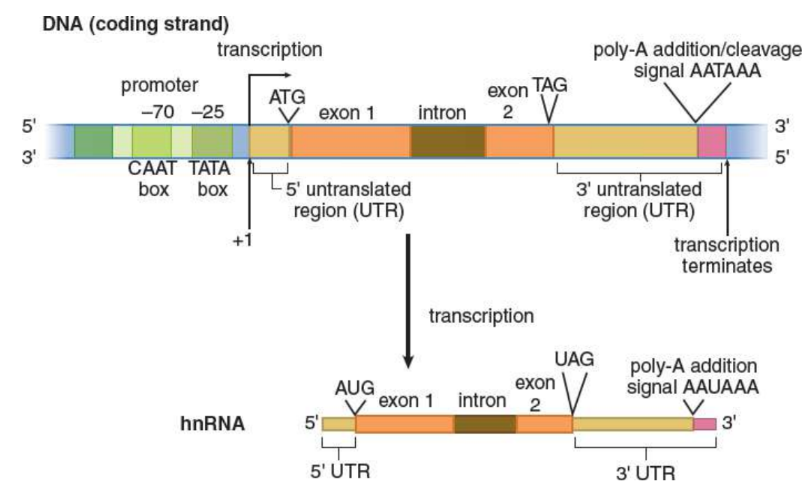
transcription
creation of mRNA from a DNA template
helicase
unzips/opens DNA during transcription
topoisomerase
releives tension in DNA strands during transcription
template (antisense) strand
antiparallel and complementary mRNA synthesized from this strand
promoter regions
RNA polymerase locates genes by searching for specialized DNA regions
RNA polymerase II
transcription enzyme in eukaryotes; produces hnRNA and small nuclear RNA (snRNA); doesn’t need primer; read 3’ to 5’, buikds 5’ to 3’; promoter region: TATA box
TATA box
binding site for RNA polymerase II in transcription; named for its high concentration of thymine and adenine bases
Transcription factors
help the RNA polymerase locate and bind to this promoter region of the DNA
RNA polymerase I
located in the nucleolus and synthesizes rRNA
RNA polymerase III
located in the nucleus and synthesizes tRNA and some rRNA
coding (sense) strand
not used as a template during transcription; identical to the mRNA transcript except that all the thymine nucleotides in DNA have been replaced with uracil in the mRNA molecule
base numbering system
identify the location of important bases in the DNA strand relative to a gene
first base transcribed = +1
upstream = negative
downstream = positive
no zero
TATA box ~ -25
heterogeneous nuclear RNA (hnRNA)
unprocessed mRNA
Posttranscriptional Processing
Intron/exon splicing
5′ cap
3′ poly-A tail
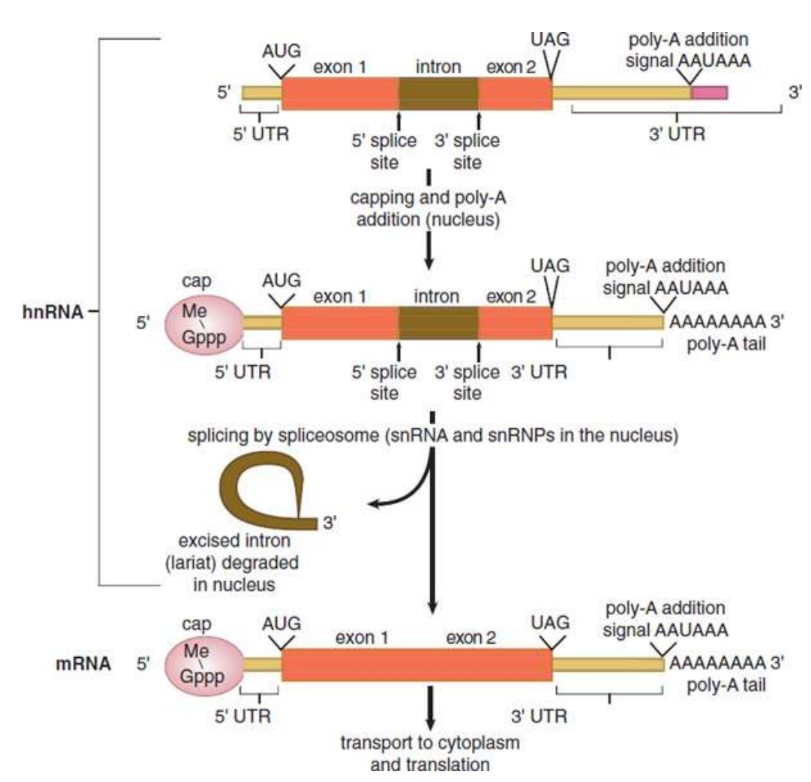
introns
noncoding sequences
exons
coding sequences
spliceosome
large snRNP/snRNA complex found primarily within the nucleus of eukaryotic cells that removes introns; recognizes both the 5′ and 3′ splice sites of the i; excised via lariat and degradedntrons
small nuclear RNA (snRNA)
a class of small RNA molecules that are found within certain complexes of the cell nucleus in eukaryotic cells
small nuclear ribonucleoproteins (snRNPs)
forms a complex with snRNA to make spliceosomes
lariat
lasso-shaped structure
7-methylguanylate triphosphate cap
5’ end of hnRNA; added during the process of transcription; recognized by the small eukaryotic ribosomal subnunit as the binding site; protects the mRNA from degradation in the cytoplasm
polyadenosyl (poly-A) tail
3’ end of hnRNA; protects the message against rapid degradation; assists with export of the mature mRNA from the nucleus
Untranslated regions (UTRs)
still exist at the 5′ and 3′ edges of the transcript because the ribosome initiates translation at the start codon (AUG) and will end at a stop codon (UAA, UGA, UAG)
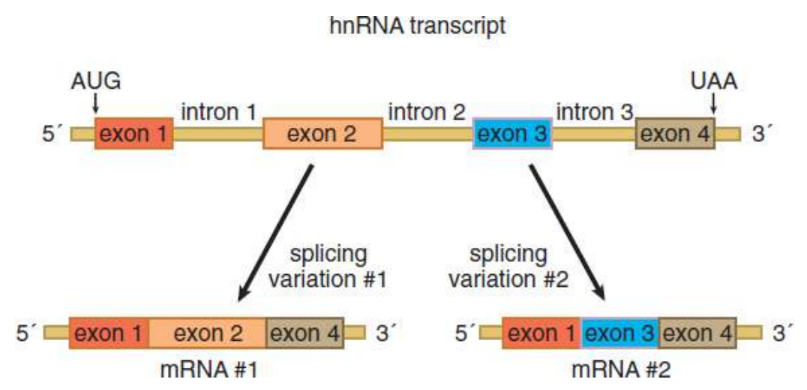
alternative splicing
the primary transcript of hnRNA may be spliced together in different ways to produce multiple variants of proteins encoded by the same original gene
nuclear pores
hole sin the nuclear membrane mRNA leaves through
translation
converting the mRNA transcript into a functional protein; requires mRNA, tRNA, ribosomes, amino acids, and energy in the form of GTP
ribosome
composed of proteins and rRNA; composed of large and small subunits that only bind together during protein synthesis; bring the mRNA message together with the charged aminoacyl-tRNA complex to generate the protein; three binding sites in the ribosome for tRNA: APE
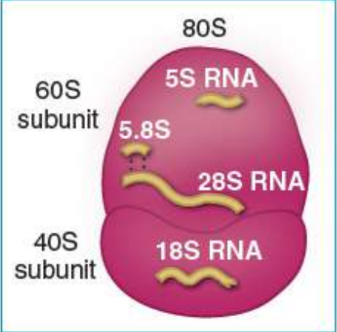
eukaryotic ribosomal subunits
28S, 18S, and 5.8S rRNAs gens in nucleolus → 28S, 18S, and 5.8S rRNAs transcribed by RNA polymerase I in one unit = 45S → processed into 18s rRNA of 40S (small) subunit and 28S and 5.8S rRNAs of the 60S (large) subunit
RNA polymerase III transcribes the 5S rRNA → 60S subunit
40S + 60S join during protein synthesis to form the whole 80S ribosome
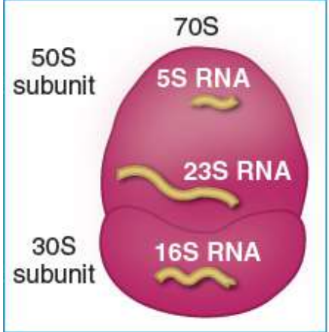
prokaryotic ribosomal subunits
50S and 30S large and small subunits create the complete 70S ribosome
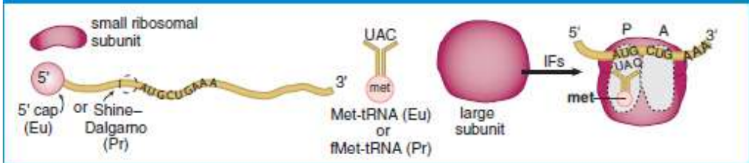
initiation
beginning of translation
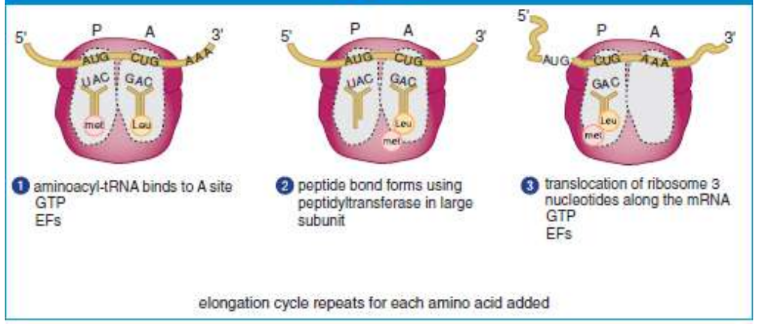
elongation
continuation of translation; three-step cycle that is repeated for each amino acid added to the protein after the initiator, methionine; moves in the 5′ to 3′ direction, amino (N-) to carboxyl (C-) terminus
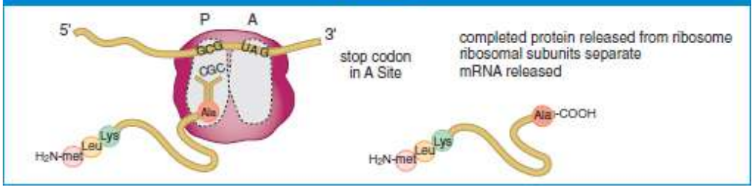
termination
end of translation; hydrolyses polypeptide chain, released from the tRNA in the P site, and the two ribosomal subunits will dissociate
Shine–Dalgarno sequence
small prokaryotic ribosomal subunit binds in the 5’ UTR of mRNA
initiator tRNA
binds to start codon
prokaryotes: N-formylmethionine (fMet)
eukaryotes: methionine
initiation factors (IF)
helps large subunit bind to small subunit, forming the completed initiation complex; not permanently associated with the ribosome
A (aminoacyl) site
holds the incoming aminoacyl-tRNA complex
P (polypeptide) site
holds the tRNA that carries the growing polypeptide chain; also where the first amino acid (methionine) binds because it is starting the polypeptide chain
peptide bond
bond between amino acids; formed as the polypeptide is passed from the tRNA in the P site to the tRNA in the A site; catalysed by peptidyl transferase
peptidyl transferase
an enzyme that is part of the large subunit; catalyses peptide bonds; uses GTP
E site
where the now inactivated (uncharged) tRNA pauses transiently before unbinding and exiting the ribosome
Elongation factors (EF)
assist by locating and recruiting aminoacyl-tRNA along with GTP, while helping to remove GDP once the energy has been used
signal sequences
designate a particular destination for the protein
secretion: ribosome → endoplasmic reticulum (ER) → translated directly into the lumen of the rough ER → Golgi apparatus → vesicle → exocytosis
other pathways: nucleus, lysosomes, cell membrane
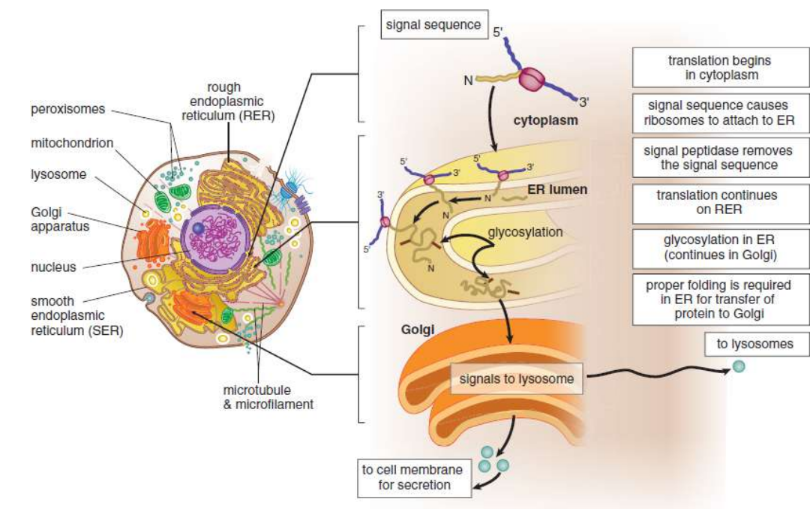
release factor (RF)
binds to the termination codon; water molecule added; allows peptidyl transferase and termination factors to hydrolyze the completed polypeptide chain from the final tRNA
chaperones
specialised class of proteins that assist in the protein-folding process
cleavsge event
protein cleaved from a larger, inactive peptide to achieve its active form
ex. insulin, signal sequence
quaternary structure
subunits come together to form the functional protein
ex. hemoglobin
Phosphorylation
addition of a phosphate group (PO42−) by protein kinases to activate or deactivate proteins; most commonly seen with serine, threonine, and tyrosine
Carboxylation
addition of carboxylic acid groups to proteins, usually to serve as calcium-binding sites
Glycosylation
addition of oligosaccharides as proteins pass through the ER and Golgi apparatus to determine cellular destination
Prenylation
addition of lipid groups to certain membrane-bound enzymes
operon
cluster of genes transcribed as a single mRNA in prokaryotes
Jacob–Monod model
used to describe the structure and function of operons; contain structural genes, an operator site, a promoter site, and a regulator gene
structural gene
codes for the protein of interest
operator site
nontranscribable region of DNA that is capable of binding a repressor protein; Upstream of the structural gene
promoter site
provides a place for RNA polymerase to bind; further upstream than operator site
regulator gene
codes for a protein known as the repressor, futher upstream than promoter
inducible systems
the repressor is bonded tightly to the operator system and thereby acts as a roadblock; an inducer must bind the repressor protein so that RNA polymerase can move down the gene
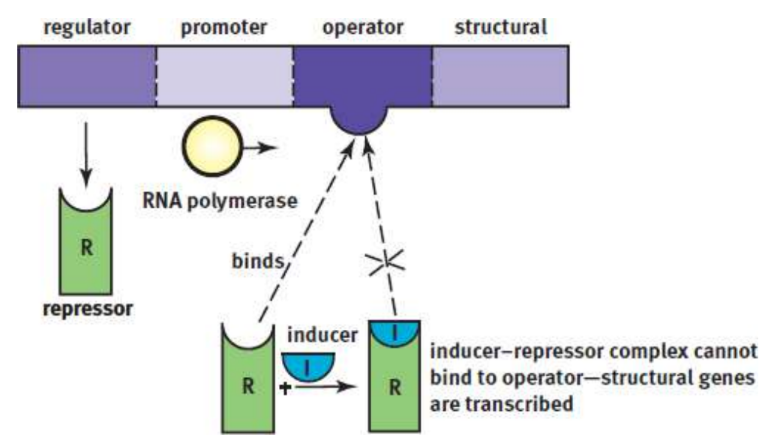
negative control mechanisms
the binding of a protein reduces transcriptional activity
lac operon
codes for lactase and lactose-specific transport proteins; induced by the presence of lactose; most efficient when lactose is high and glucose is low
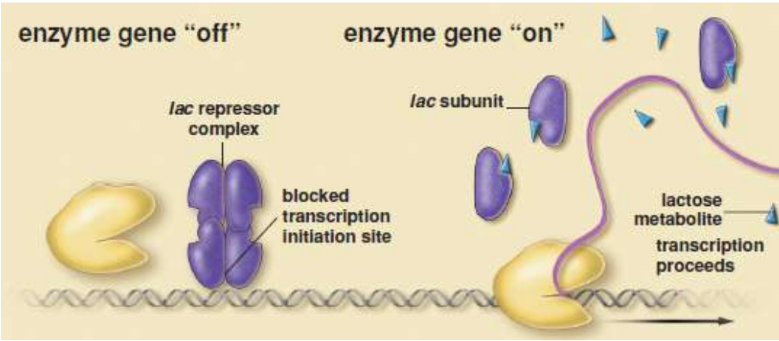
catabolite activator protein (CAP)
transcriptional activator used by E. coli when glucose levels are low; Falling levels of glucose → increase in cAMP → binds to CAP → conformational change → bind the promoter region of the operon → increasing transcription of the lactase gene
positive control mechanisms
the binding of a molecule increases transcription of a gene
Repressible systems
allow constant production of a protein product; repressor made by the regulator gene is inactive until it binds to a corepressor; often engative feedback
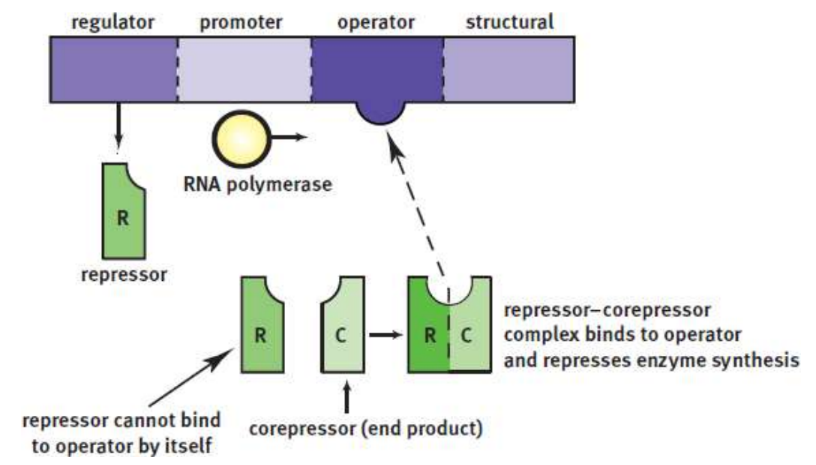
trp operon
tryptophan corepressor; stops production of tryptophan in cell if too much made/in local environment
Transcription factors
transcription-activating proteins that search the DNA looking for specific DNA-binding motifs
DNA-binding domain
binds to a specific nucleotide sequence in the promoter region or to a DNA response element to help in the recruitment of transcriptional machinery
response element
a sequence of DNA that binds only to specific transcription factors
activation domain
allows for the binding of several transcription factors and other important regulatory proteins, such as RNA polymerase and histone acetylases, which function in the remodeling of the chromatin structure
amplified
increased
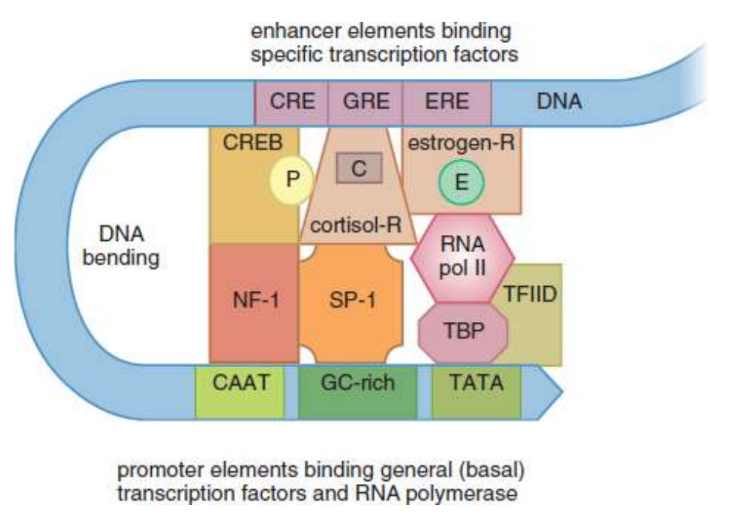
enhancer
Several response elements grouped together; allows for the control of one gene’s expression by multiple signals; up to 1000 base pairs away from the gene they regulate and can even be located within an intron
ex. cyclic AMP (cAMP) and cyclic AMP response element-binding protein (CREB); cortisol and glucocorticoid (cortisol) receptor; estrogen and estrogen receptor
Gene Duplication
increase the expression of a gene product; duplicated on same chromosome or in parallel (repeated replication)
Heterochromatin
tightly coiled DNA that appears dark under the microscope; its tight coiling makes it inaccessible to the transcription machinery, so these genes are inactive
Euchromatin
looser and appears light under the microscope; the transcription machinery can access the genes of interest, so these genes are active
histone acetylases
acetylate lysine residues found in the amino terminal tail regions of histone proteins