MCAT Biochemistry Chapter 7: RNA and the Genetic Code
1/103
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
104 Terms
Central Dogma of Biology: chain of transformation of genetic information starting from nucleus
(replication) -> DNA -> (transcription/reverse transcription) ➝ RNA -> (translation)➝ Protein

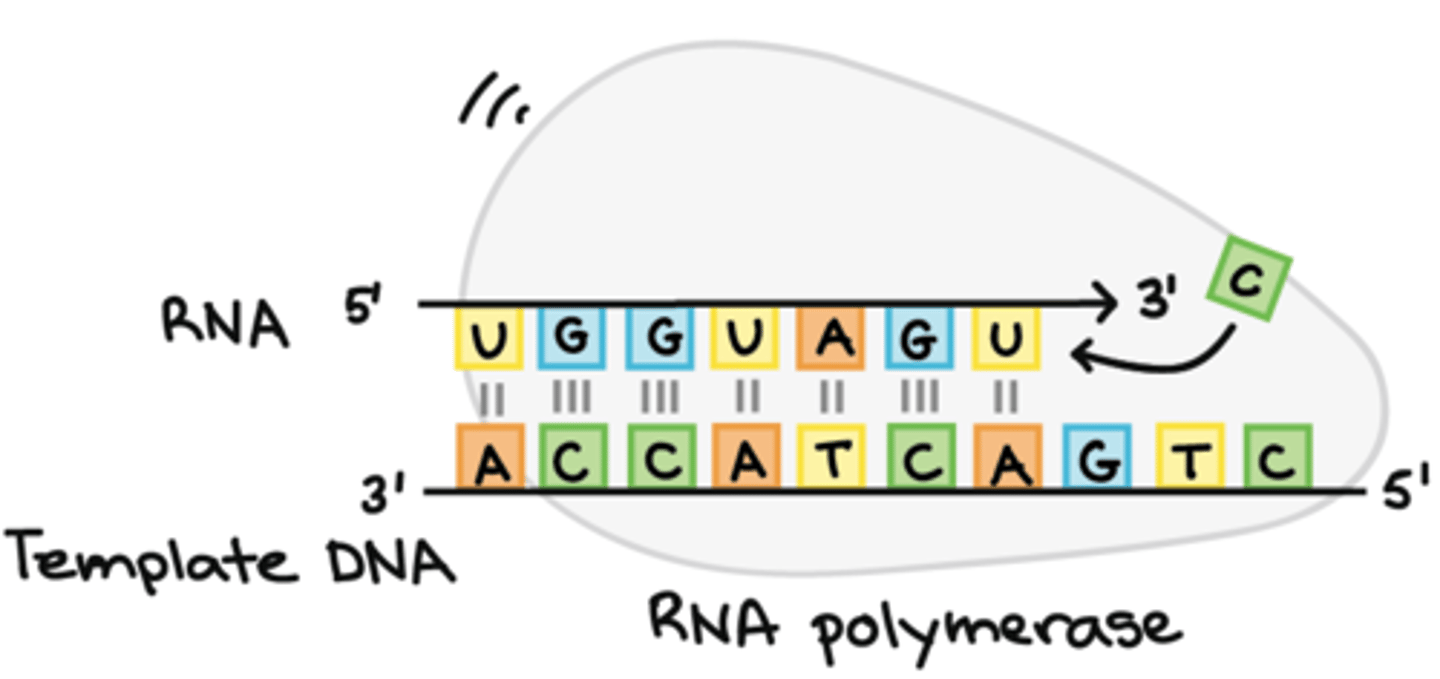
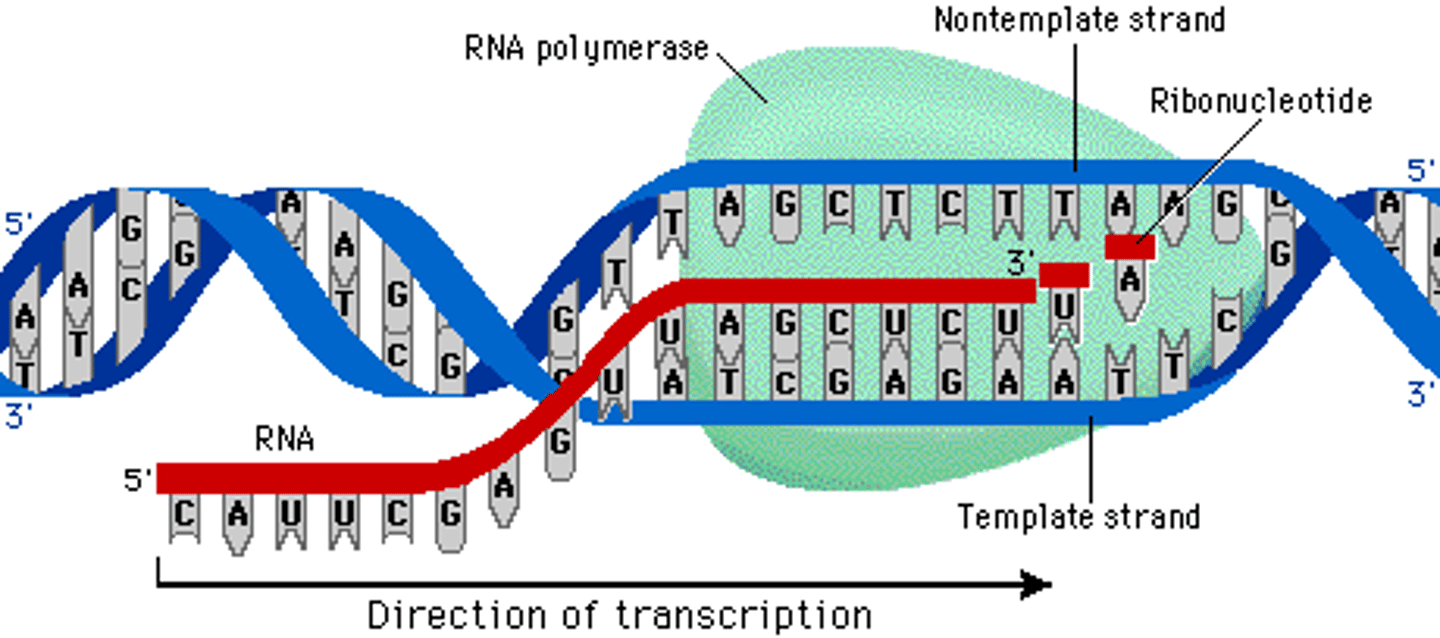
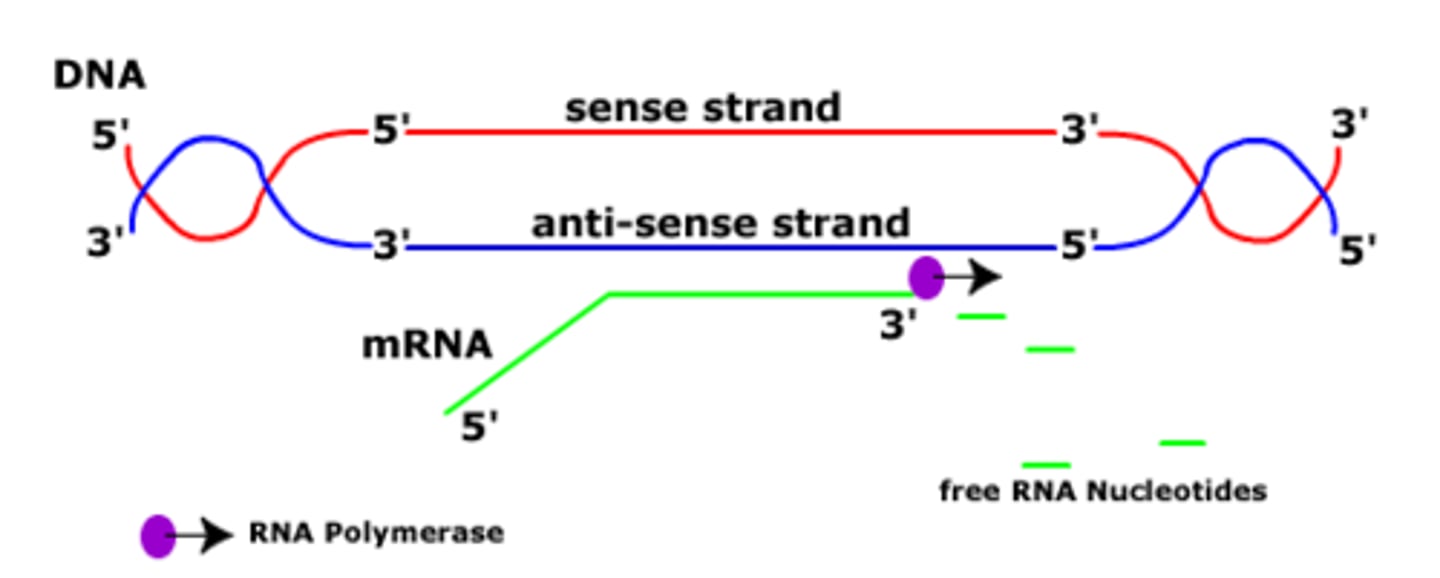
Direction of transcription
5' ➝ 3', building a complementary strand to the DNA
It synthesizes the RNA strand in the 5' to 3' direction, while reading the template DNA strand in the 3' to 5' direction.
Direction of translation of the RNA
The mRNA is single-stranded and therefore only contains three possible reading frames, of which only one is translated. The codons of the mRNA reading frame are translated in the 5′→3′ direction into amino acids by a ribosome to produce a polypeptide chain.
Direction of synthesis of a protein: ______ to ______ term
(N-terminal/C-terminal)
N to C term
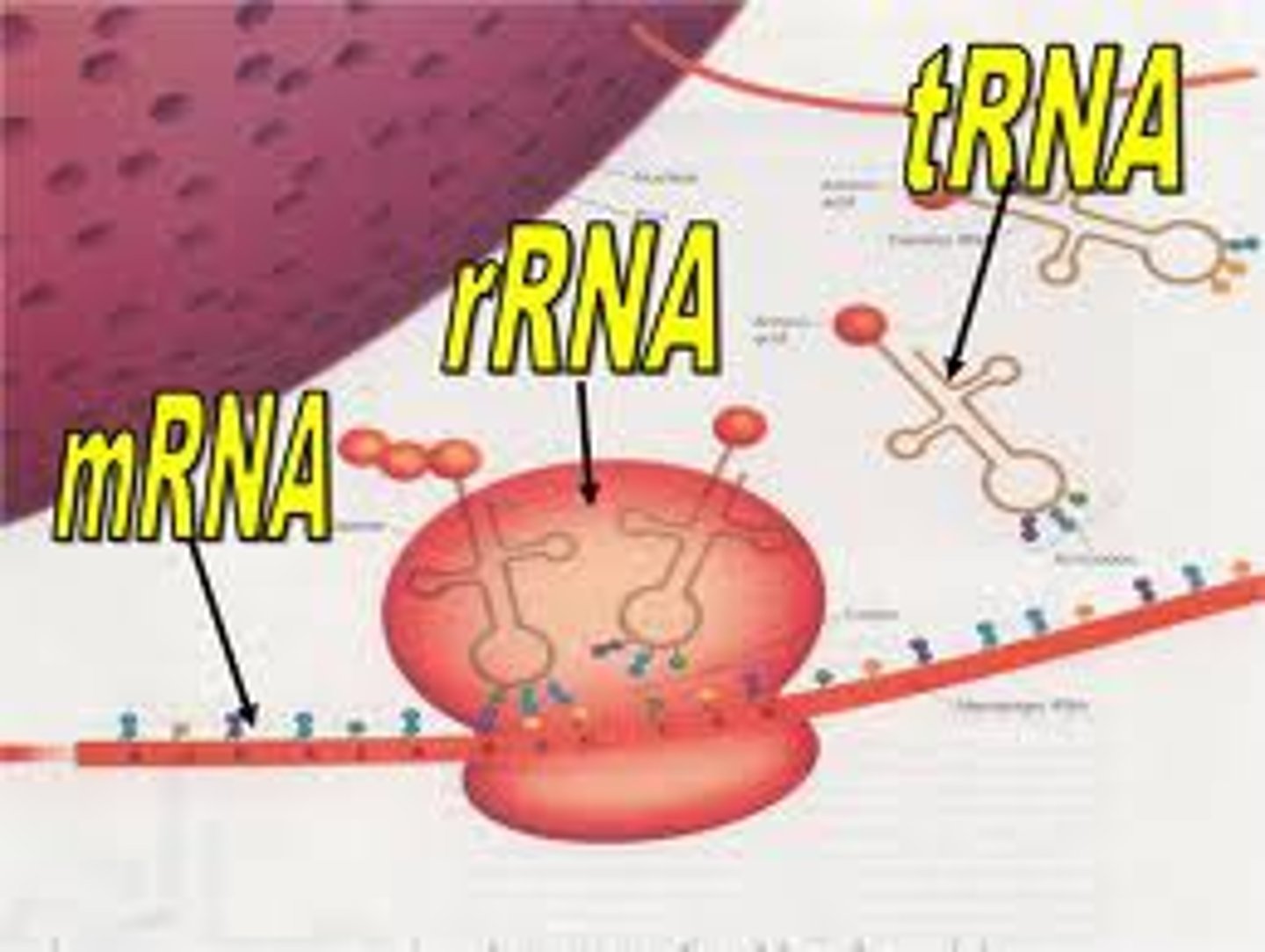
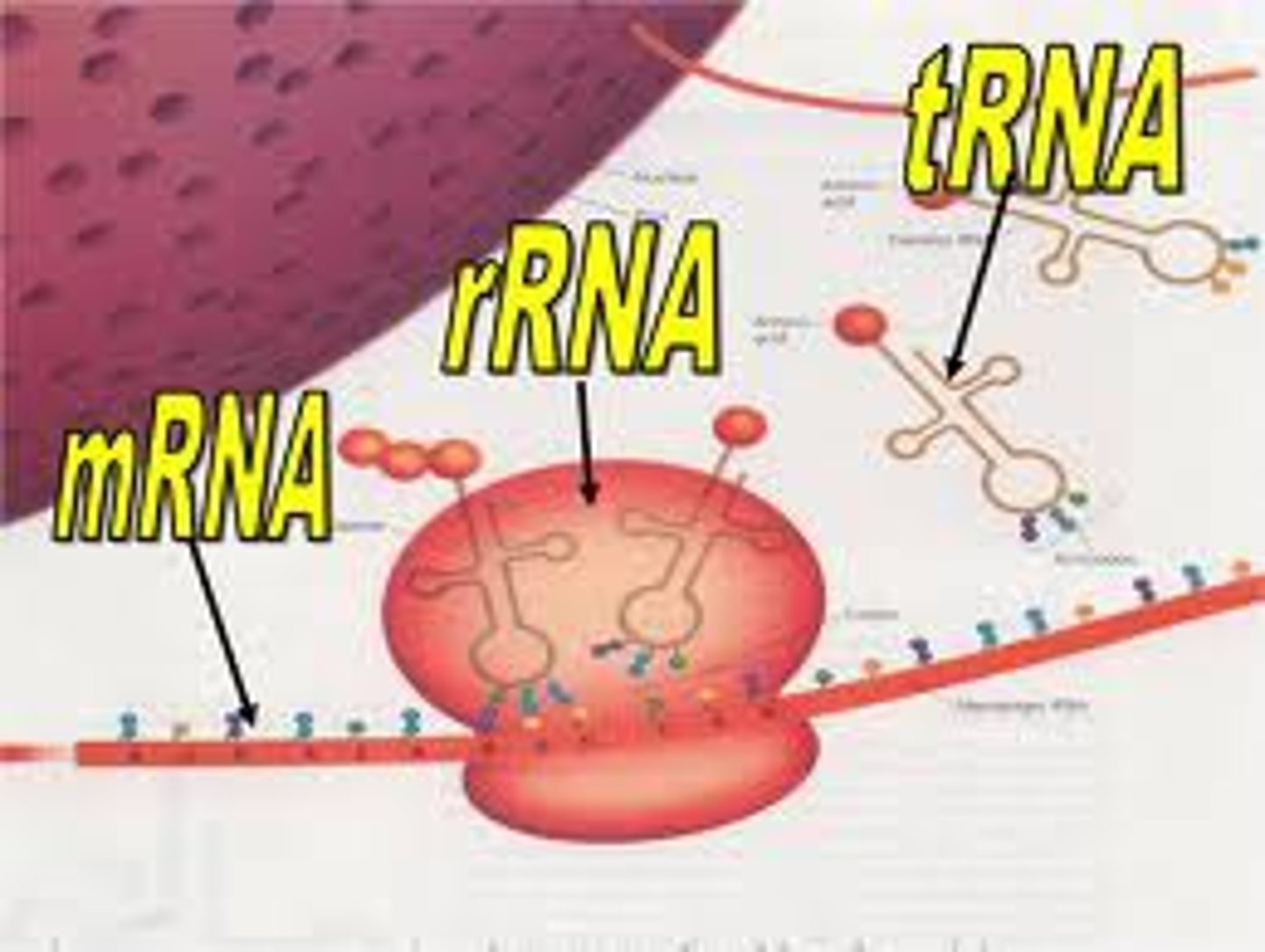
Main 3 types of RNA
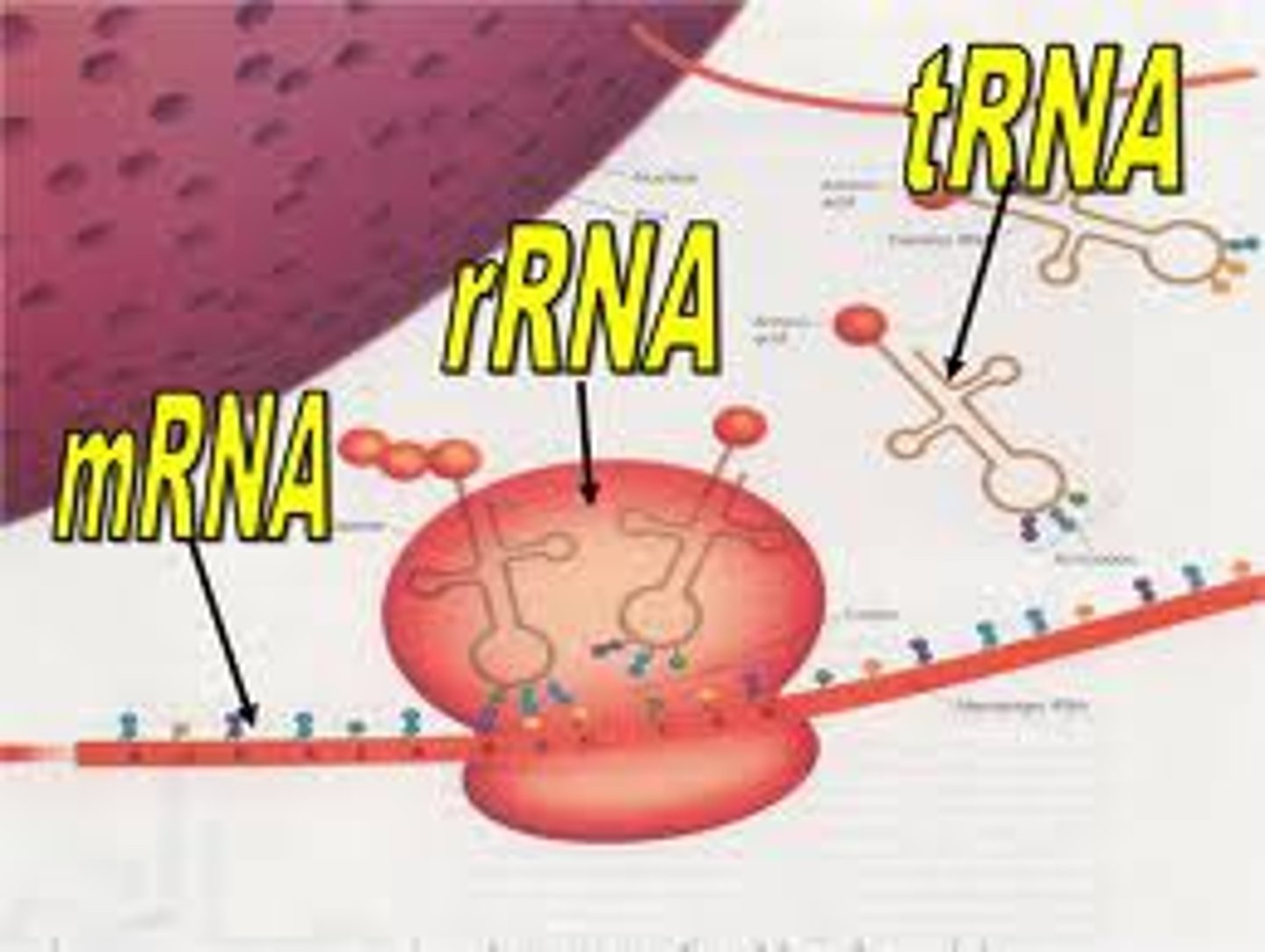
mRNA
Messenger RNA; is the ONLY info that translates DNA to protein via ribosome
RNA Polymerase function?
the polymerase that builds RNA off a parent DNA molecule
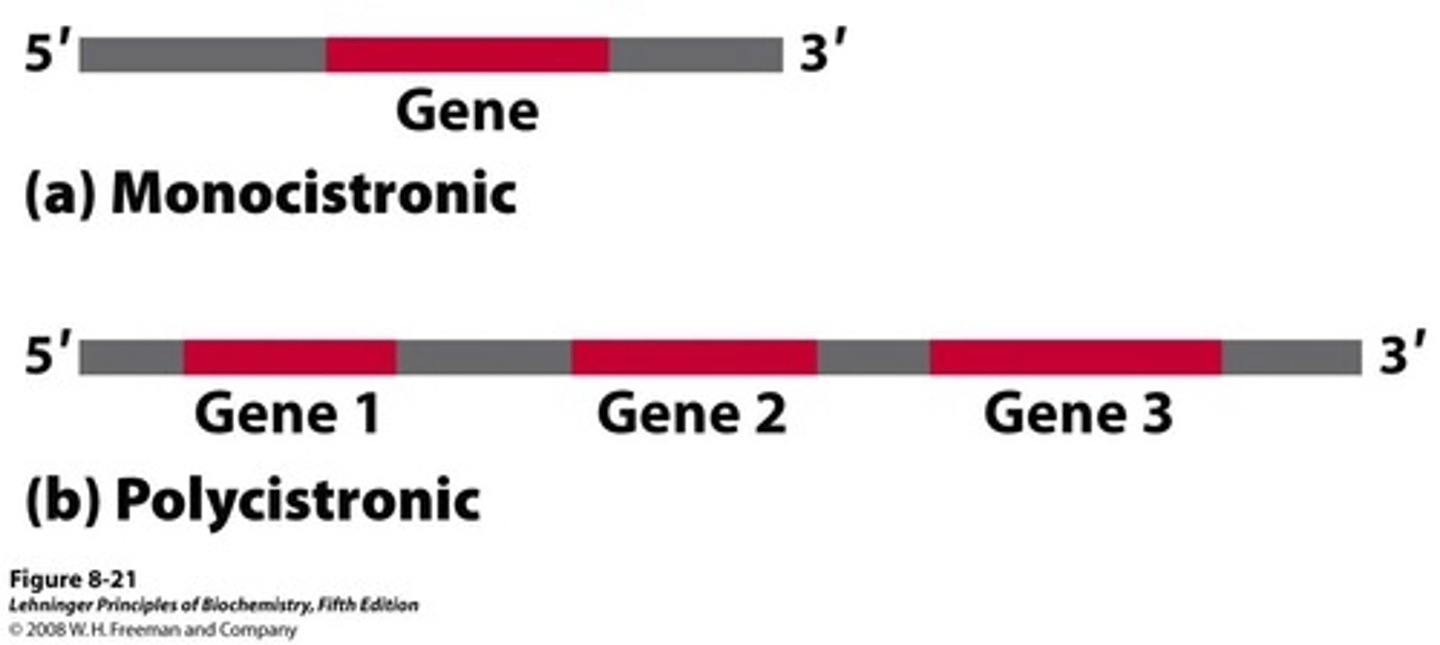
In eukaryotes, RNA is _________________, meaning 1 RNA translates into 1 protein only
monocistronic
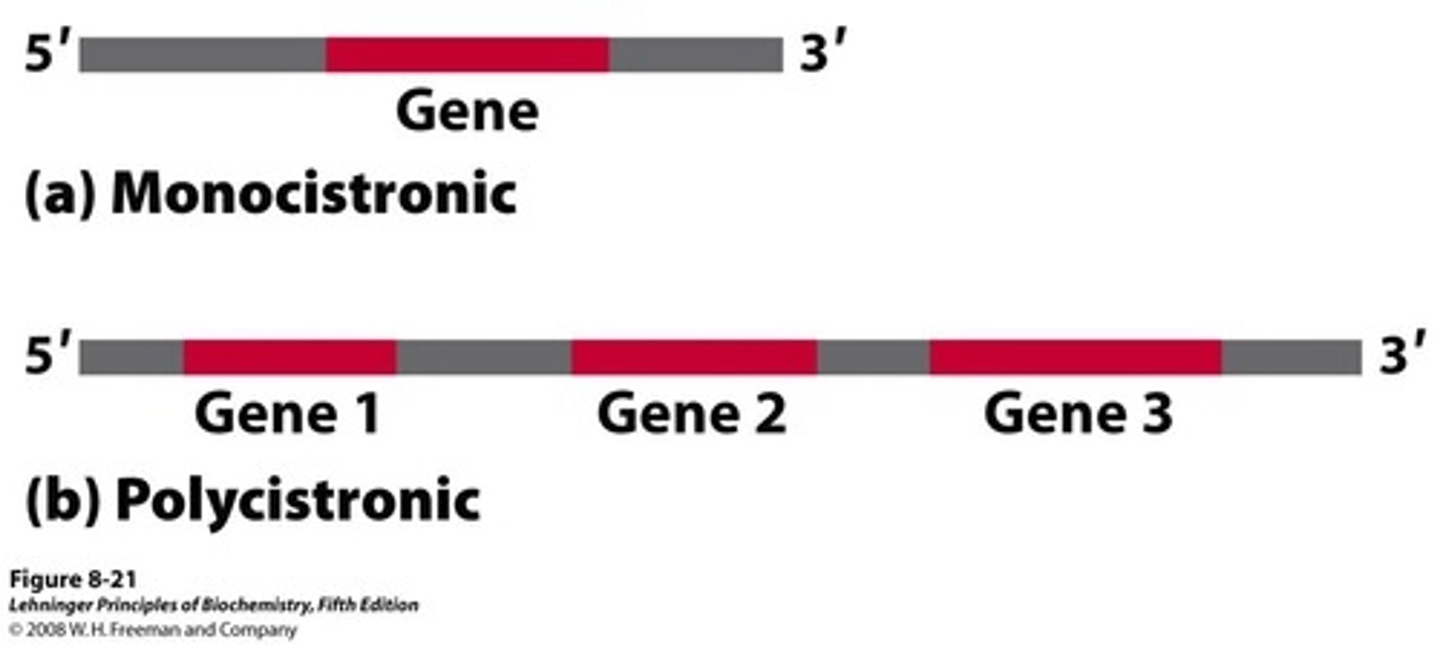
In prokaryotes, RNA is _________________, meaning 1 RNA translates into many proteins depending on where translation begins
polycistronic
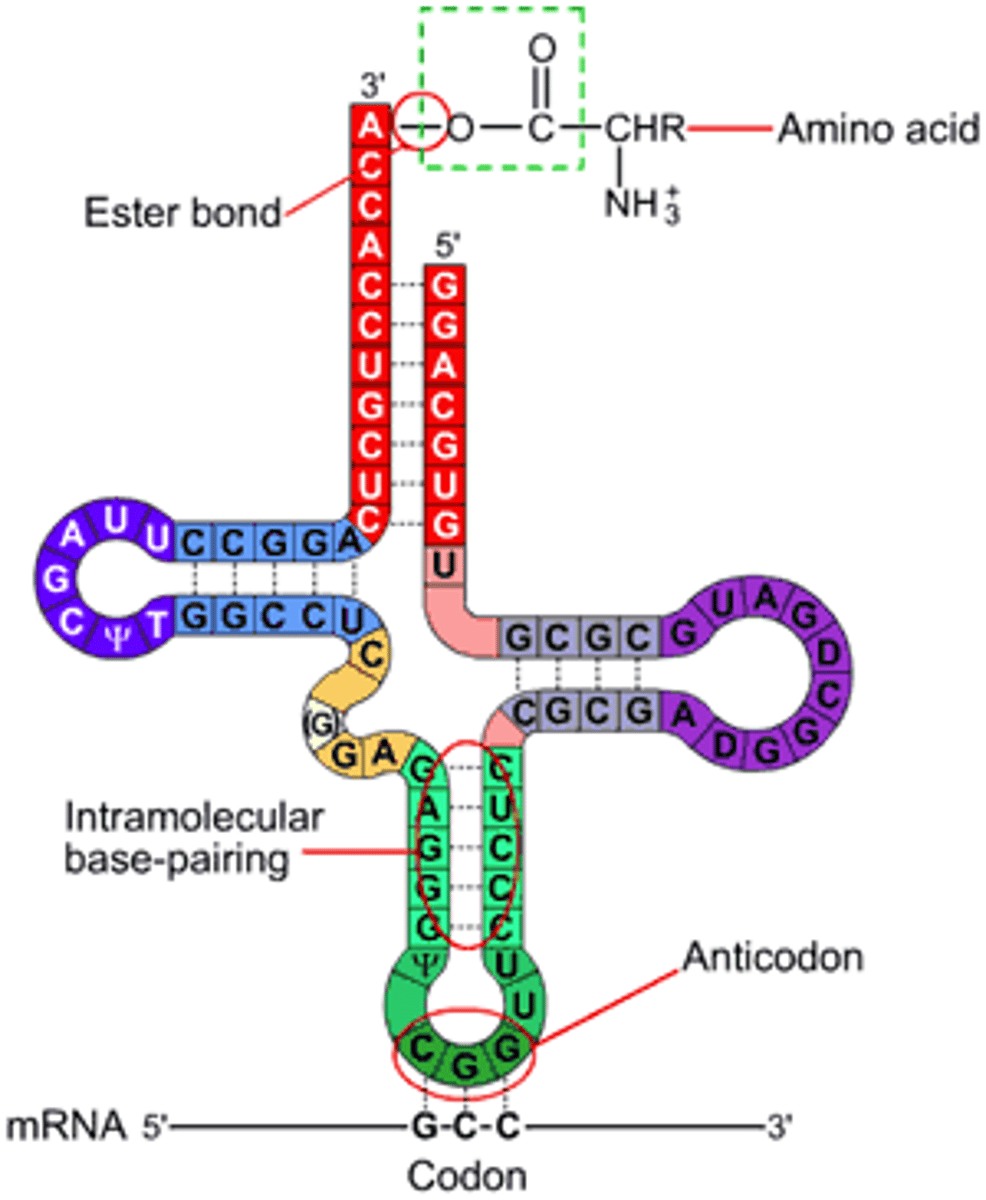
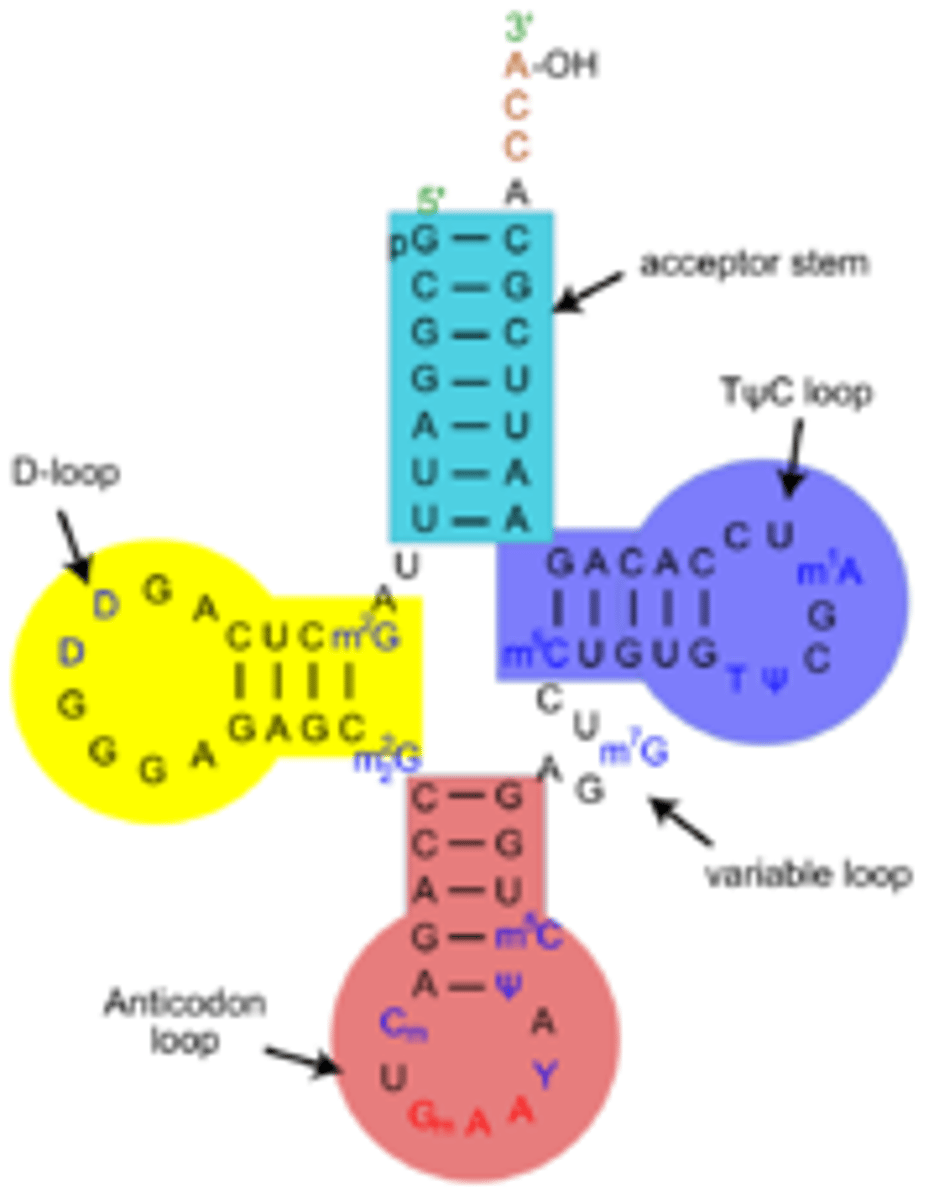
tRNA
Transfer RNA; a converting factor for amino acids into peptides; has a 3-nucleotide Anticodon that recognizes a corresponding Codon on the mRNA, and the tRNA carries a corresponding amino acid
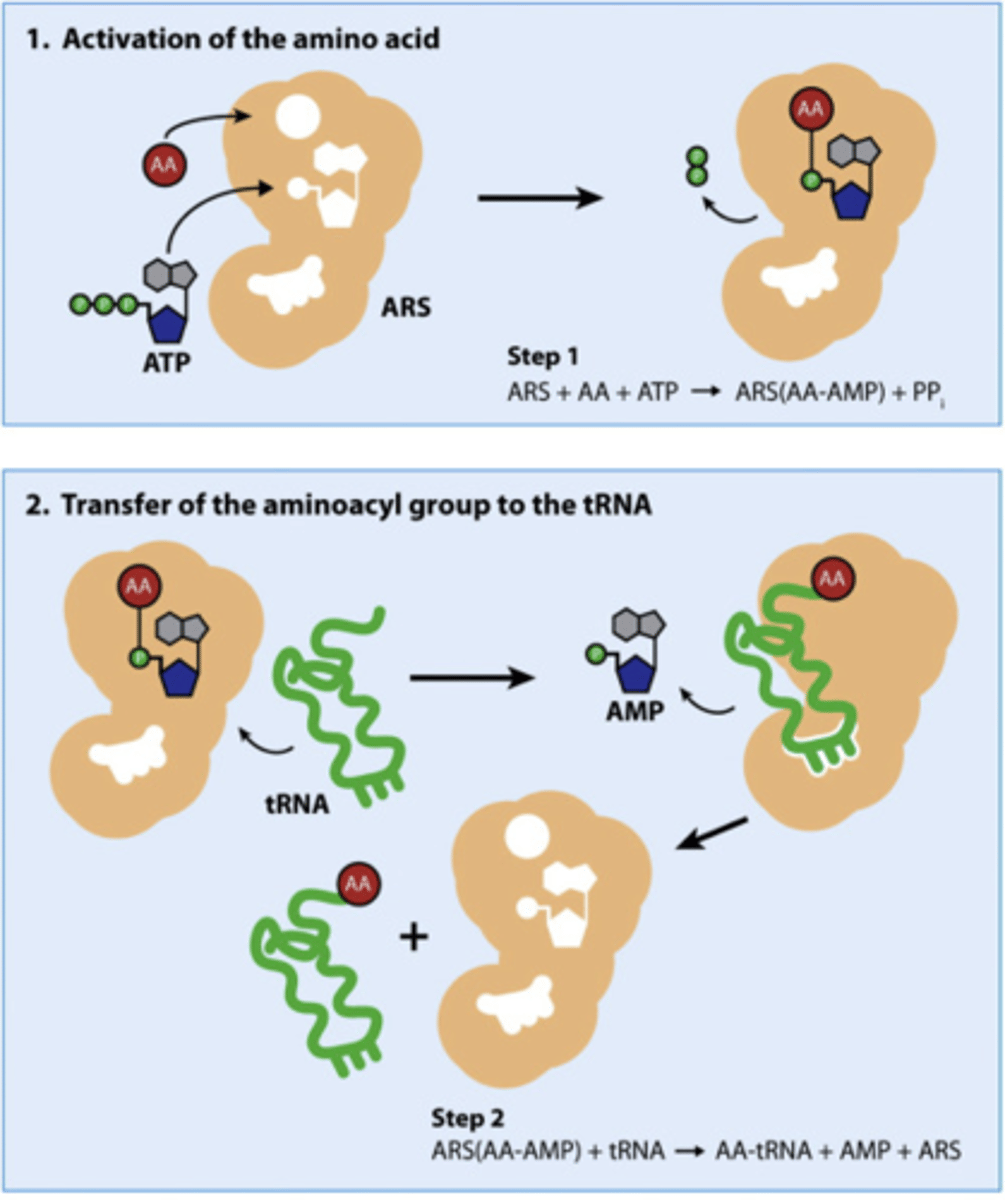
How is a tRNA activated?
the tRNA is 'charged' or activated by adding the corresponding Amino Acid; 'aminoacyl-tRNA'
# of amino acids : # of mRNA codons that code it
each AA has at least 1 codon that codes it; many have more!!
Where are mature tRNA (bound to AA) found?
in the cytoplasm
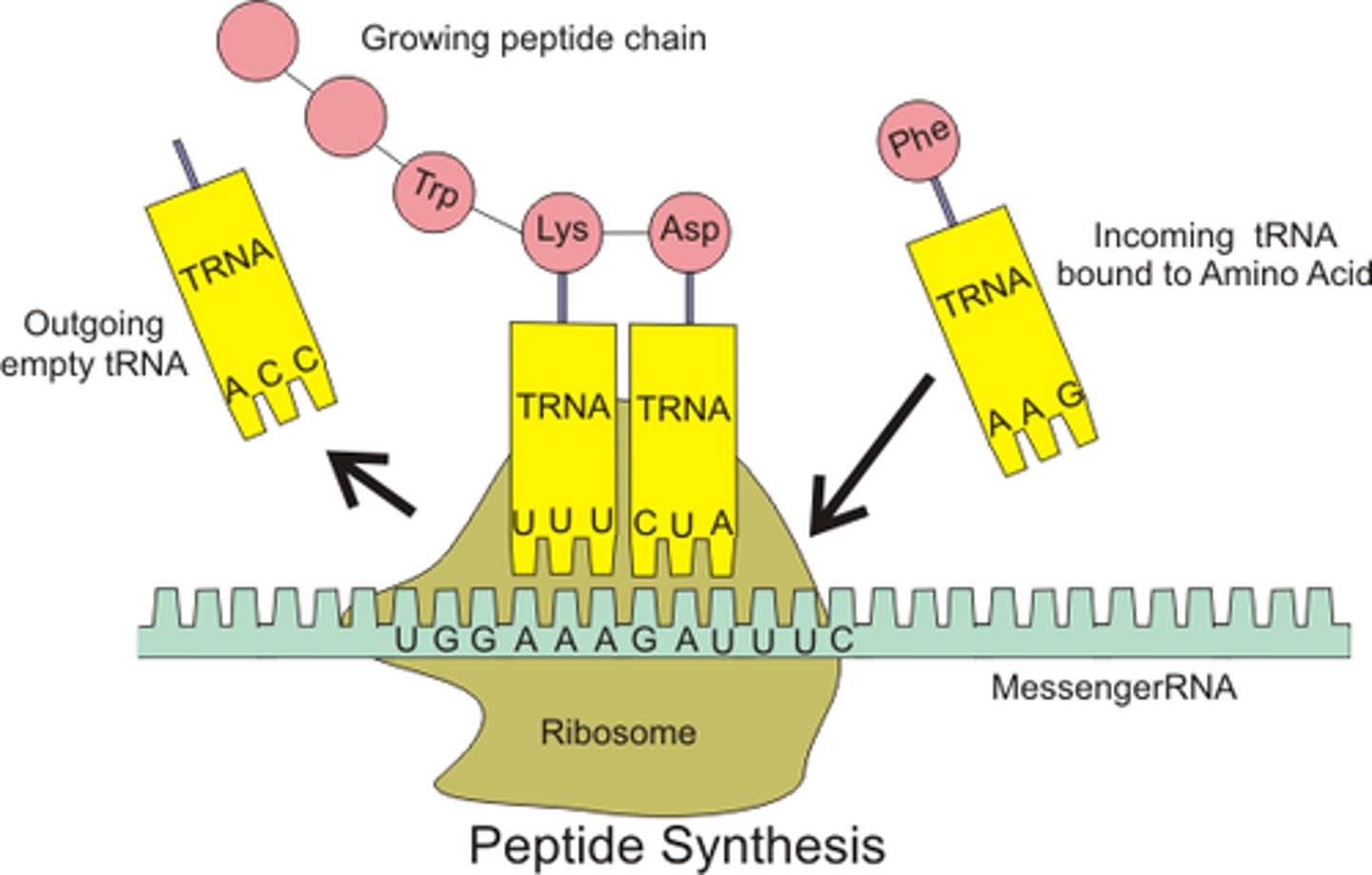
Aminoacyl-tRNA Synthetases (ARS) function?
AKA: -able to bind to different tRNAs that code for specific amino acid
-catalyzes covalent attachment to tRNA -> aminoacyl tRNA released from enzyme
-available to deliver AA to growing polypeptide chain of ribosome
the many different synthetases that add different AAs to different tRNAs; do so by adding 2 high-energy bonds from ATP (AA adding is energy-rich) to the 3' end of tRNA
To which end of the tRNA is the AA attached? What is the signifying nucleotide triplet there?
the 3' end; CCA
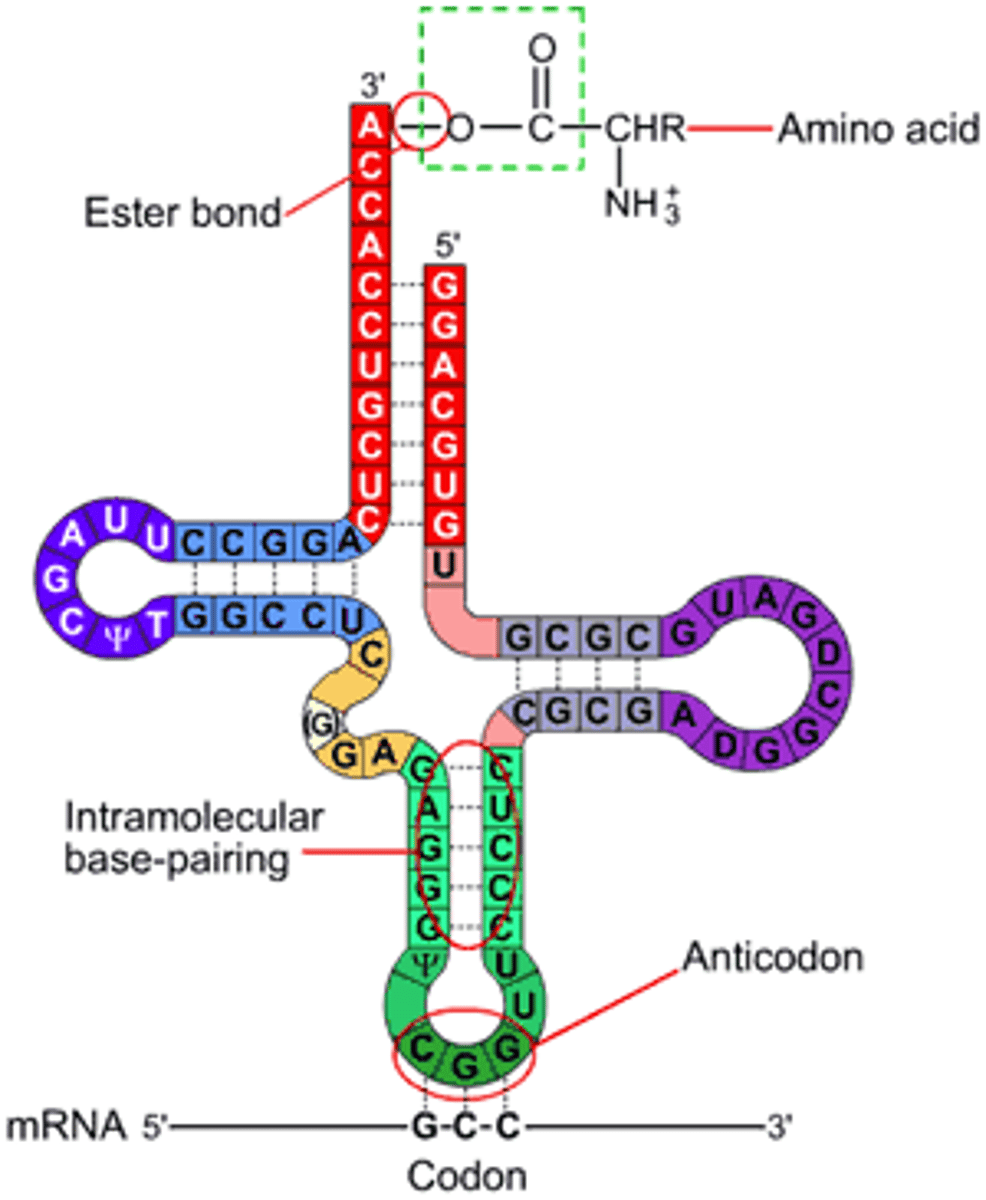
Purpose of AA-tRNA bond being high energy
this energy is used to fuel the formation of a peptide bond
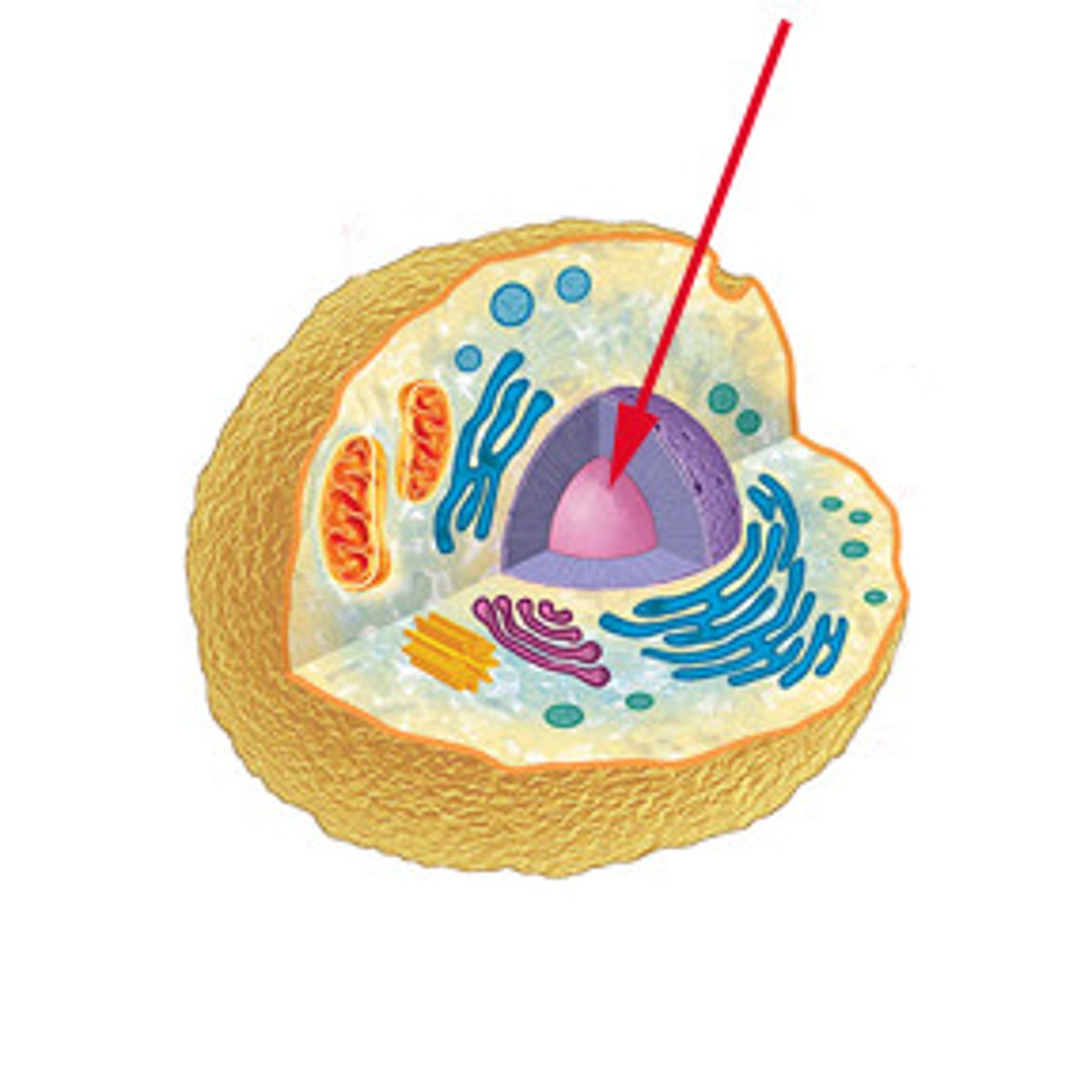
rRNA? synthesized where? 2 functions?
Ribosomal RNA; synthesized in the nucleolus
usually Ribozymes (enzymes made of rRNA) that do important reactions like translation (peptide bonds) & splicing out introns
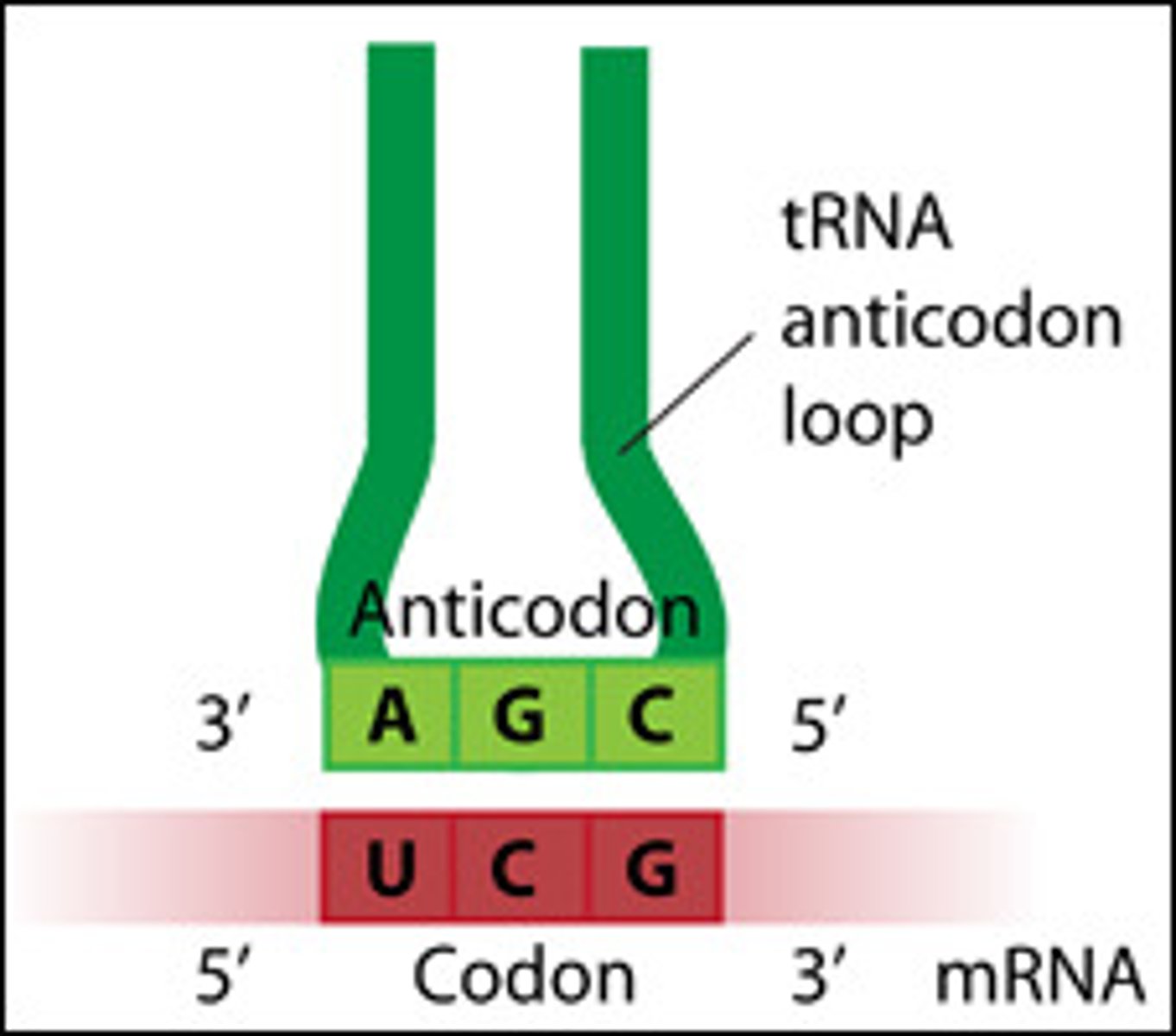
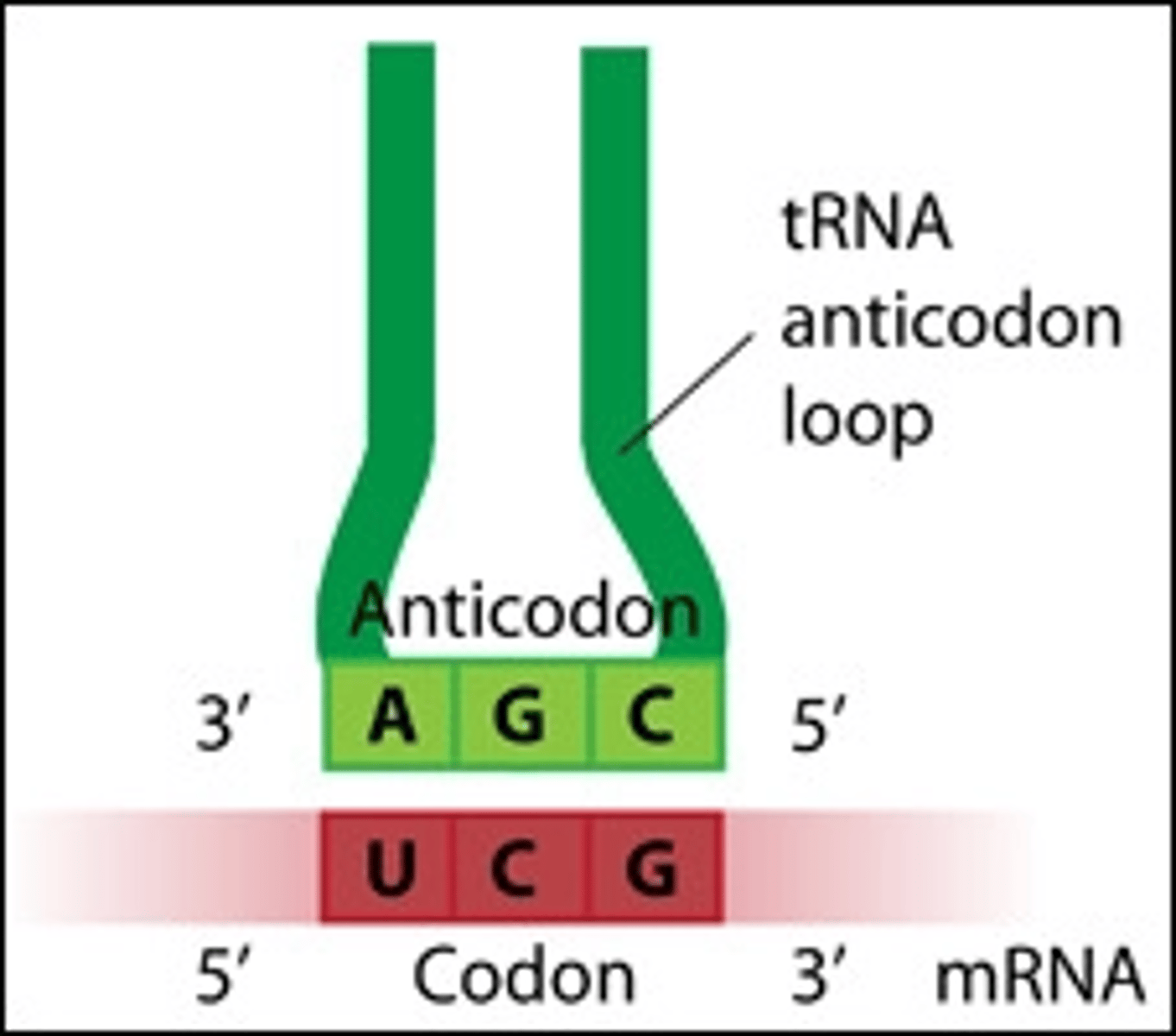
Important notes on reading an RNA codon
1) all codons are 5' ➝ 3',
2) all codons refer to what is written in the mRNA!!!,
3) each codon corresponds to ONE AA alone
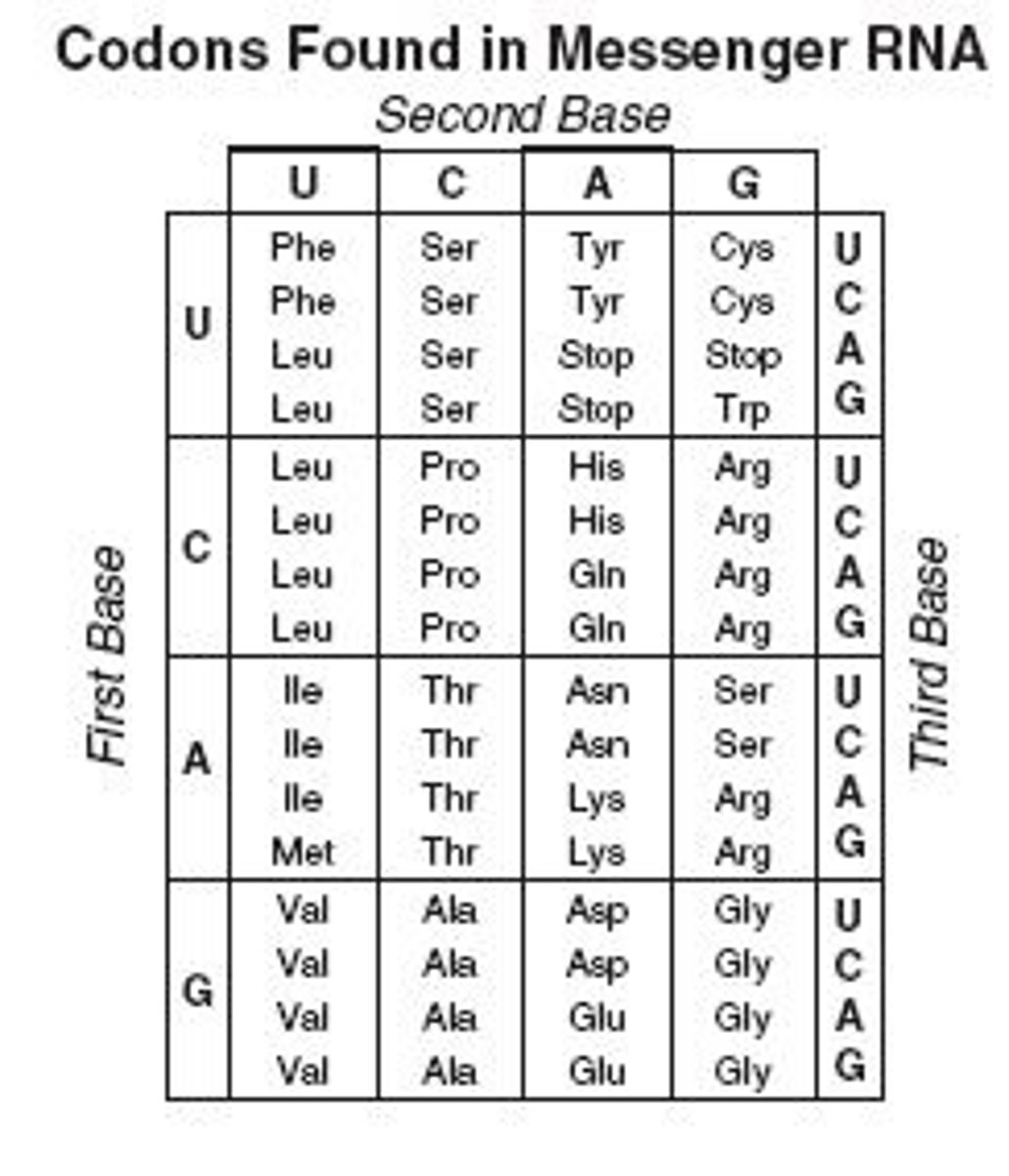
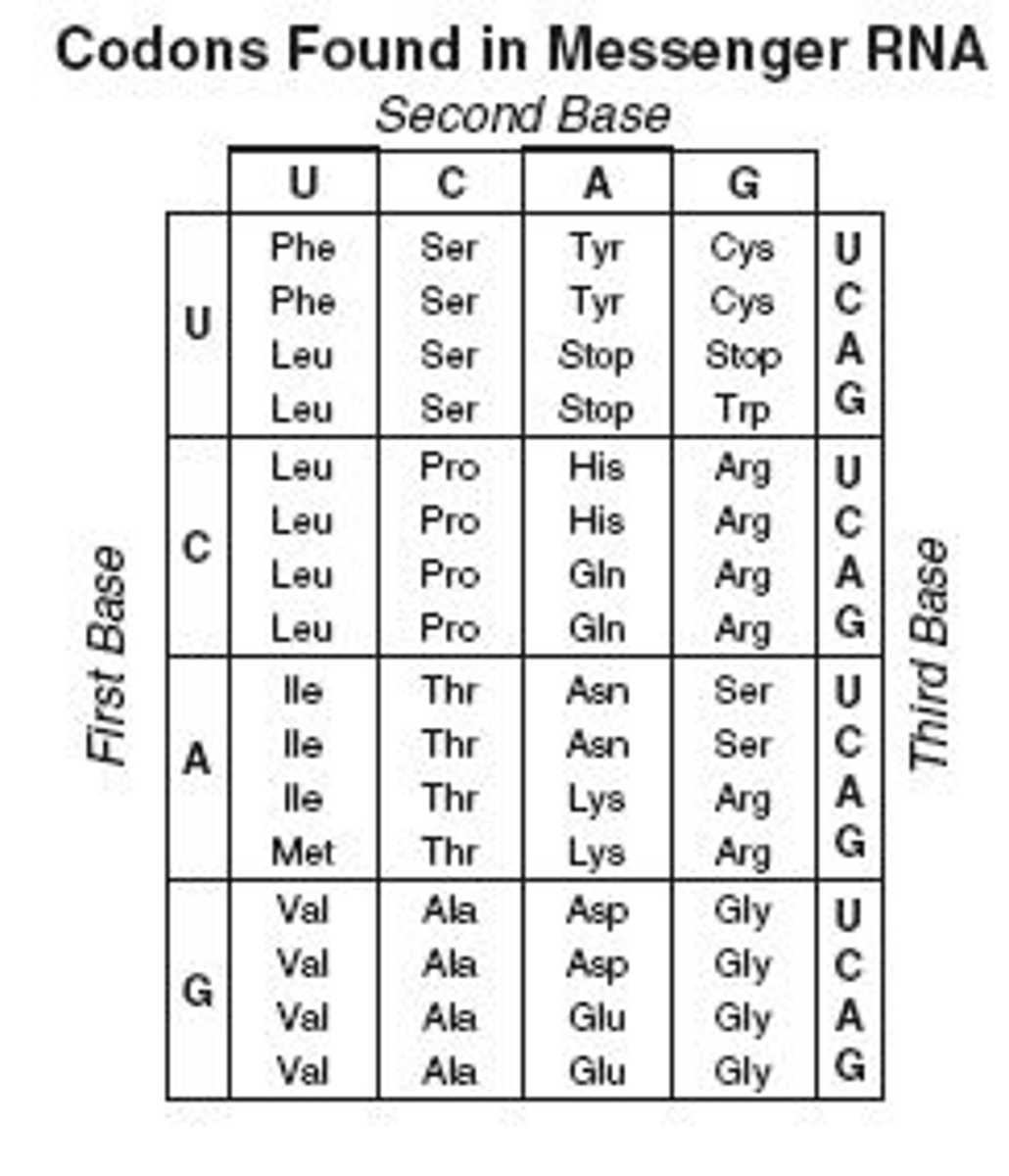
Number of codons overall encoded into amino acids? What are the other codons encoded into?
64 codons; 61 are for the 20 AA, 3 are for stop
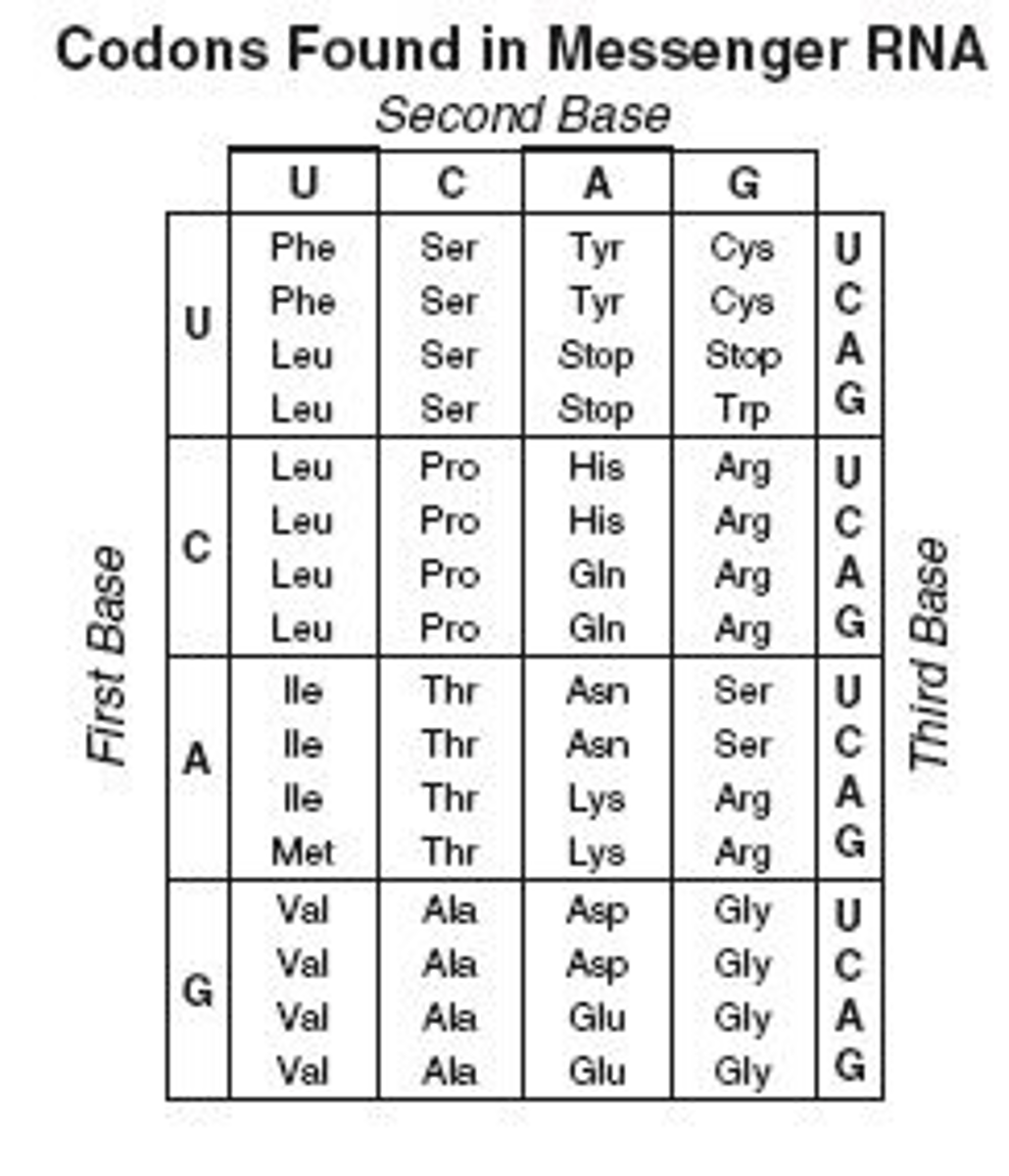
T/F: The mRNA codons used are universal to all life
TRUE: UNIVERSAL; we all use the same codons

mRNA-tRNA interaction in translation? which has the codon? anticodon? parallel or anti-parallel?
the codon and anticodon are anti-parallel and opposite, binding to add the AA
Eukaryotic start codon & start AA
AUG; Methionine! All eukaryotic proteins start with this, and AUG is the START codon
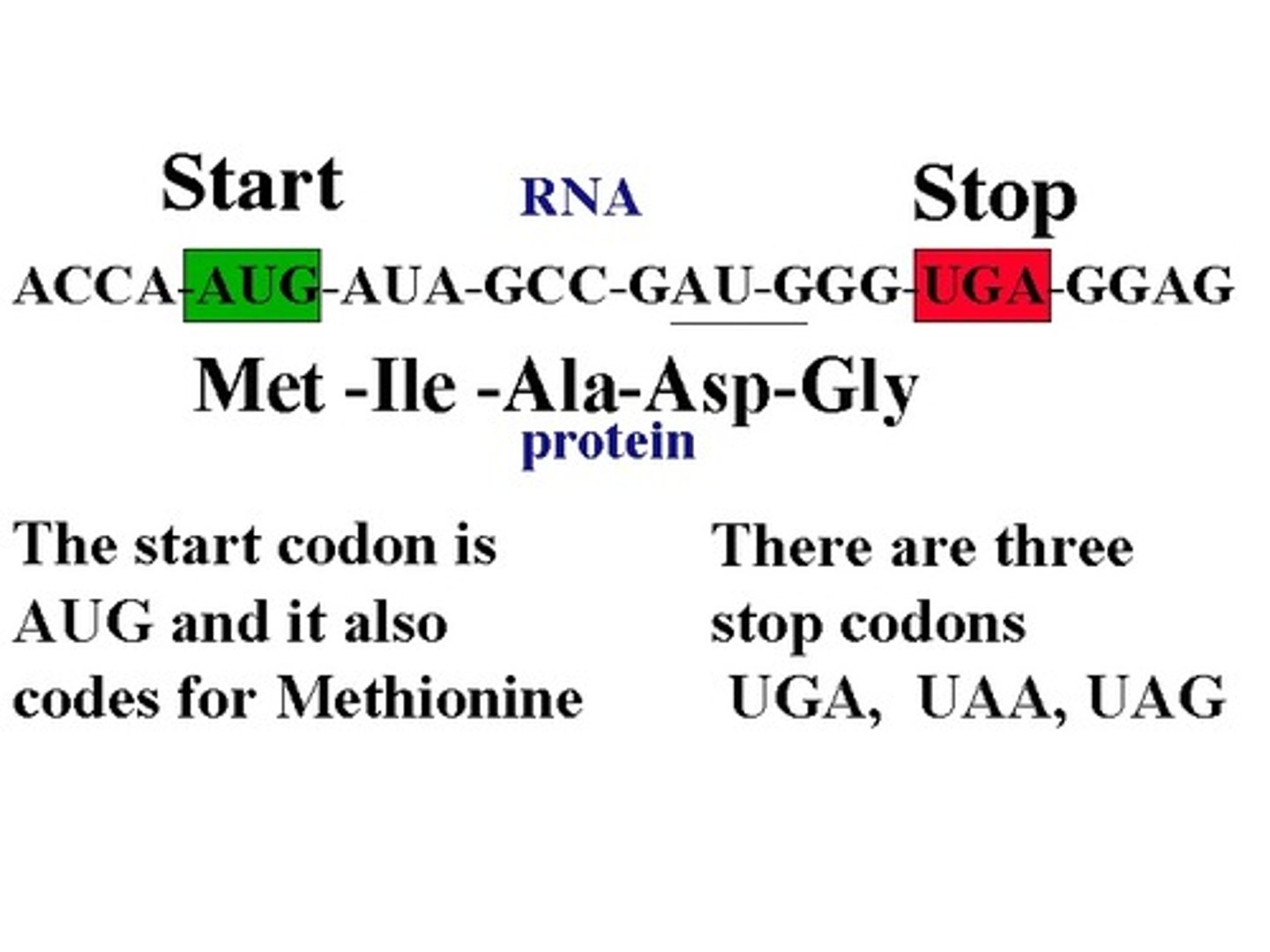
3 Stop Codons (mnemonic)
UAA, UGA, UAG (U are annoying, U go away, U are gone)

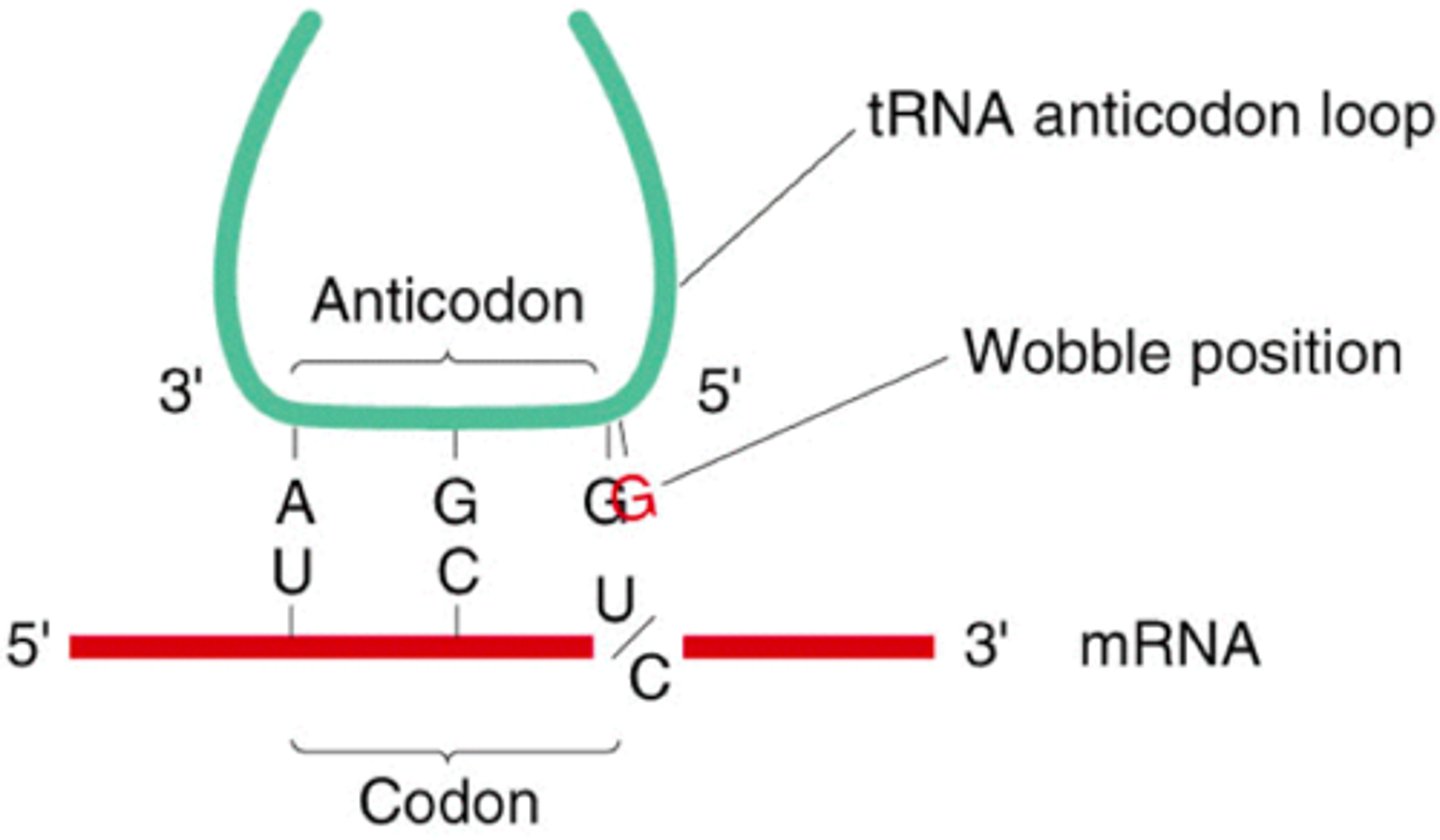
Degeneracy of the genetic code
refers to the fact that 1+ codons can code for an AA (all but Met and Trp); usually the first 2 bases are the same for the same AA, and the third 'wobbly base' varies; in general this protects us against single-base mutations
Wobble Position in codons? what aids damage from this?
the 3rd/final base in a codon: usually for codons that code for the same AA, the first 2 bases are the same, and this third one is different: ADVANTAGE IS THAT IT PROTECTS AGAINST MUTATION AS A MUTATION IN IT WOULD BE SILENT
Point Mutation
when a mutation affects 1 base; they are Expressed Mutations (can affect primary sequence)
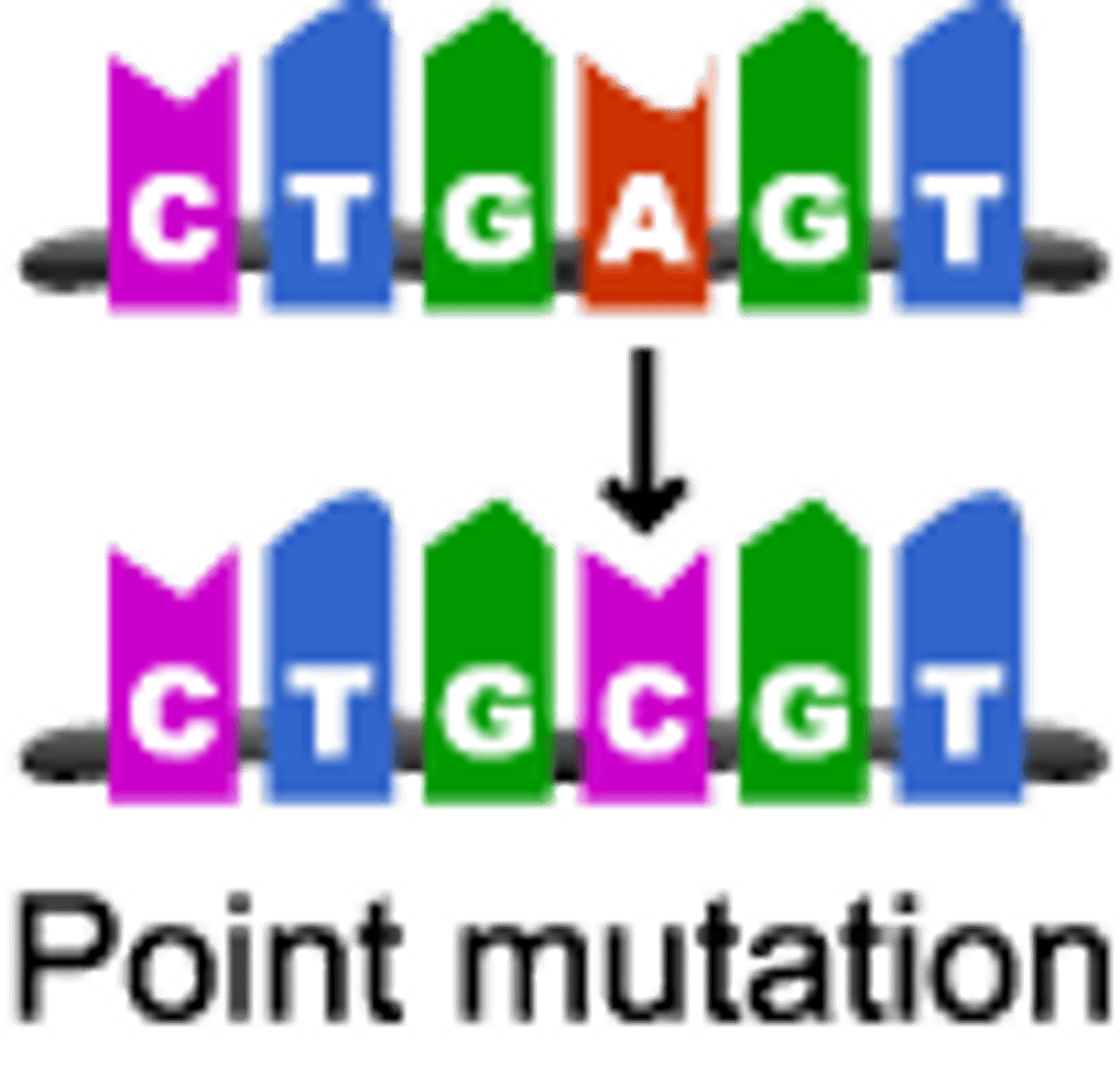
Expressed Mutations
mutations that affect 1˚ structure of a protein; 2 categories: Missense and Nonsense
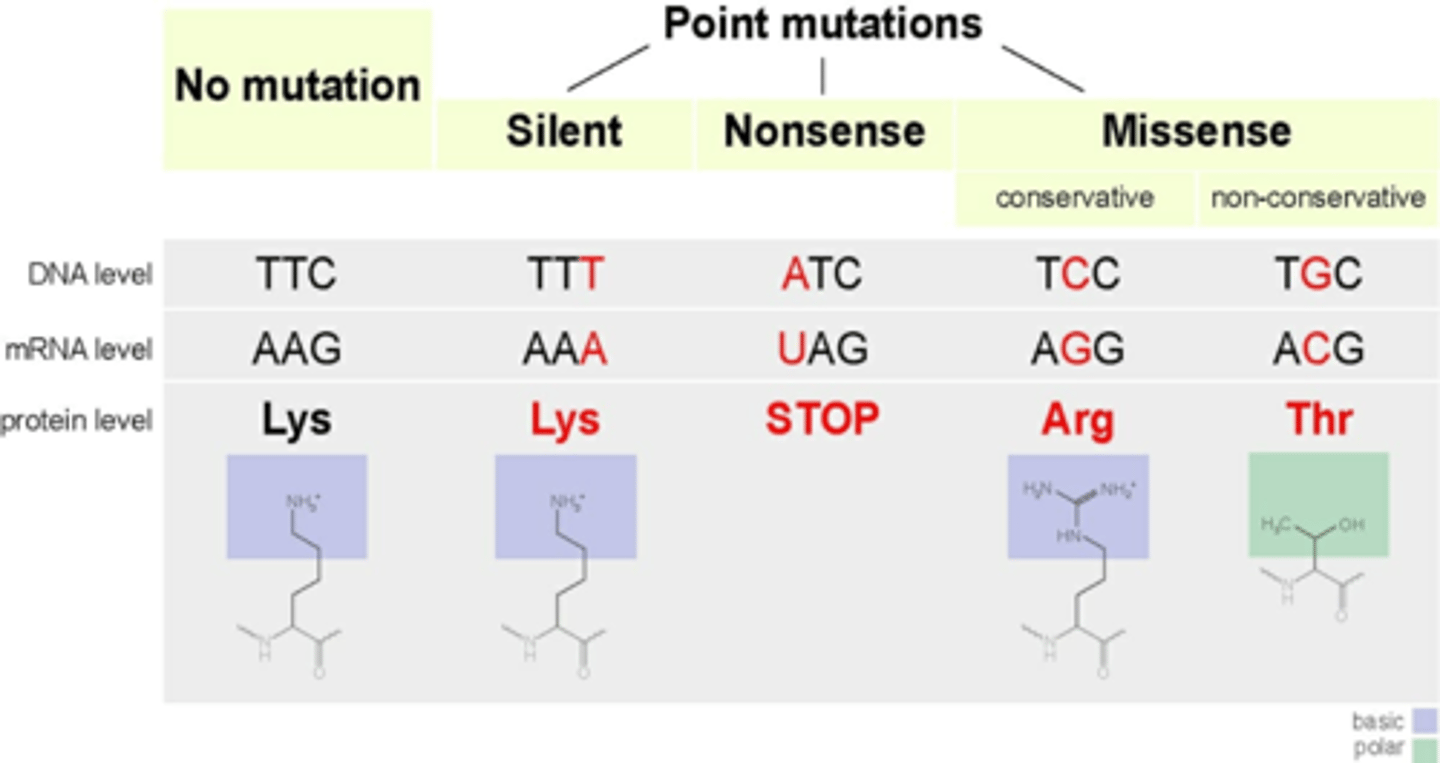
Missense Mutations
an expressed mutation where 1 AA substitutes for another
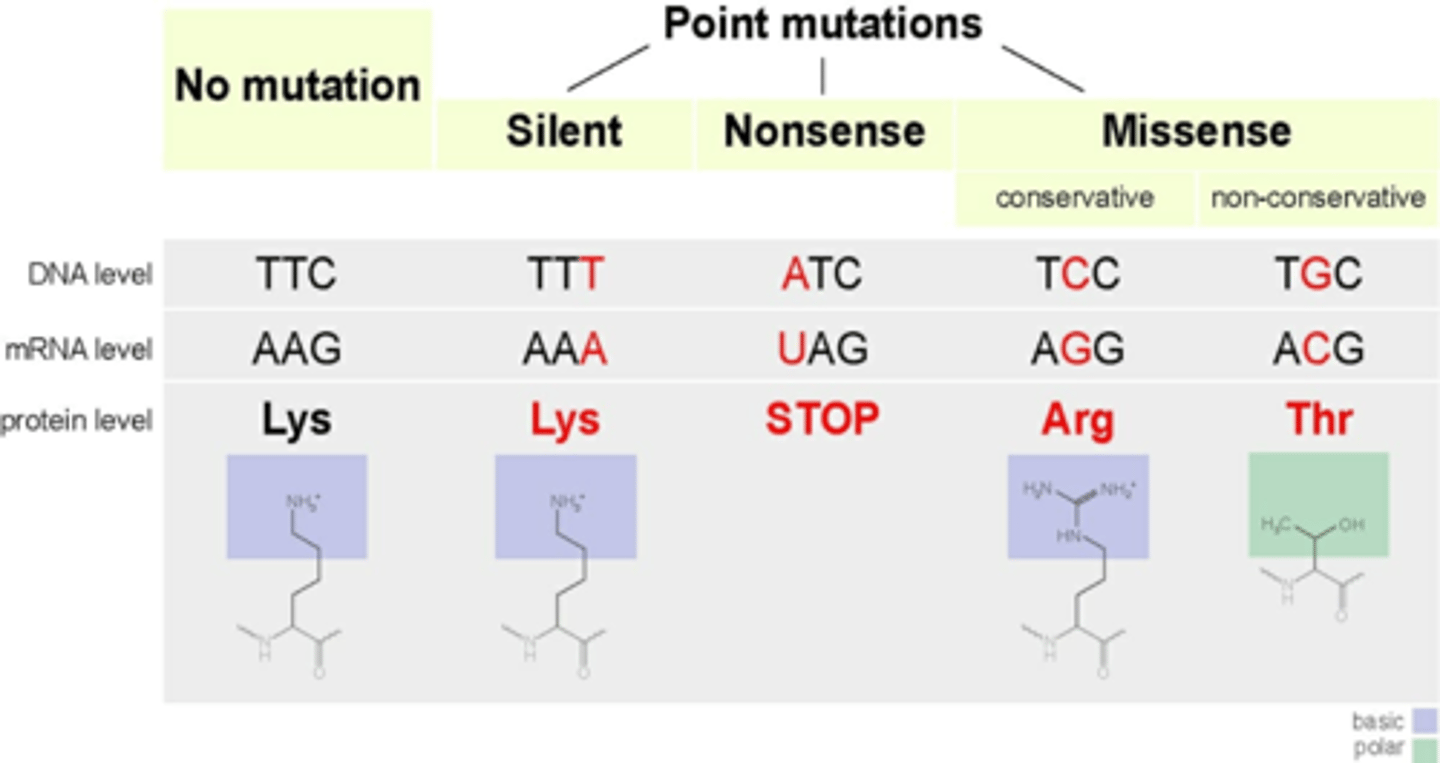
Nonsense Mutations
aka Truncation; an expressed mutation where an AA is replaced with a premature STOP (UAG, UGA, UAA)
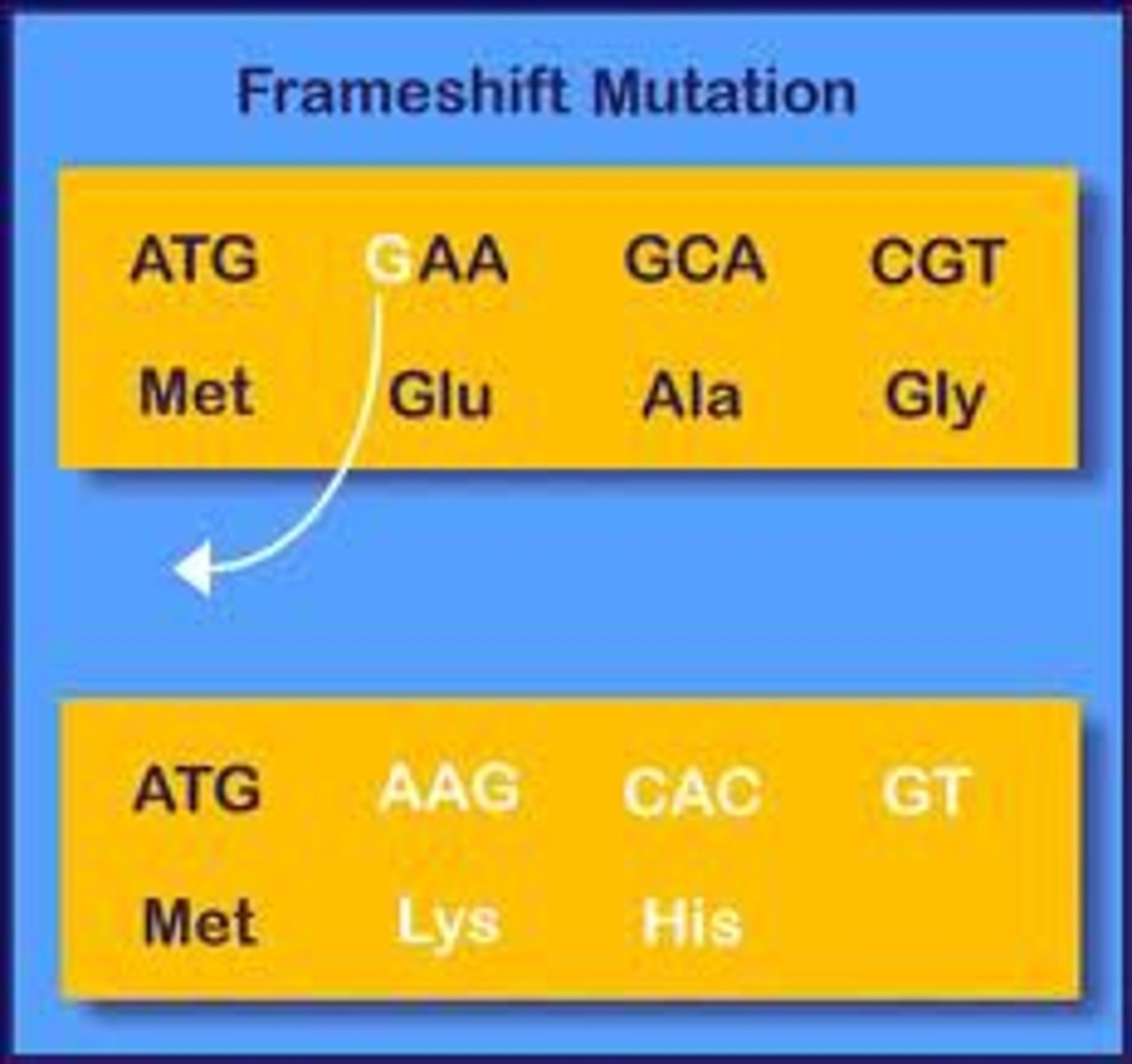
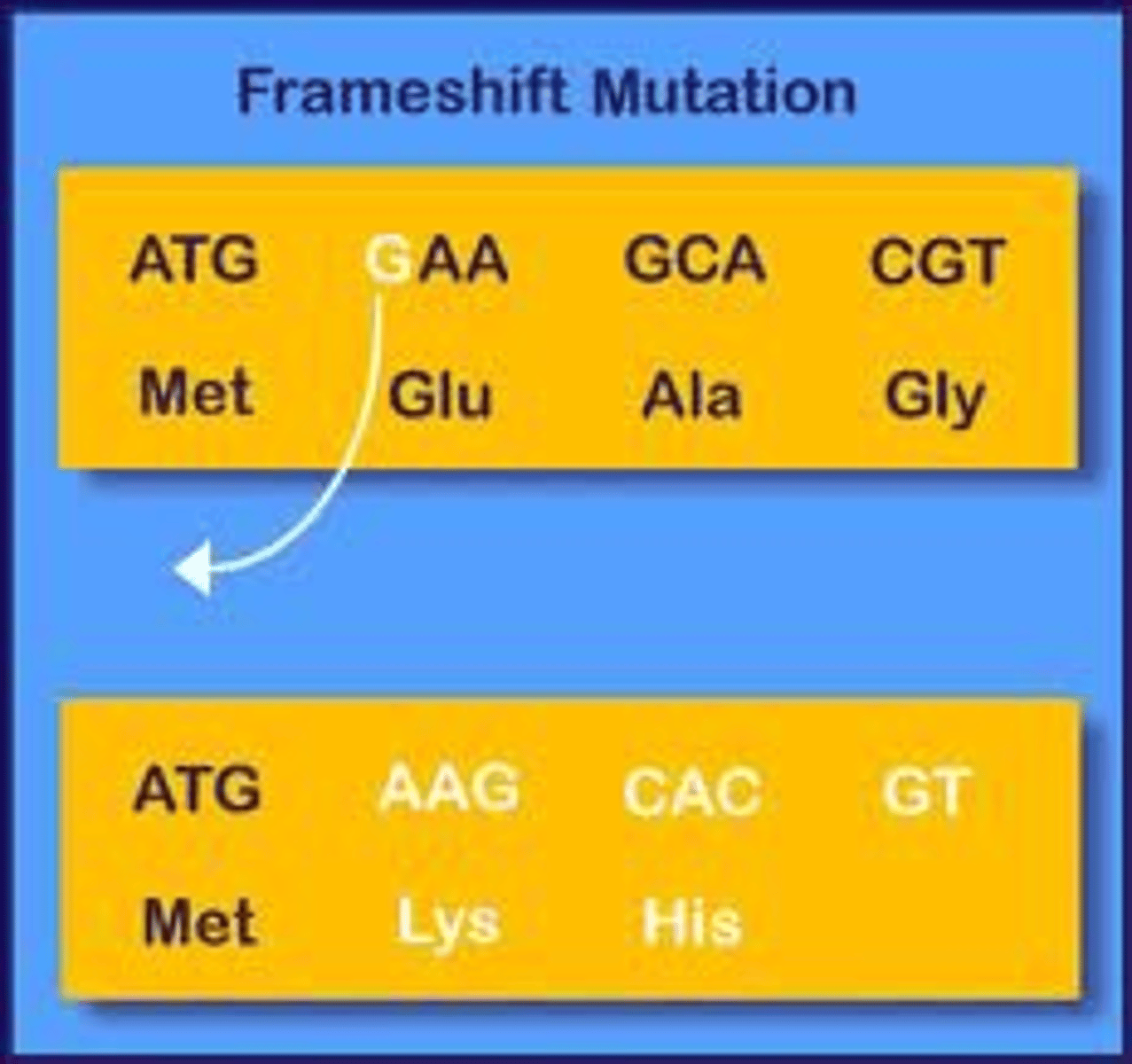
Frameshift Mutations
when a number of nucleotides (that are not a multiple of 3) are lost/gained, resulting in a shift of reading from of the mRNA that dramatically changes AAs or truncates the protein
Which is more serious: frameshift or missense mutations? Why?
Frameshift, they tend to change WAAAAAY MORE AAs, but in the end this depends on where the AA is and how vital it is to the protein
Where are the ribosomes location (for translation)?
in the CYTOPLASM

Transcription
the creation of mRNA from DNA
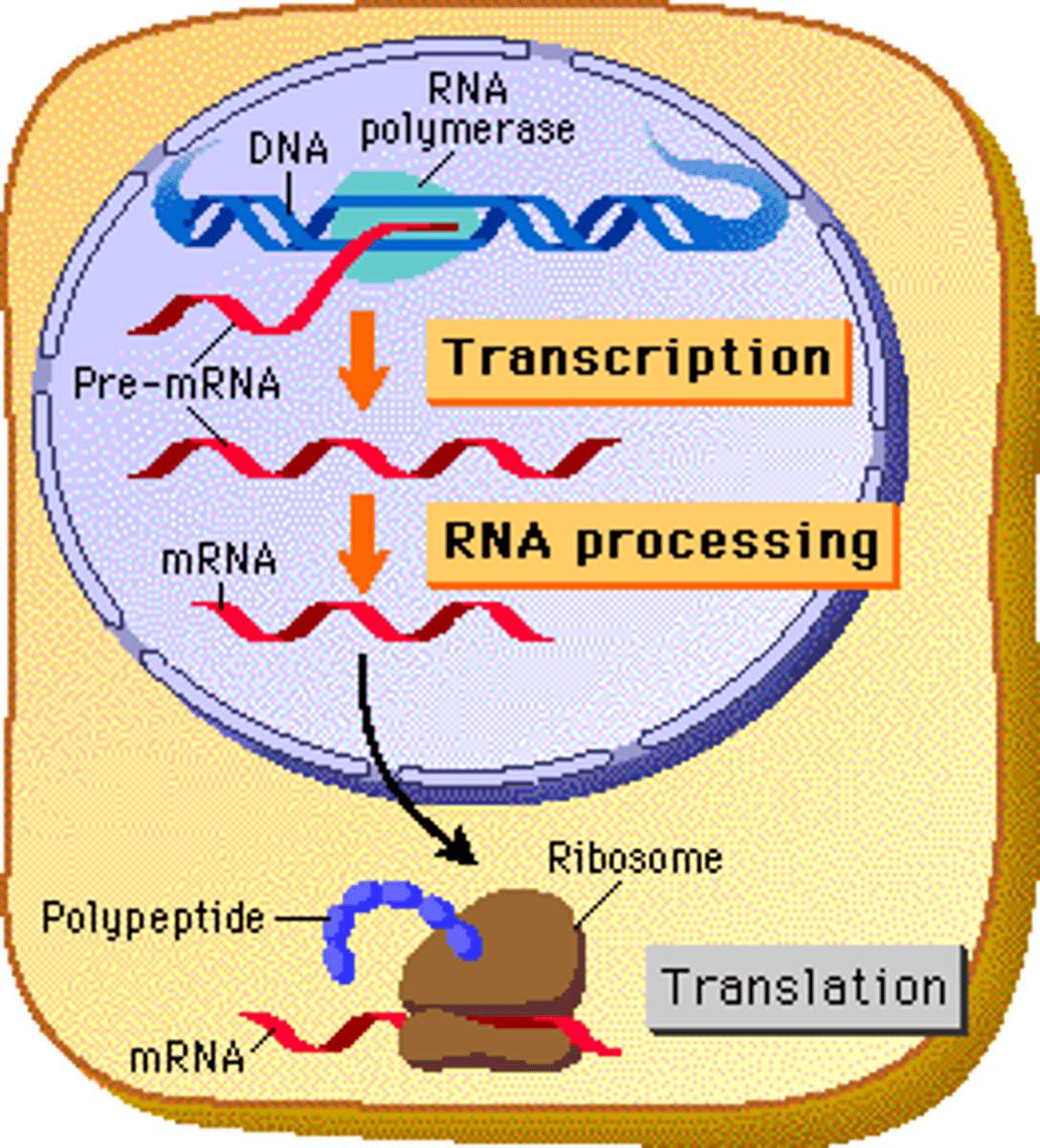
How transcription happens
helicase unwinds DNA, topoisomerase relieves supercoils, template/antisense strand synthesize 1 antiparallel sense strand of mRNA from the template strand via DNA-dependent RNA polymerase (binds to TATA box)
During transcription, RNA polymerase II searches for ___________ ___________, often signaled by the ______ ______ binding site
promoter regions; TATA Box (high T and As)
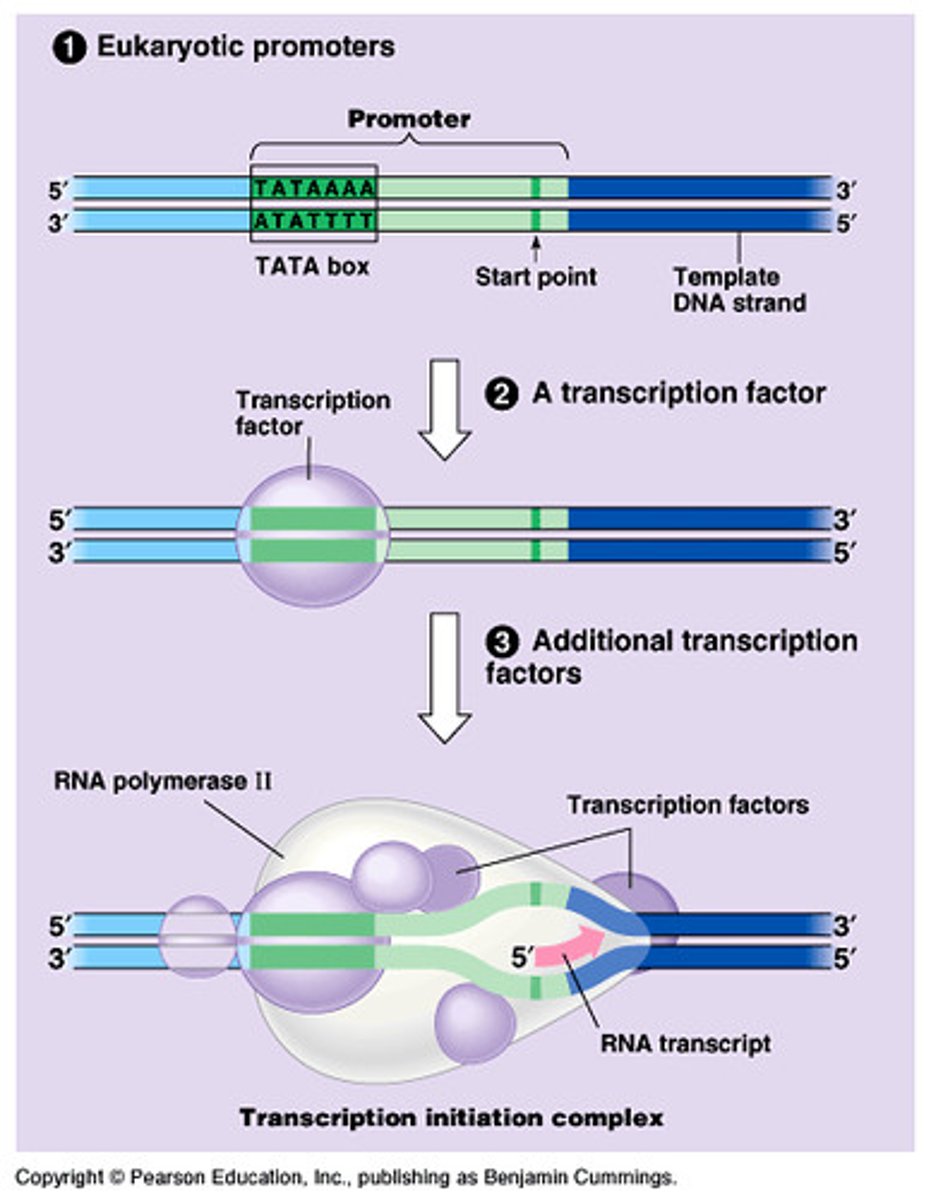
___________ ____________ help RNA polymerase locate the correct binding site to start transcription
Transcription Factors
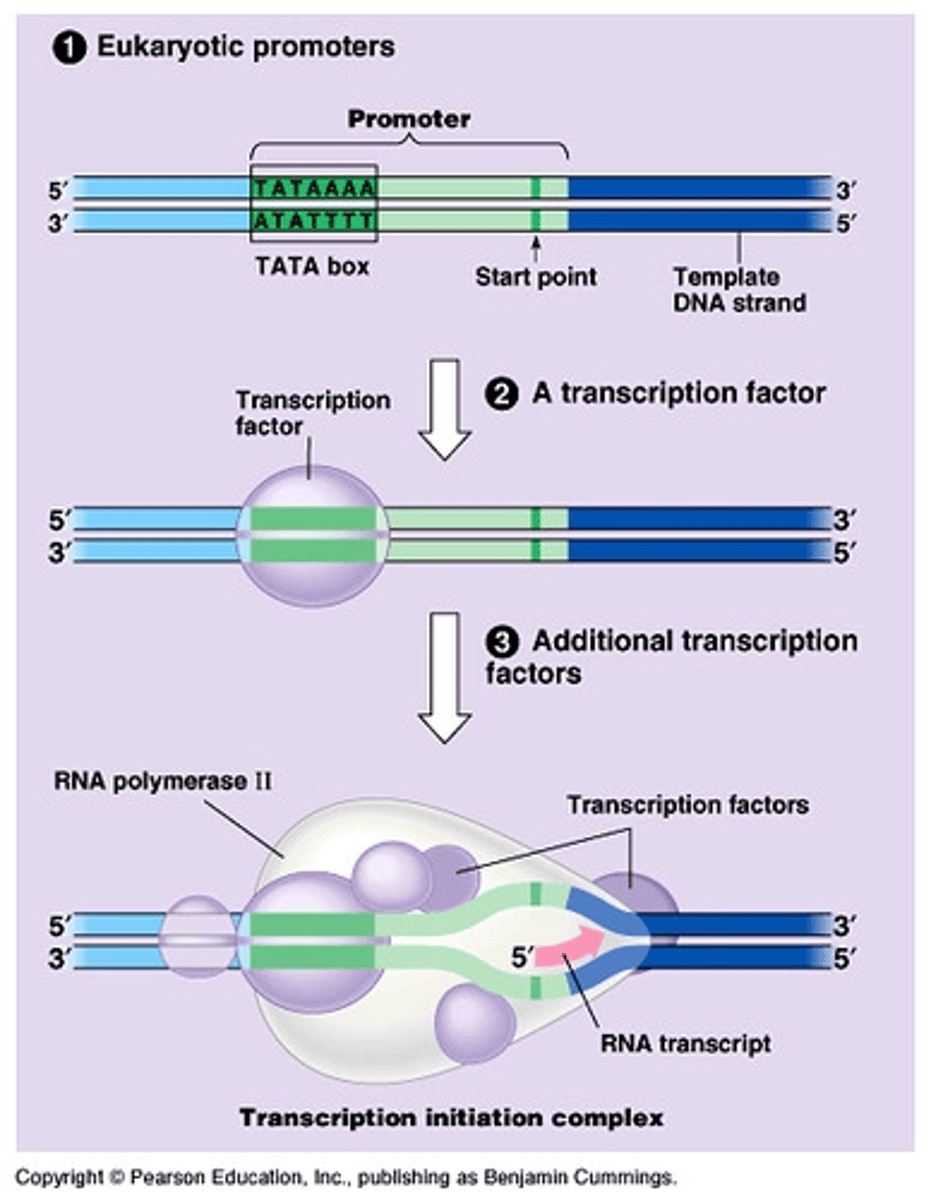
Does RNA polymerase require a primer to start transcription?
NO! Note that DNA polymerase does
3 RNA polymerases in eukaryotes & what they do/make?
RNA pol I: nucleolus, synthesizes rRNA
RNA pol II: nucleus, synthesizes hnRNA (pre-processed) (& small nuclear RNA) (snRNA)
RNA pol III: nucleus, synthesizes tRNA (& some rRNA)
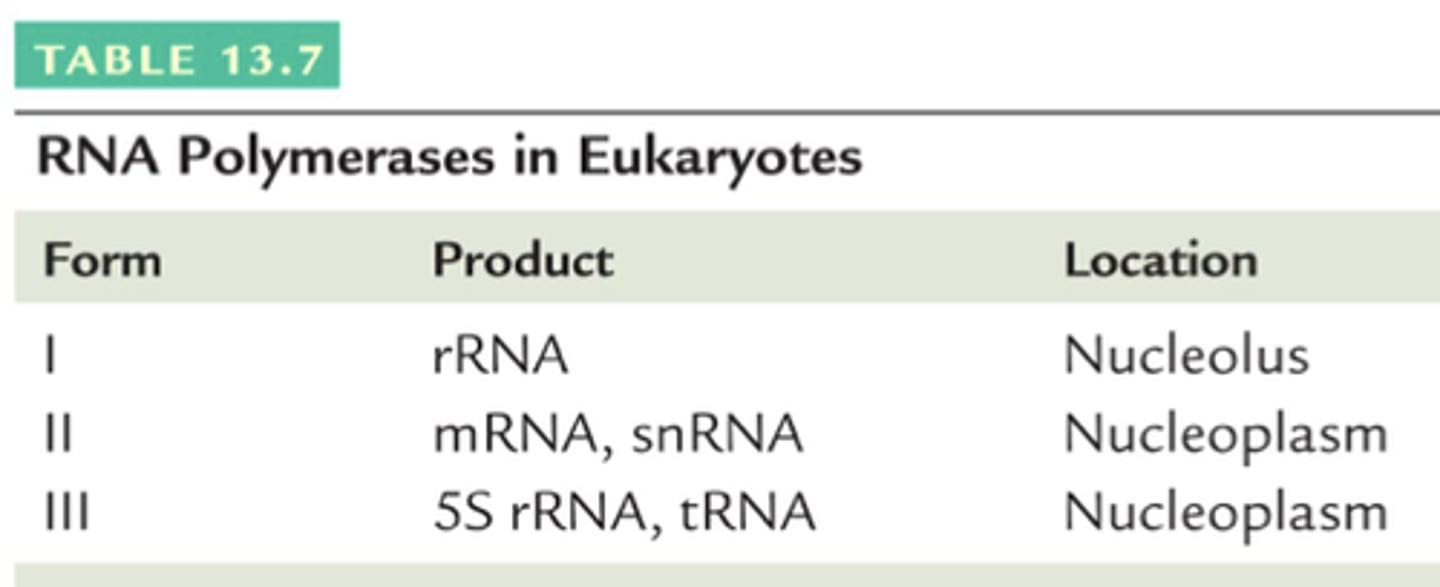
RNA Polymerase II
the eukaryotic RNA polymerase that does transcription (hnRNA) & small nuclear RNA (snRNA)
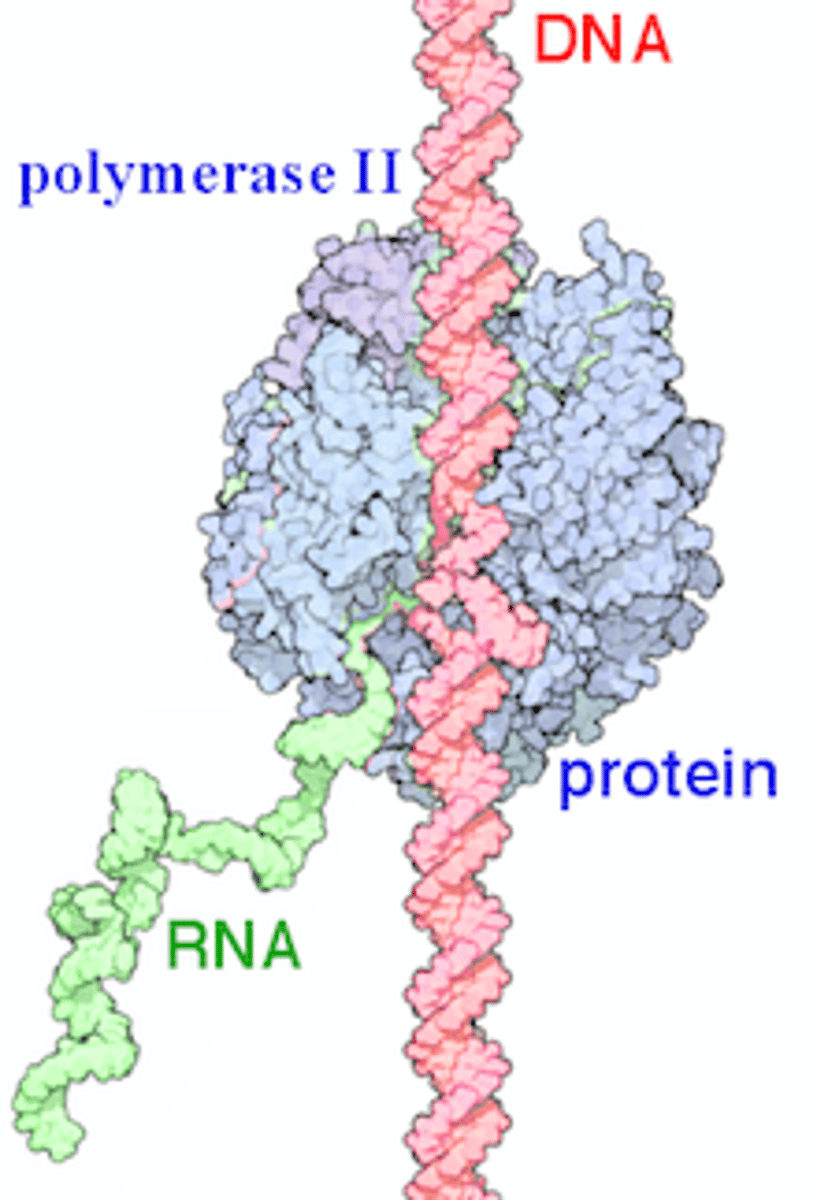
RNA Polymerase I
the eukaryotic RNA polymerase that synthesizes rRNA in the nucleolus
Ribosomal ribonucleic acid (rRNA) is a noncoding type of RNA that acts as the primary building block for ribosomes and the assembly line on which protein synthesis occurs in those ribosomes, essential to all living organisms.
RNA Polymerase III
the eukaryotic RNA polymerase that synthesizes tRNA (& some rRNA) in the nucleus
All RNA polymerases are located in...
the nucleus!
The __________ strand is used as the template DNA for RNA, whereas the ___________ is not
(antisense/sense)
antisense (template); sense (not)
Does RNA polymerase proofread?
NOPE
Numbering of bases in transcription
numbering of DNA BASES:
+1 is the first base transcribed
upstream to the 5' end (prior) is labelled -1, -2 etc., and upstream to 3' end (after) is labelled increasing: NO NUCLEOTIDE IS LABELLED 0
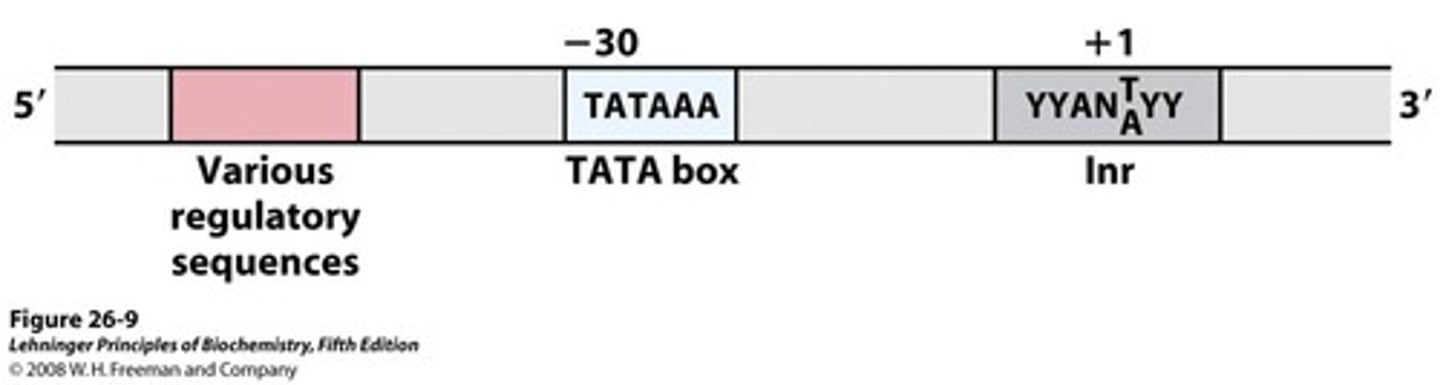
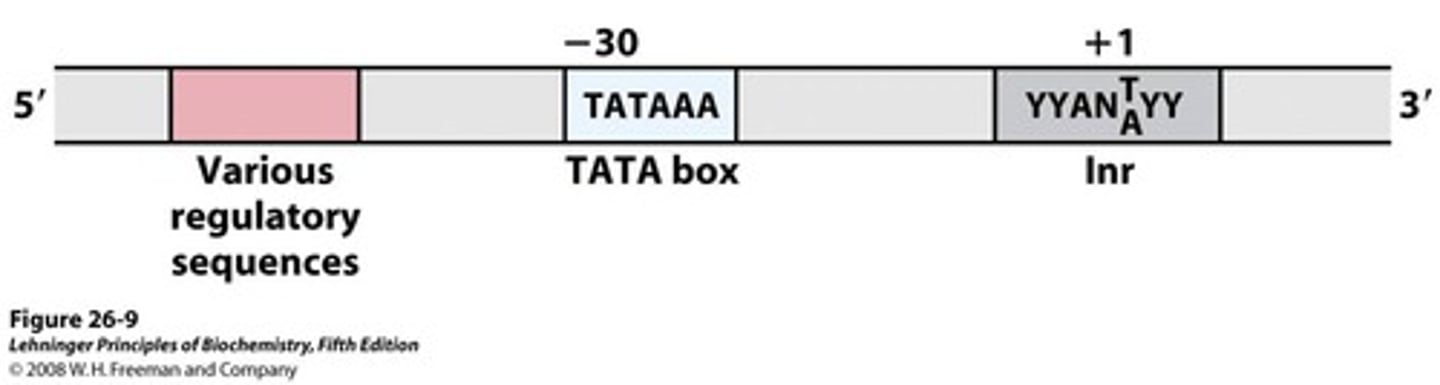
Location of the TATA box (# nucleotides behind +1)
about -25; this is where RNA pol II binds
hnRNA
heterogeneous nuclear RNA; has just been released from DNA and not processed yet
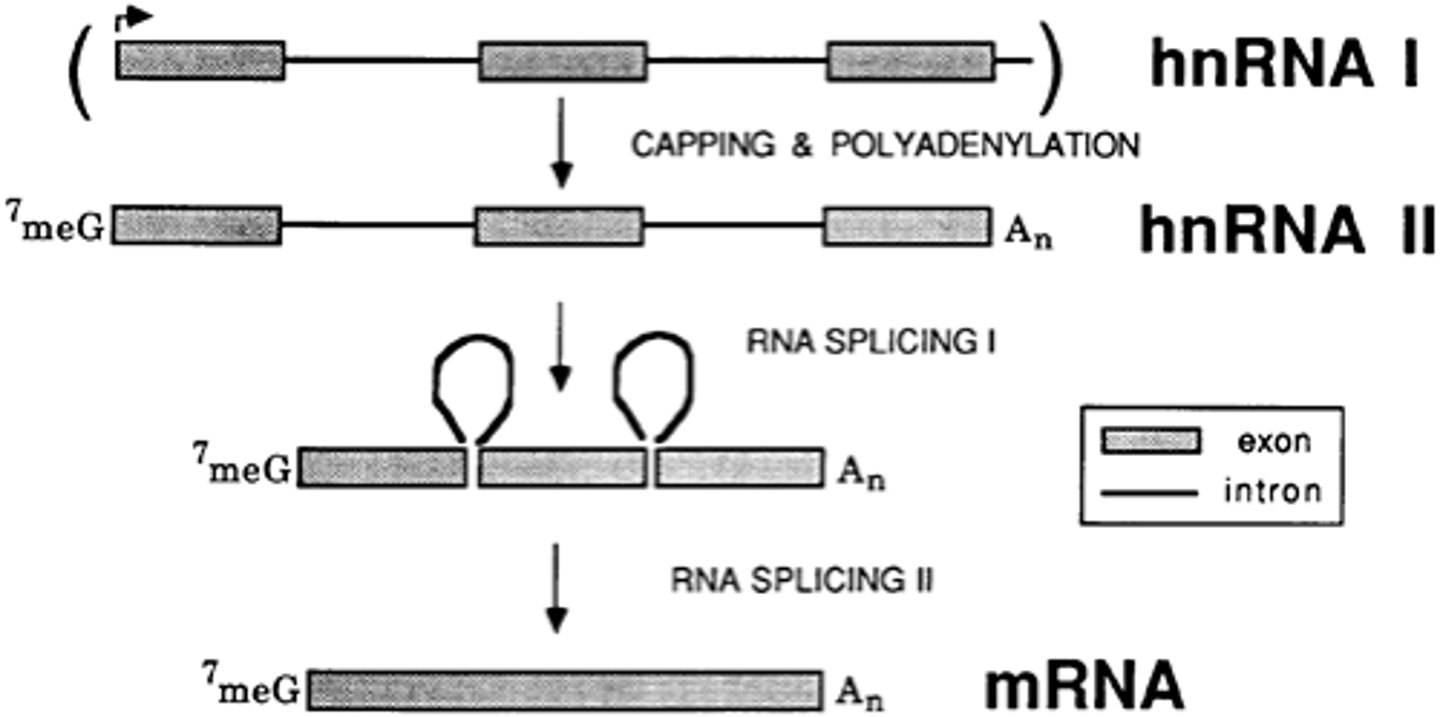
3 Biggest post-transcriptional modifications of hnRNA:
1) intron-exon splicing, 2) 5' cap, 3) 3' poly-A tail
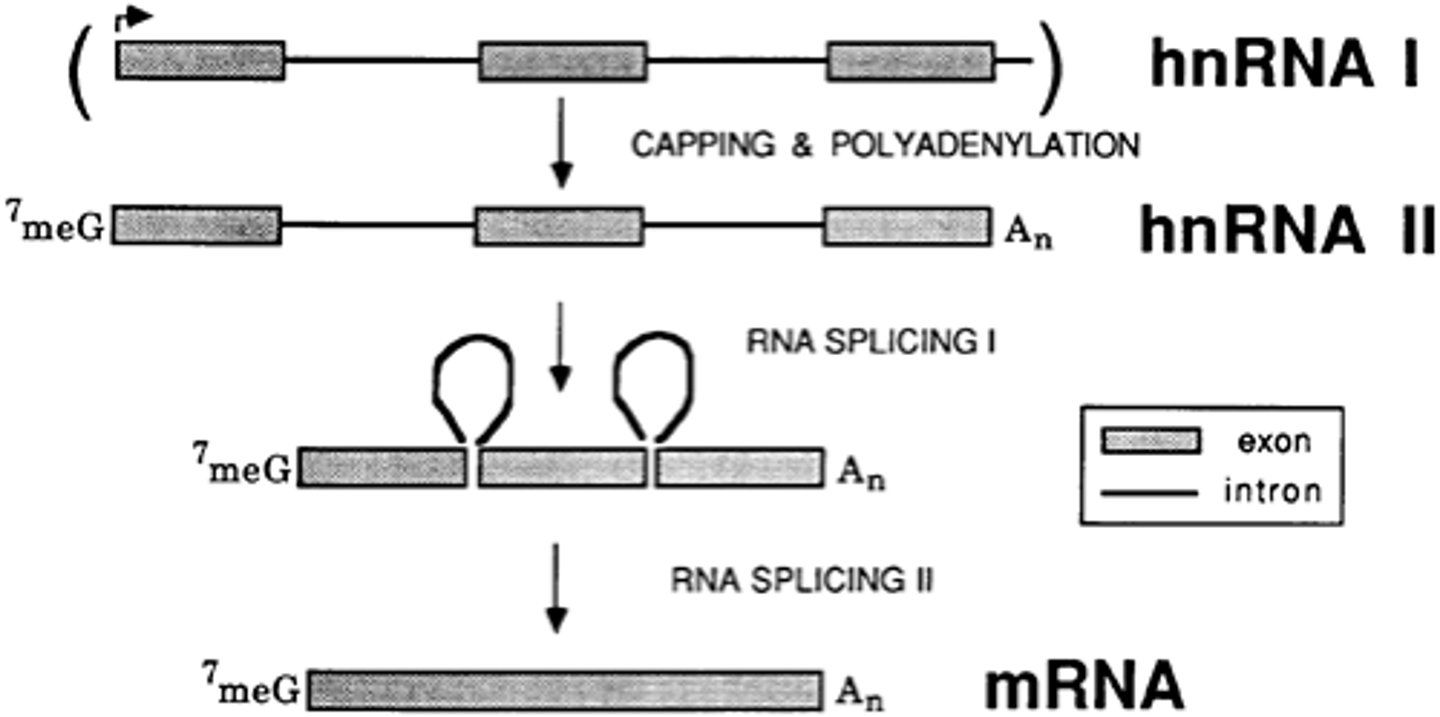
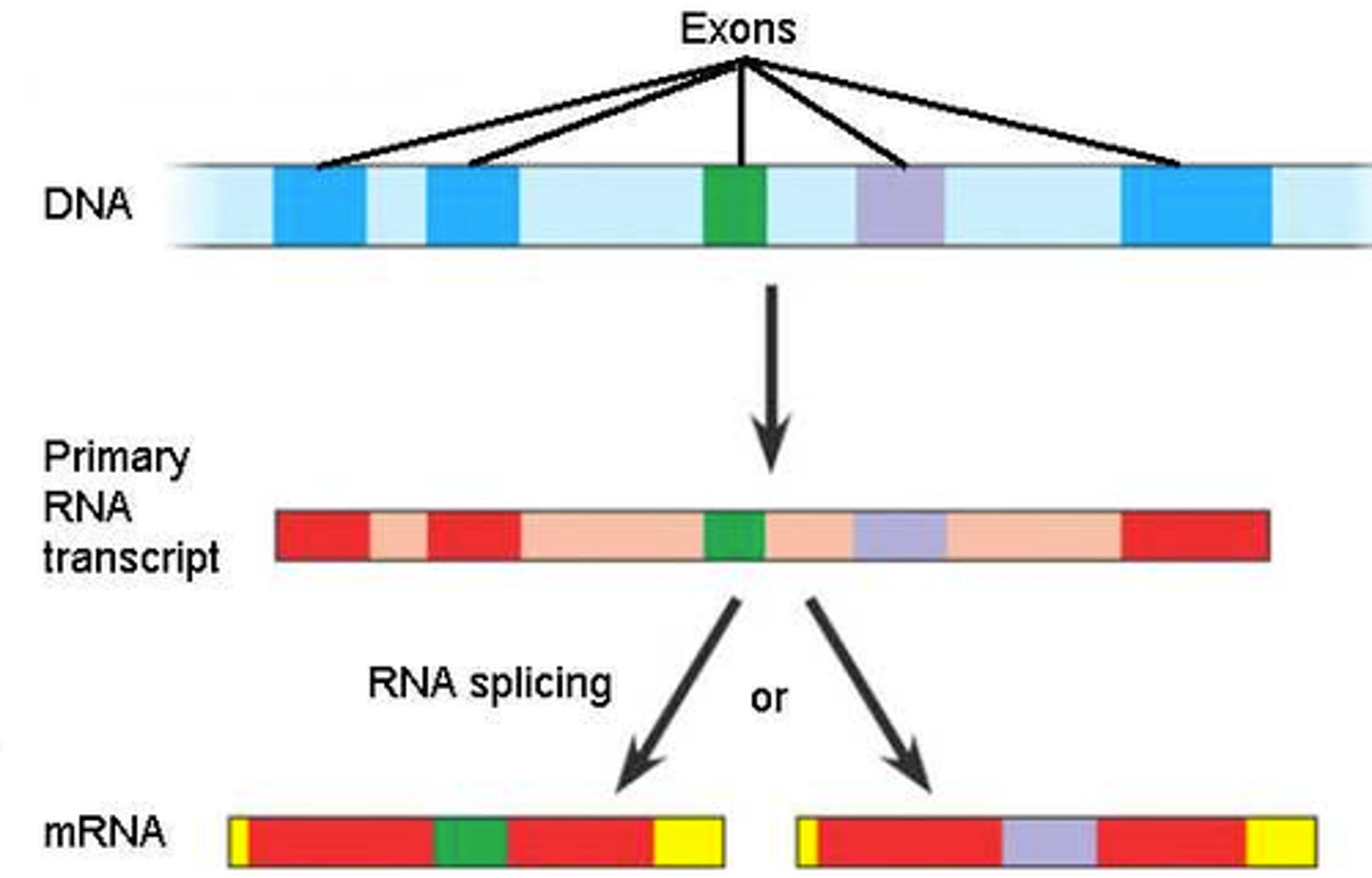
Post-transcriptional modification of hnRNA: intron-exon splicing
introns are non-coding regions thus you want to remove them and paste exons together
the Spliceosome contains snRNA and small nuclear ribonucleoproteins (snRNPs): snRNP-snRNA recognizes 5' and 3' of the splice site, forms them into a lariat (lasso), excises it, and degrades it
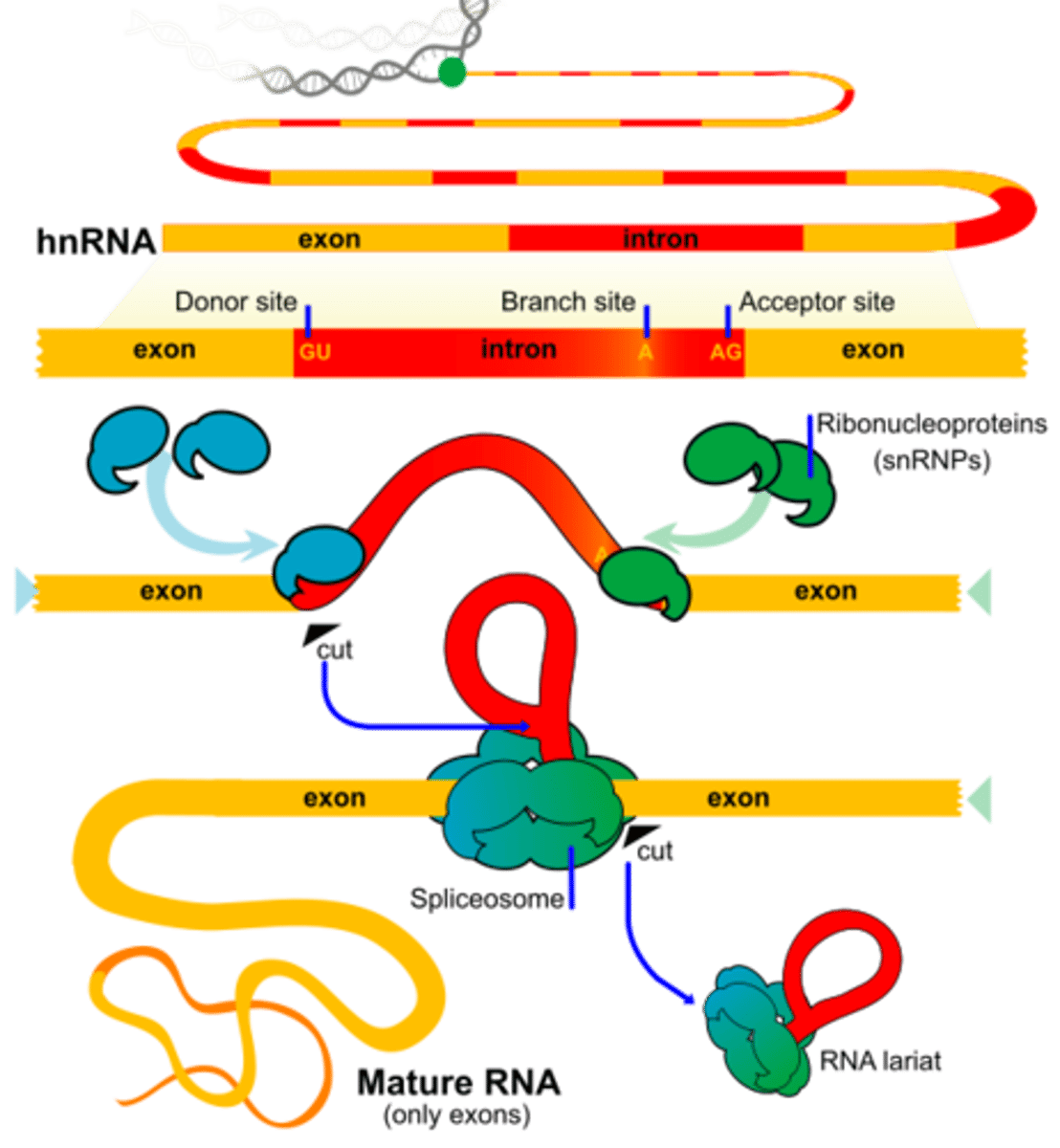
Spliceosome Structure
the complex that removes introns from hnRNA; contains snRNA & small nuclear ribonucleoproteins (snRNPs): snRNP-snRNA recognizes 5' and 3' of the splice site, forms them into a lariat (lasso), excises it, and degrades it
Evolutionary theory of why introns exist (3)
probably
1) to regulate gene expression
2) maintain genome size
3) allows rapid protein evolution due to many shared 'modular' peptide sequences between proteins that can be swapped in and out
Post-transcriptional modification of hnRNA: 5' Cap? purpose?
7-methylguanylate triphosphate cap added; added during TRANSCRIPTION ITSELF after ribosome recognizes it to bind; protects against 5' end degradation of mRNA
AKA: The 5' cap is added to the first nucleotide in the transcript during transcription. The cap is a modified guanine (G) nucleotide, and it protects the transcript from being broken down. It also helps the ribosome attach to the mRNA and start reading it to make a protein.
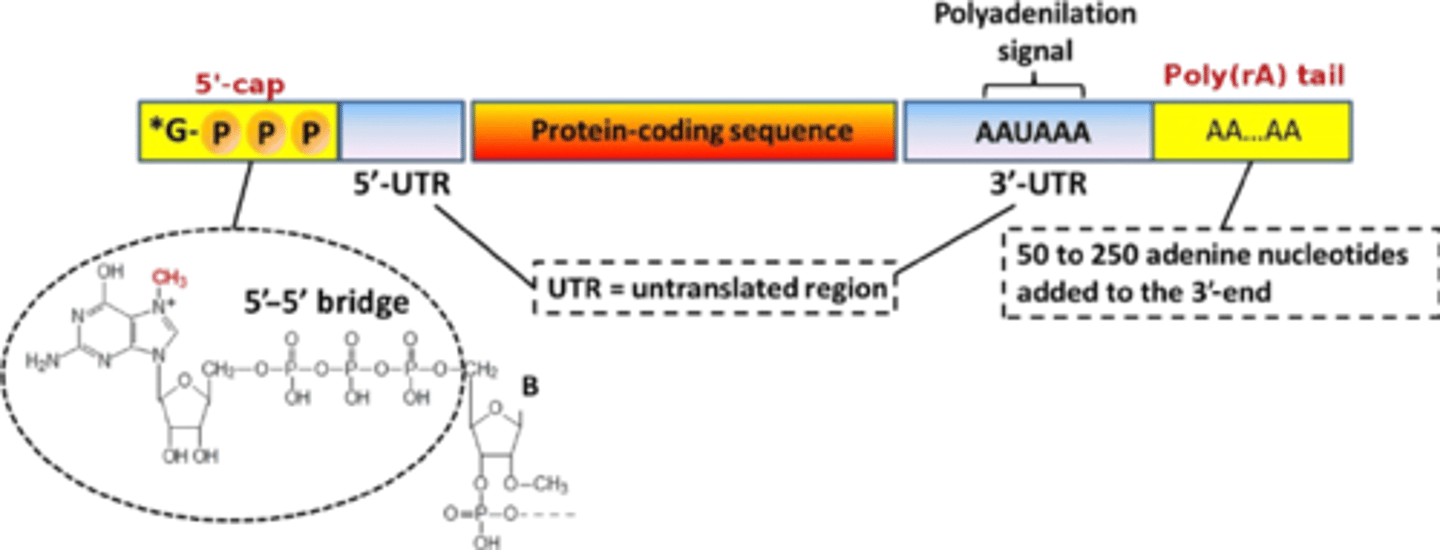
Post-transcriptional modification of hnRNA: 3' Poly-A Tail? purposes?
polyadenosyl tail is added that protects against mRNA degradation; acts as a 'fuse' for a bomb: as soon as mRNA leaves nucleus, this end starts to get degraded (the longer the tail is, the longer mRNA lives) & allows transport out of nucleus
AKA: In nuclear polyadenylation, a poly(A) tail is added to an RNA at the end of transcription. On mRNAs, the poly(A) tail protects the mRNA molecule from enzymatic degradation in the cytoplasm and aids in transcription termination, export of the mRNA from the nucleus, and translation.
Alternative Splicing
the concept that splicing an hnRNA in different ways can produce different mRNAs for different proteins for regulation & diversity
known since humans produce WAY MORE proteins than distinct genes (100k proteins : 25k genes), meaning something like this must happen
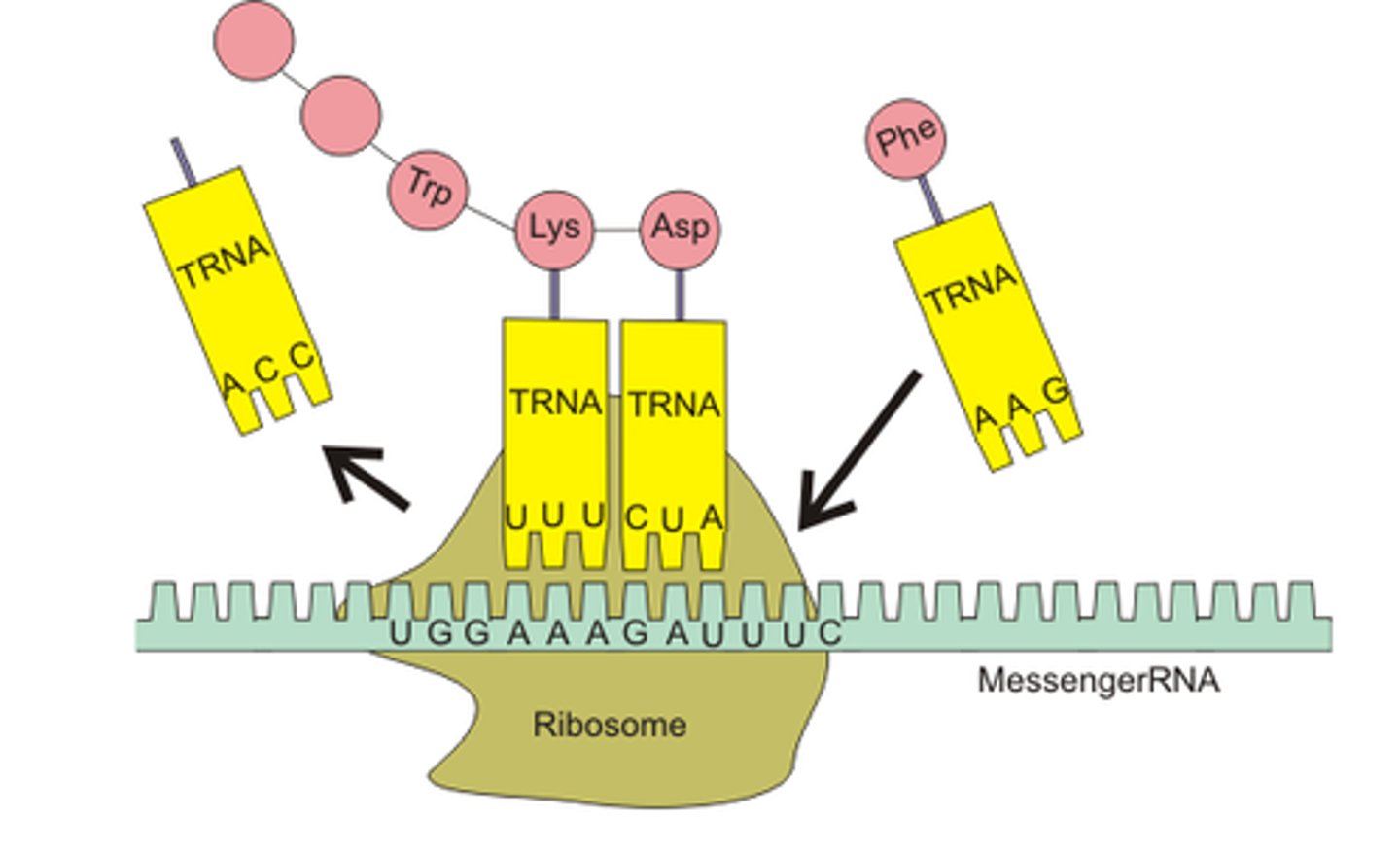
Translation (genetics)
converting mRNA to protein
The energy molecule that powers translation is _______________
GTP
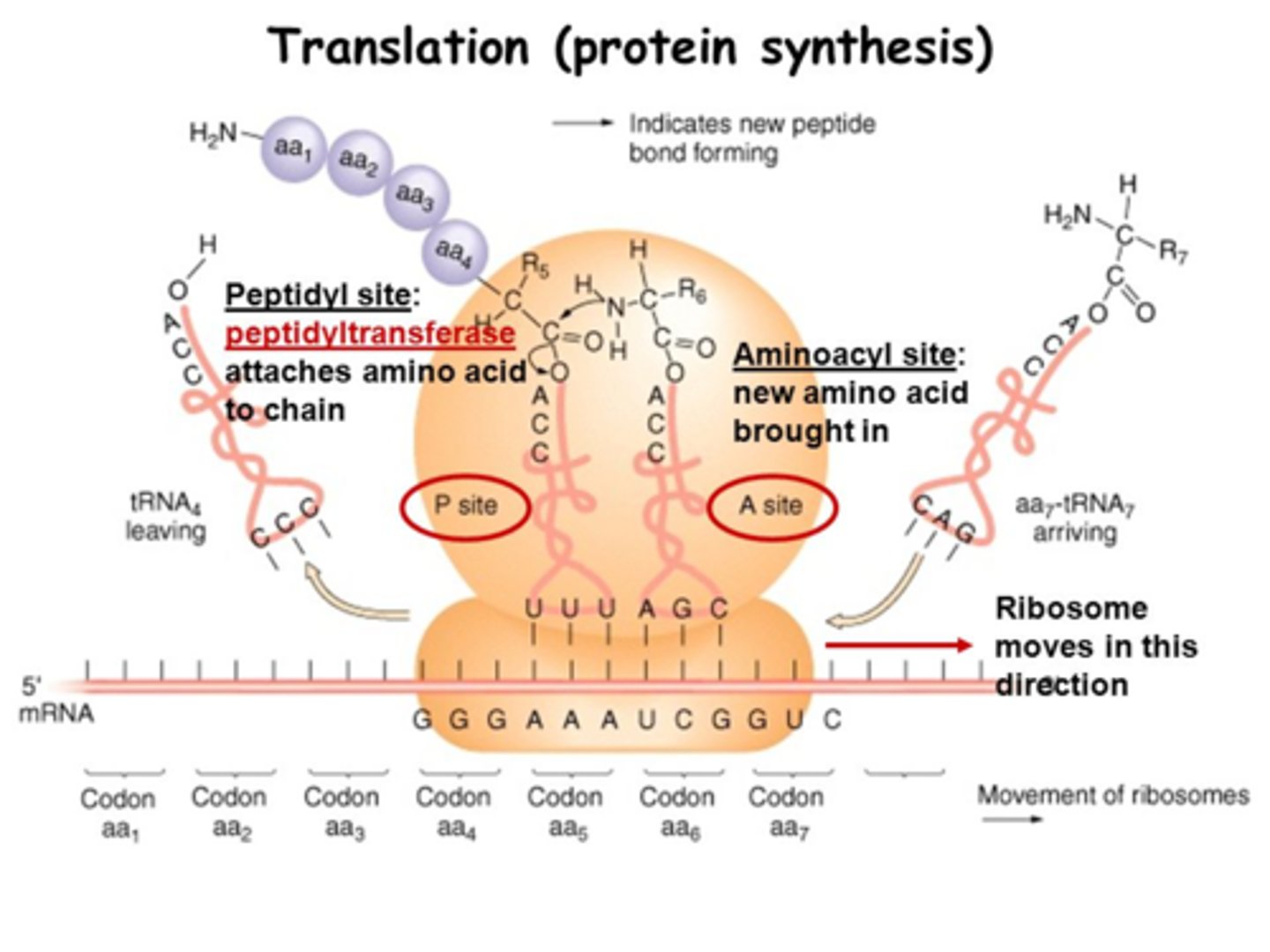
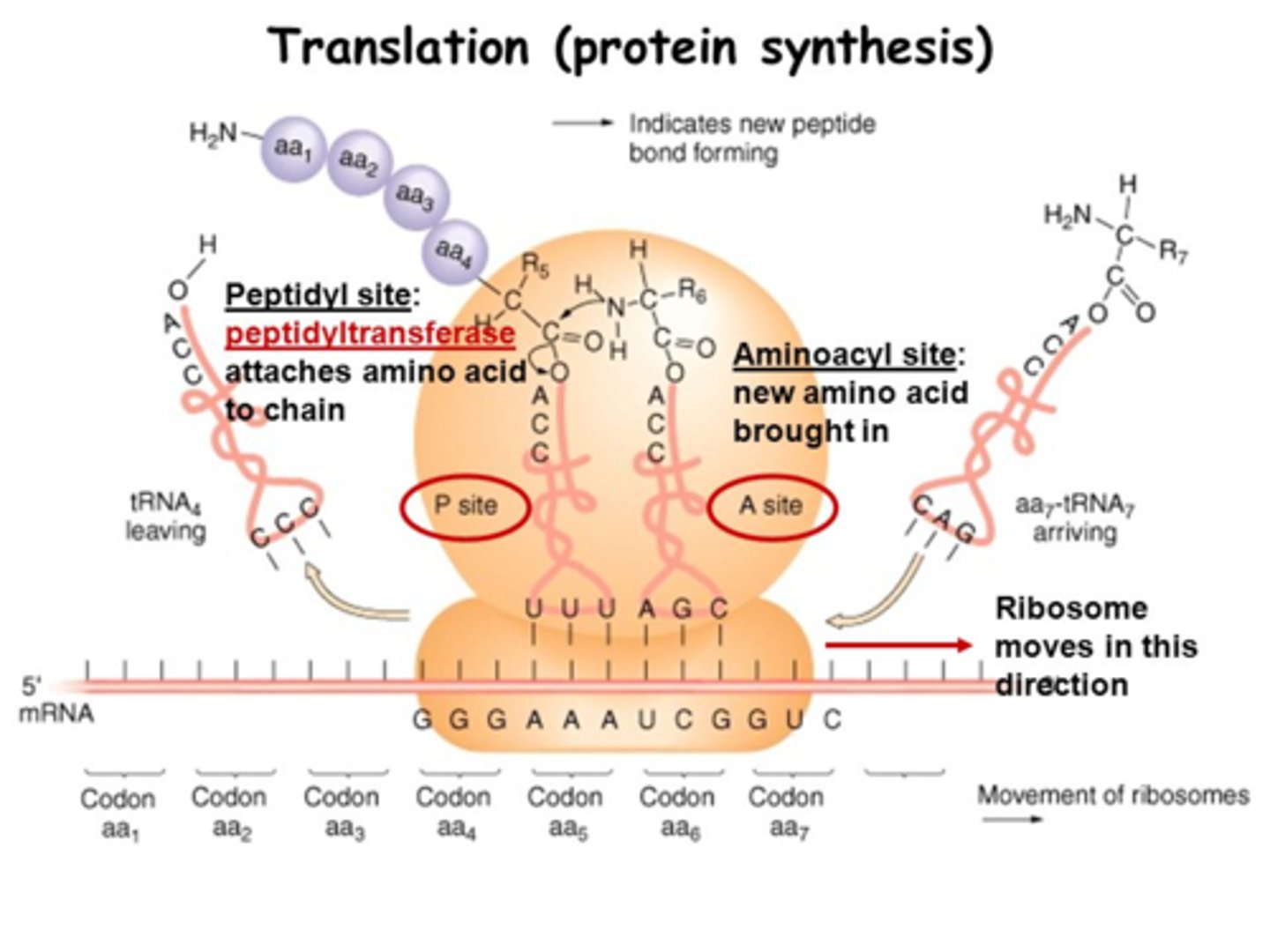
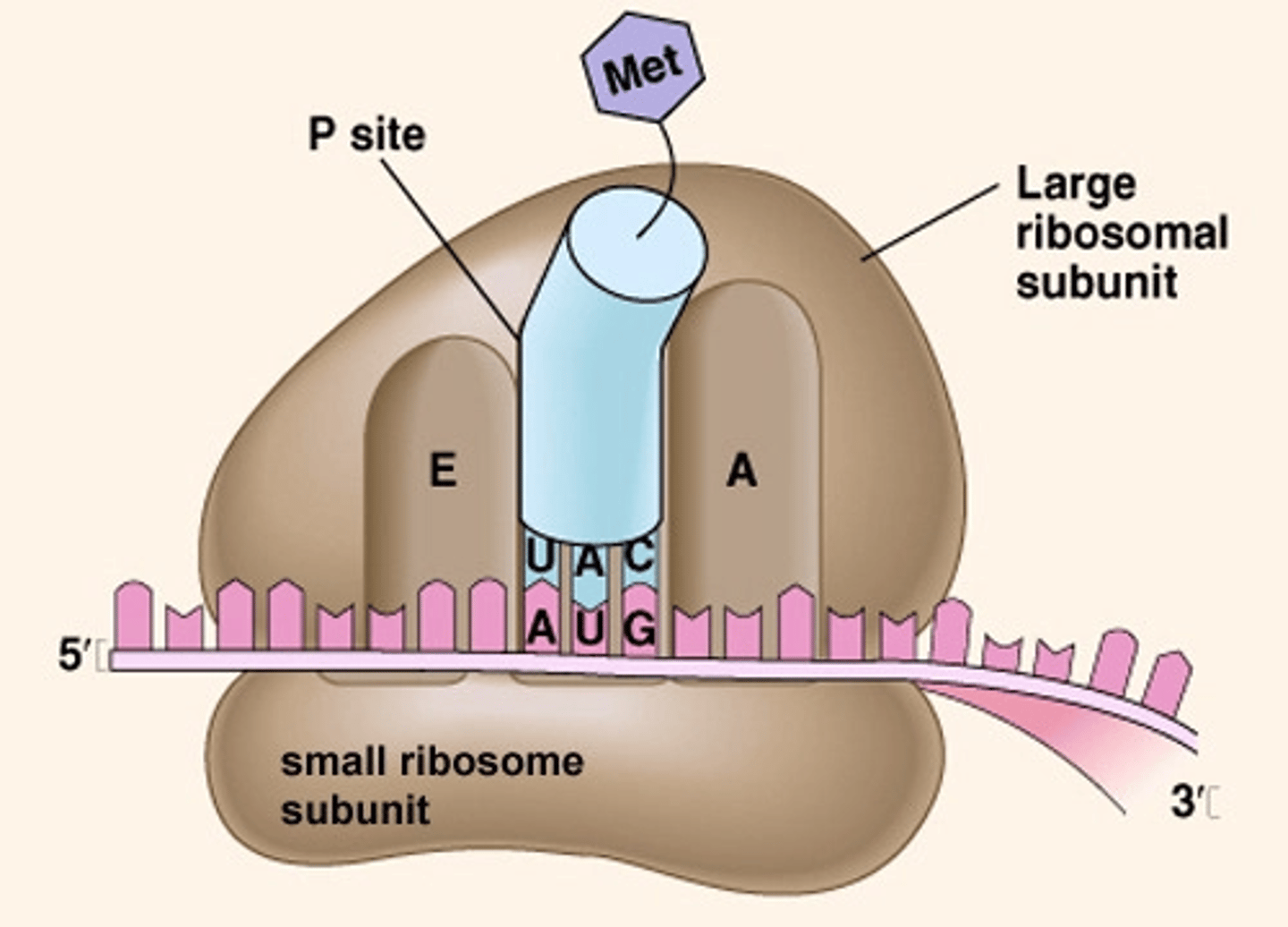
Ribosome structure? sites? subunits?
protein + rRNA; a small & large subunit (in both euk and pro) which only bind together during translation; 3 sites: A (aminoacyl), P (peptidyl), E (exit)
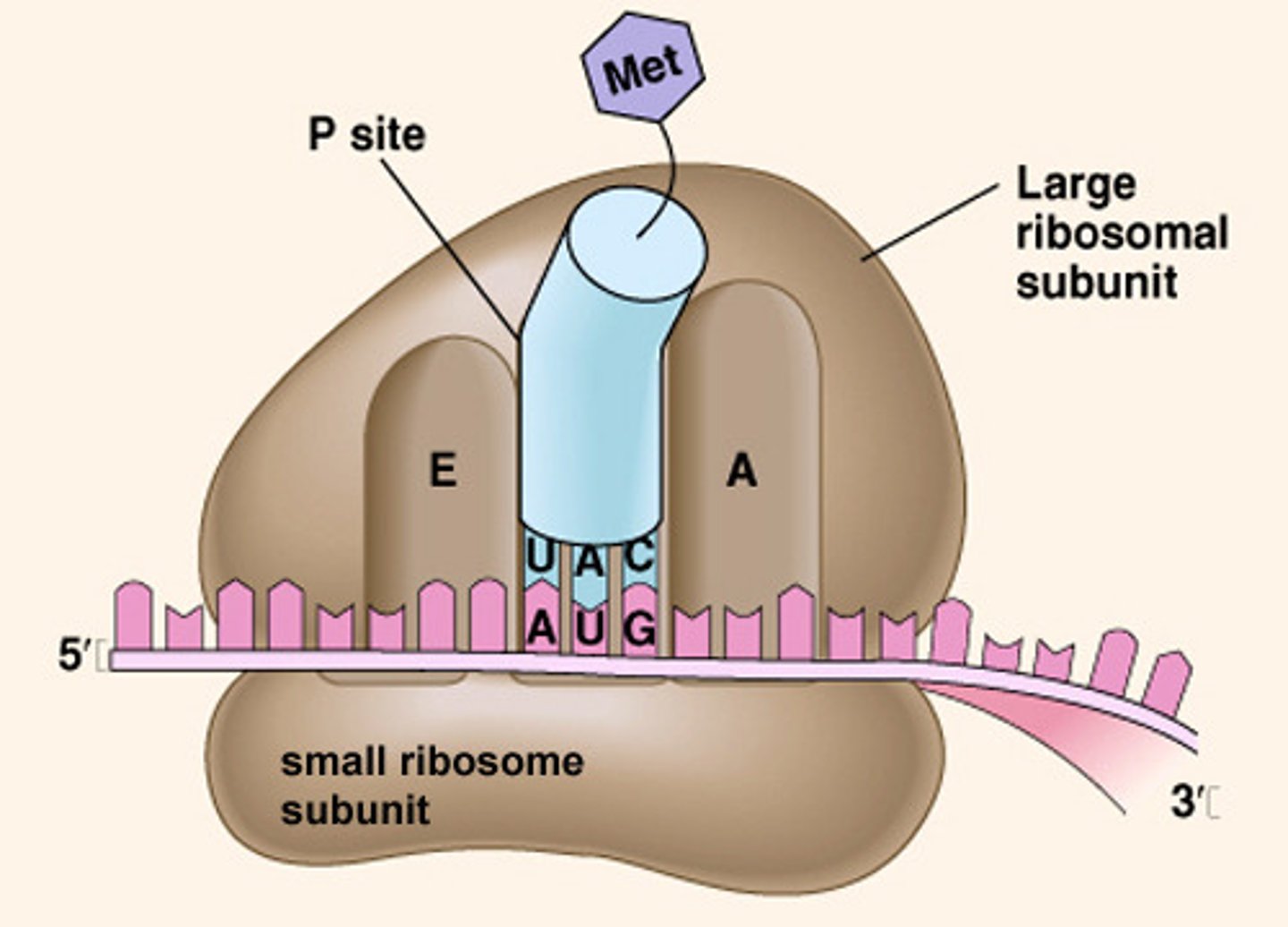
3 sites of a ribosome:
A (aminoacyl), P (peptidyl), E (exit)
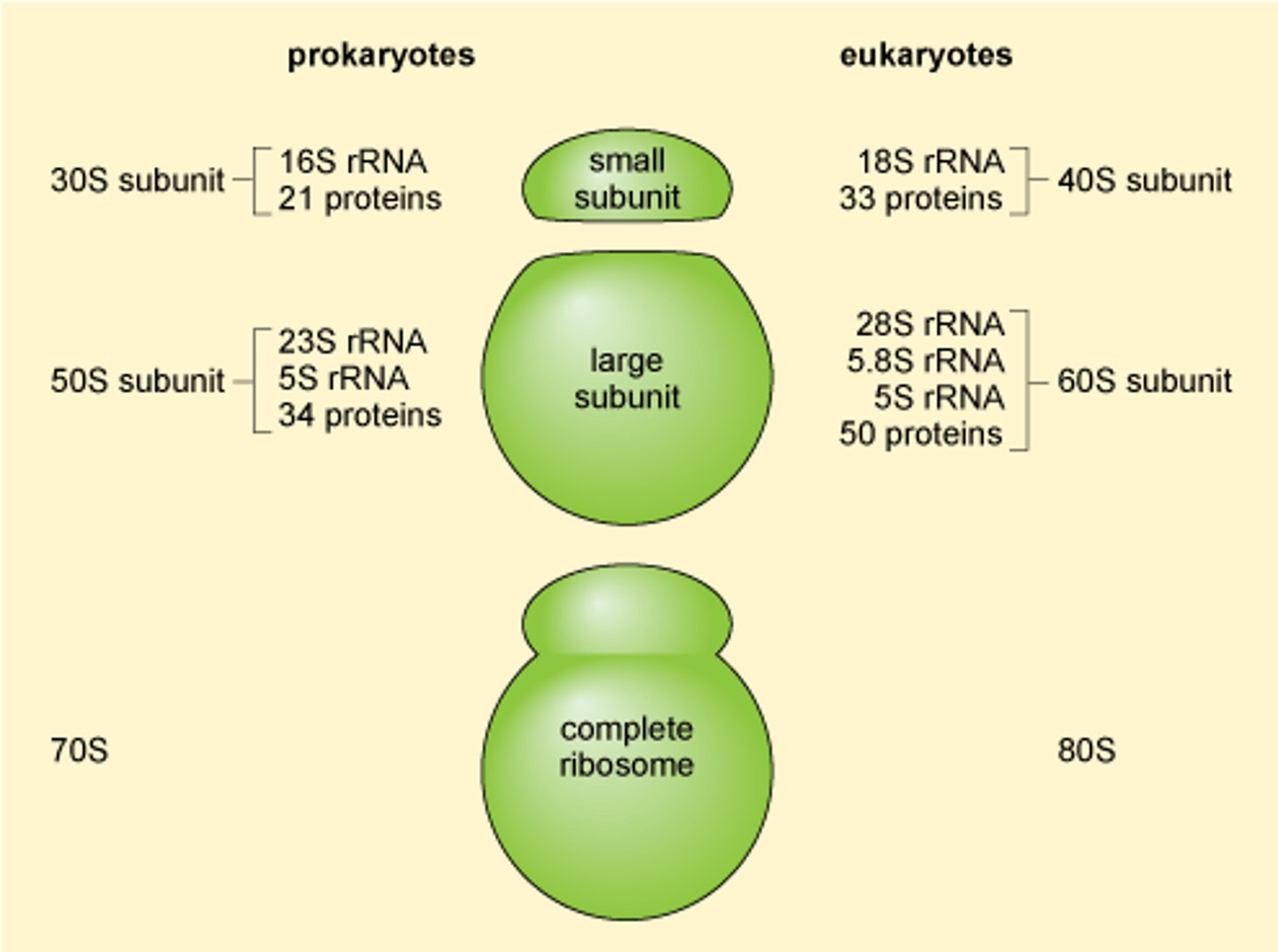
Eukaryotic rRNA in the ribosome (# & names):
4 strands: 28S, 18S, 5.8S, 5S (where the numbers indicate their size)
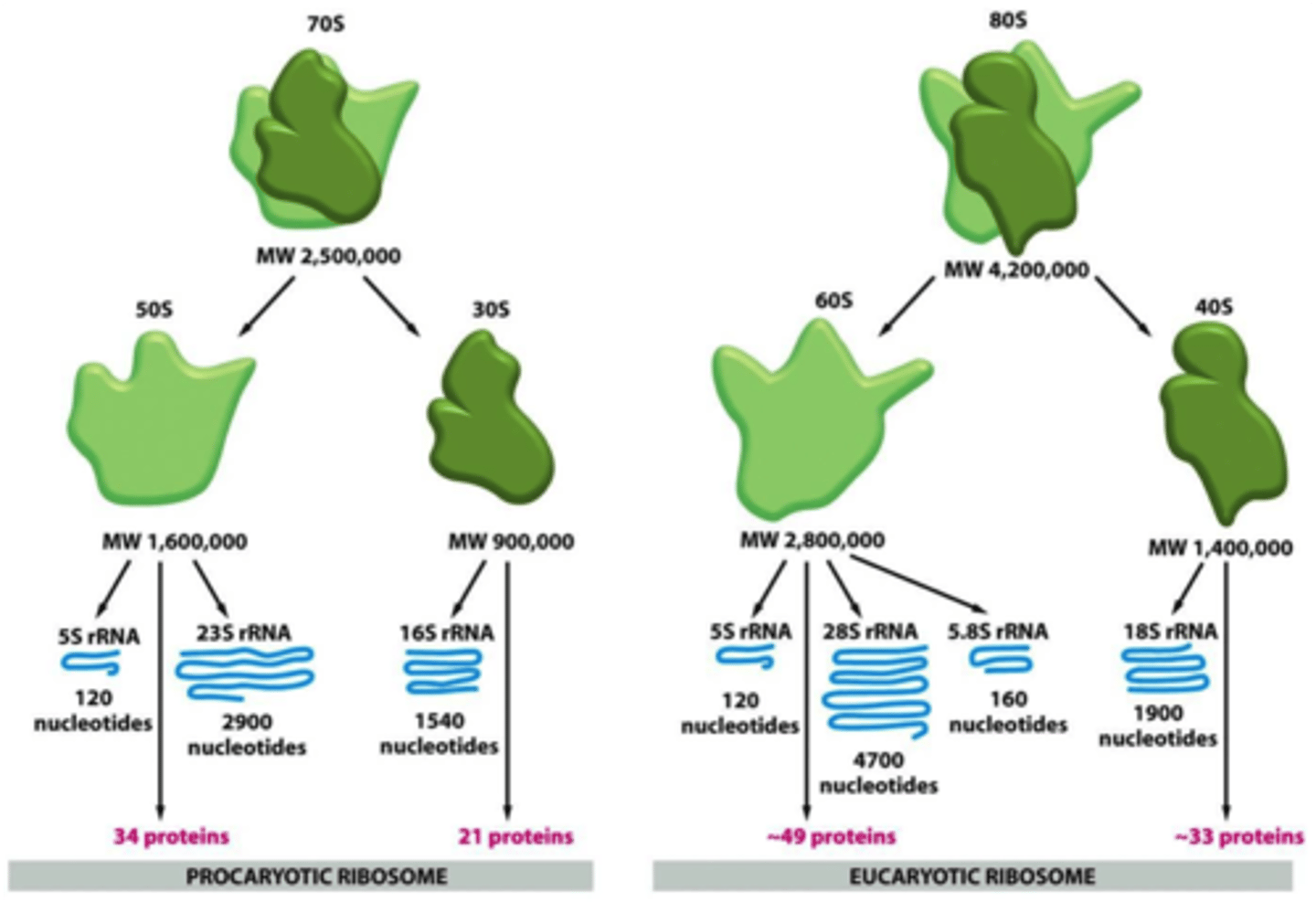
How the 28S, 18S, 5.8S rRNA, & 5S for the ribosome are synthesized? which is in the large? which is in the small subunit?
RNA pol I synthesizes them as 1 45S rRNA in the nucleolus ➝ processed to 18S (40S small subunit) & 28S + 5.8S (60S large subunit)
5S is synthesized by RNA pol III outside of nucleolus and placed in the 60S subunit
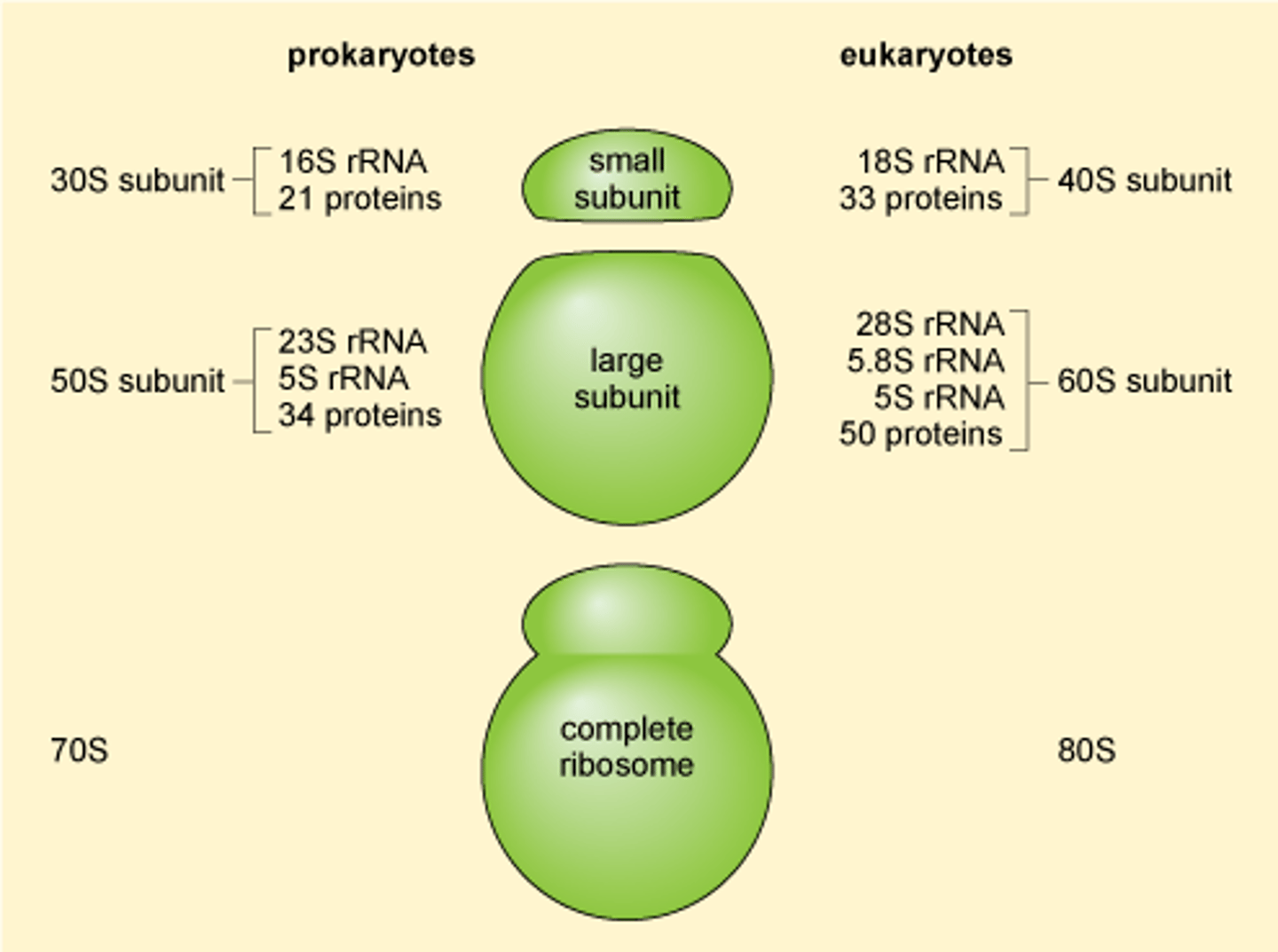
Overall 'S' of the eukaryotic ribosome and its 2 subunits? what does the S mean?
80S; made up of 60S (28S + 5.8S + 5S) and 40S (18S) subunits, with each of those sub-subunits
it is a value determined experimentally based on SIZE and SHAPE of the rRNAs; THUS THEY ARE NOT ADDITIVE (e.g., the 80S eukaryotic ribosome is made up of 40S + 60S subunits)
'S' of the prokaryotic ribosome + its subunits? what does the S mean?
70S ribosome: 50S + 30S subunits
it is a value determined experimentally based on SIZE and SHAPE of the rRNAs; THUS THEY ARE NOT ADDITIVE (e.g., the 80S eukaryotic ribosome is made up of 40S + 60S subunits)
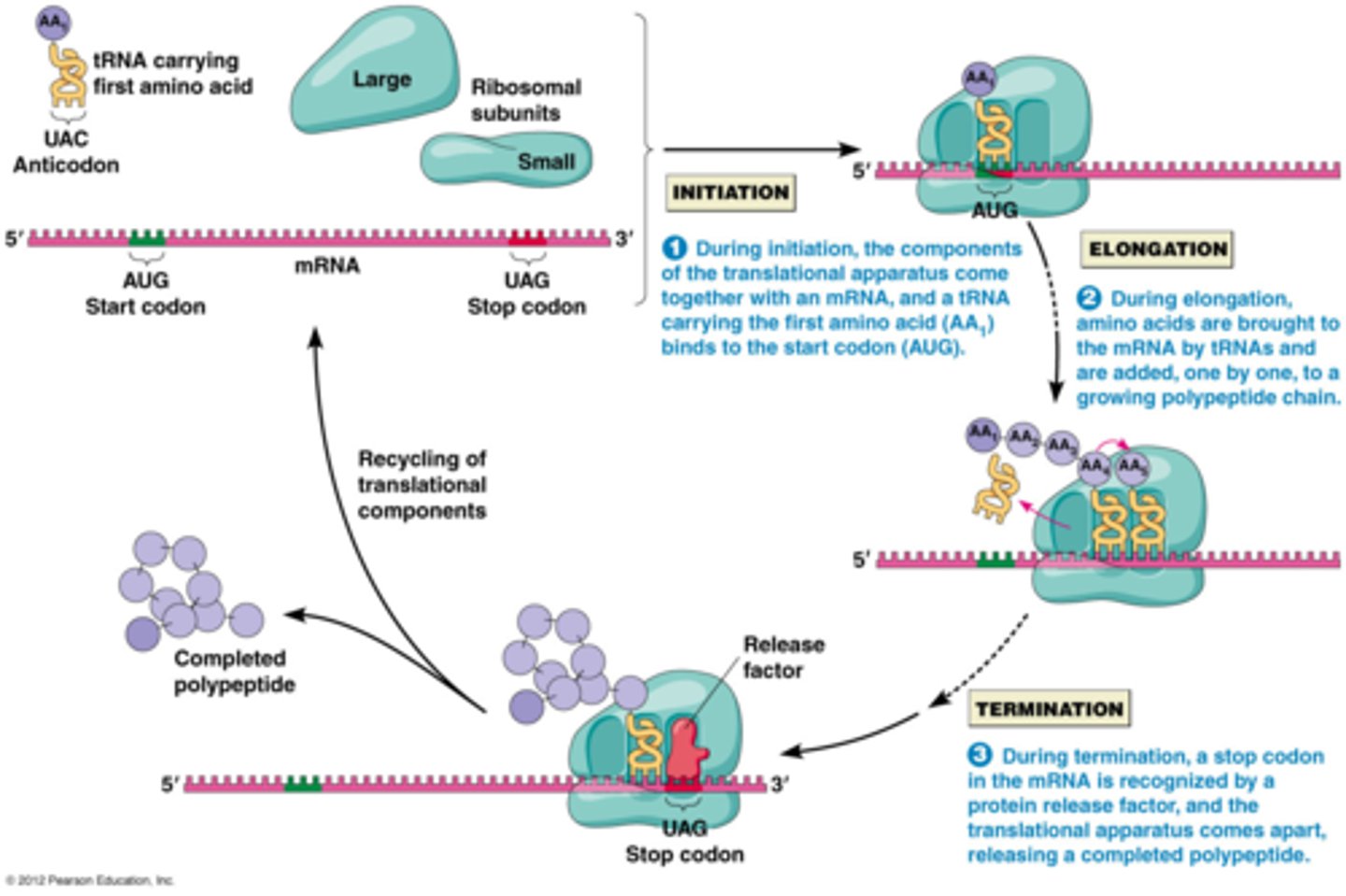
How translation happens
in the cytoplasm:
1) initiation,
2) elongation,
3) termination occur in sequence with 'factors' corresponding to their names ('initiation factors' for example), using GTP to push
In eukaryotes, translation occurs __________ transcription, and in prokaryotes, translation occurs ___________ transcription
(before/during/after)
after; during (prokaryotes have no nucleus so ribosome starts working right away; not compartmentalized)
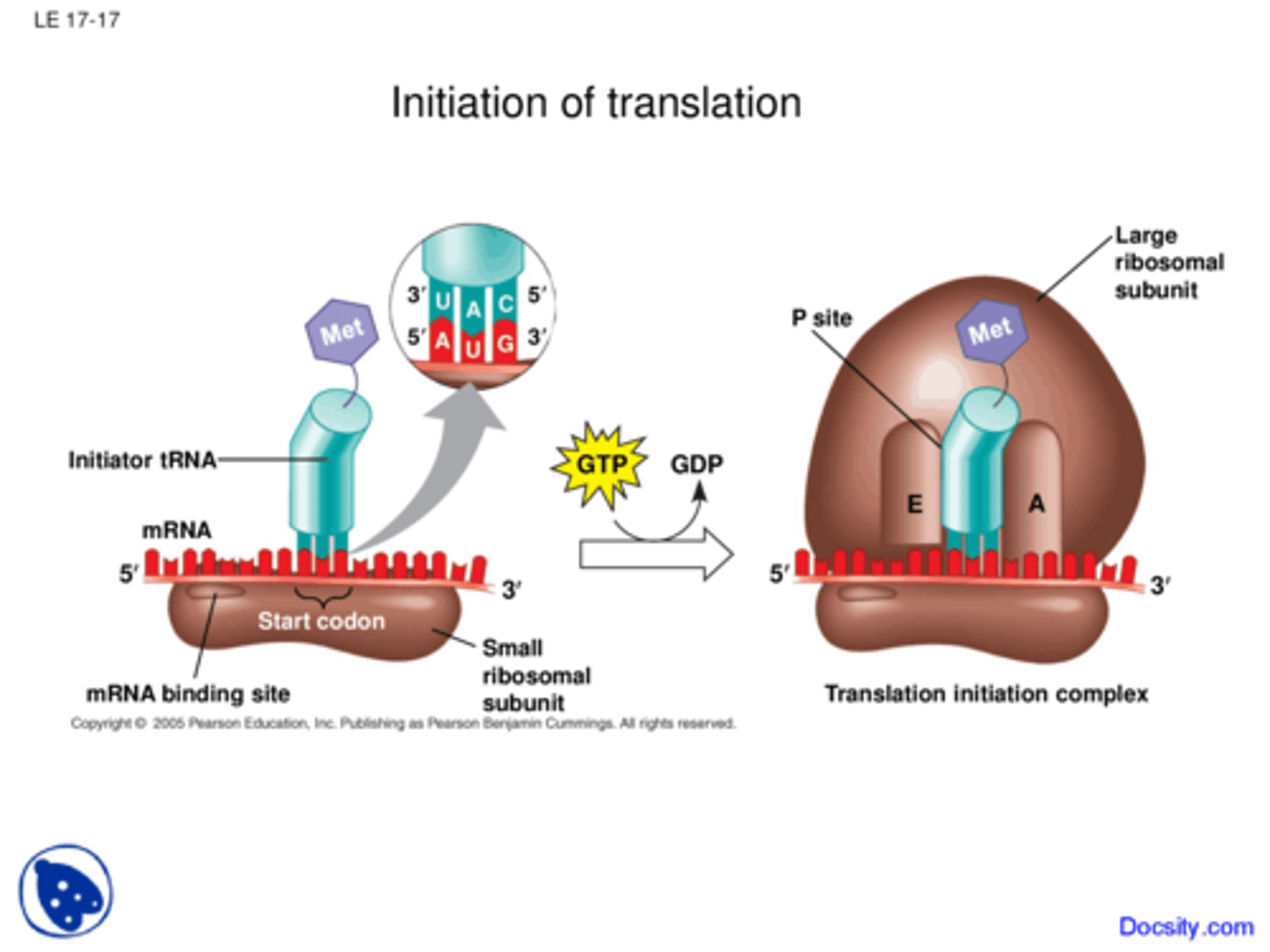
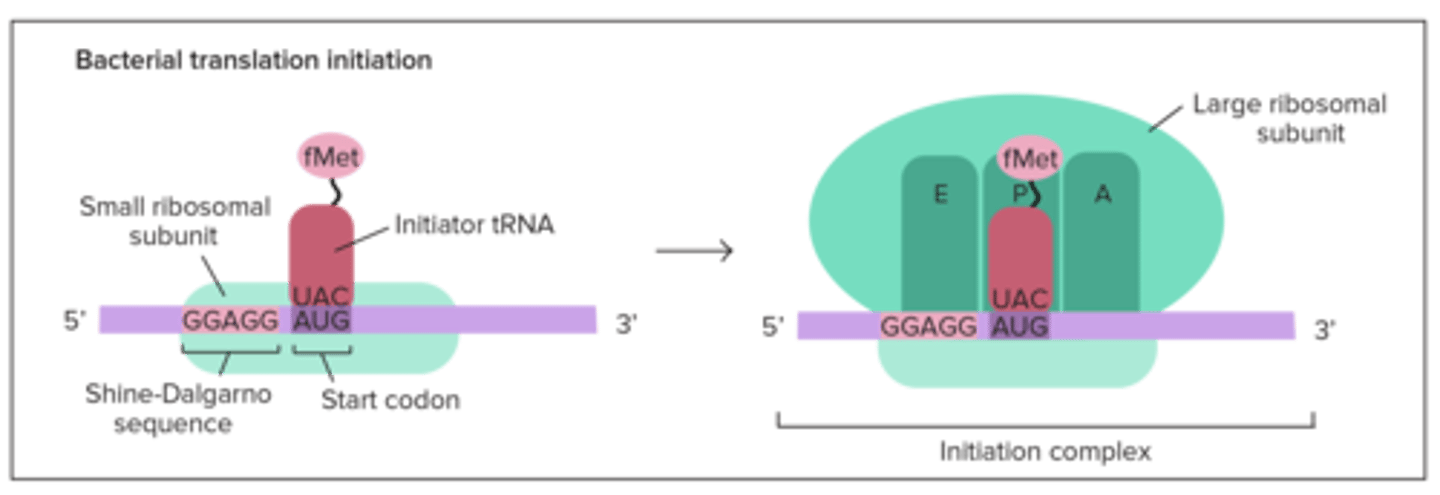
Translation: Initiation? prokaryotes? eukaryotes?
small ribosome binds to mRNA,
prokaryotes have it bind to Shine-Dalgarno Sequence in 5' UTR &
in eukaryotes, ribosome binds to 5' Cap; charged tRNA binds to mRNA's AUG start codon in the P site; large subunit then binds down onto small using initiation factors fleetingly
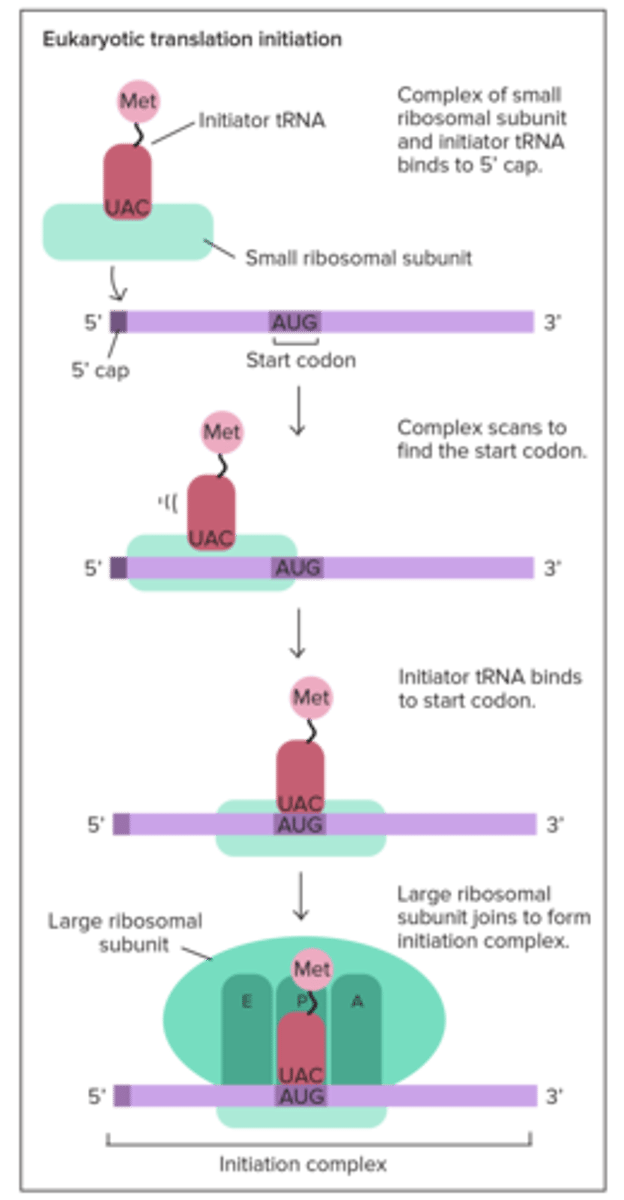
In translation initiation, the small ribosome subunit first binds to the ____________________________ in prokaryotes, and the _____________ in eukaryotes
Shine-Dalgarno Sequence; 5' cap
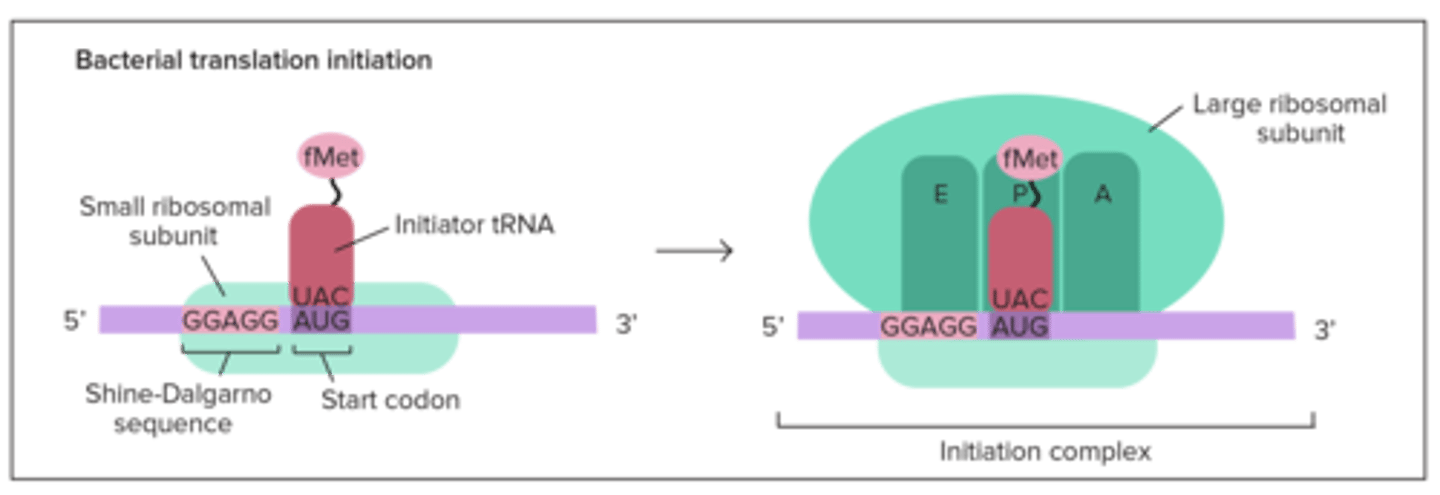
In prokaryotes, the AUG start codon codes for ________, and in eukaryotes it codes for _________
fMet (N-formylmethionine); Met (normal)
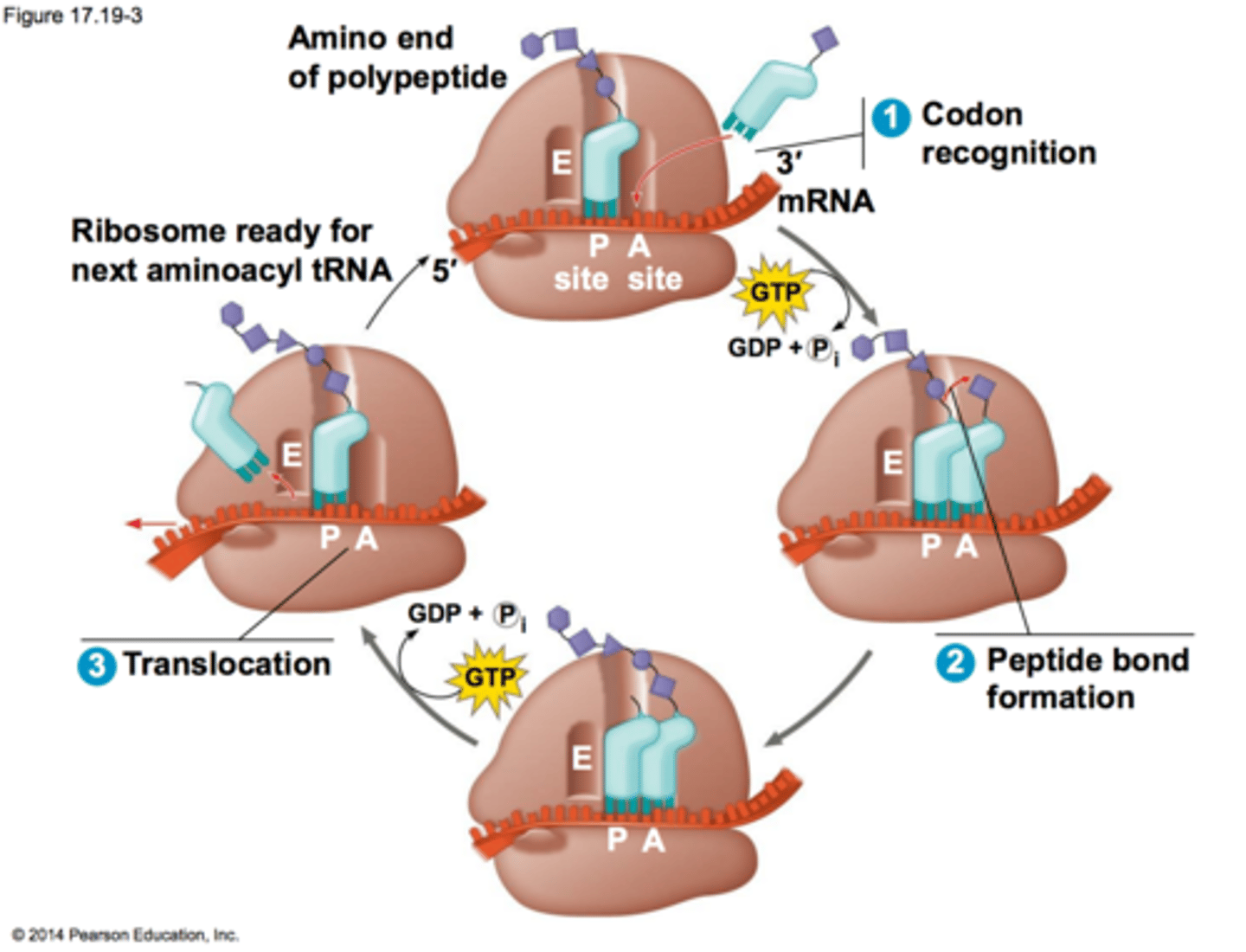
Translation: Elongation? steps?
3-step cycle that is repeated in translation:
ribosome moves 5' to 3' on mRNA
1) A site holds the aminoacyl-tRNA depending on what mRNA codon enters the A site,
2) the P site holds a tRNA that is carrying the polypeptide chain, which forms peptide bonds as the peptide moves from P to A site using Peptide Transferase in the large subunit, using GTP to power the bond's formation;
3) E site is where uncharged tRNA with no AA sit before they are released, and it unbinds from mRNA; Elongation Factors recruit AA-tRNA and GTP and remove the GDP
In what site does the first AA in a peptide enter? Where do all the rest enter?
the P site is where Met first enters, but all the other AA-tRNA enter at the A site
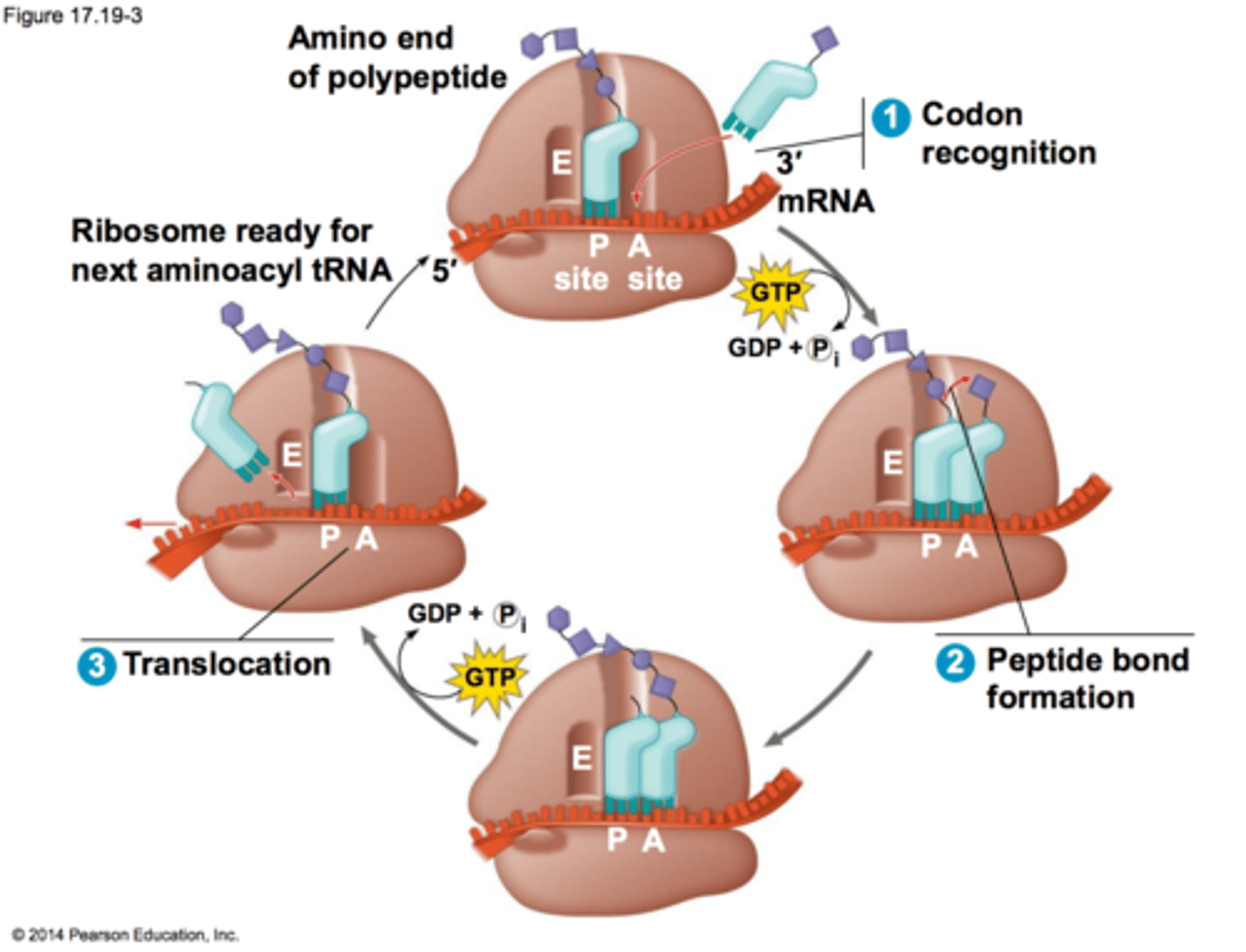
What do translation elongation factors do?
they recruit the correct AA-tRNA for transcription along with GTP to power peptide synthesis, and remove the GDP after
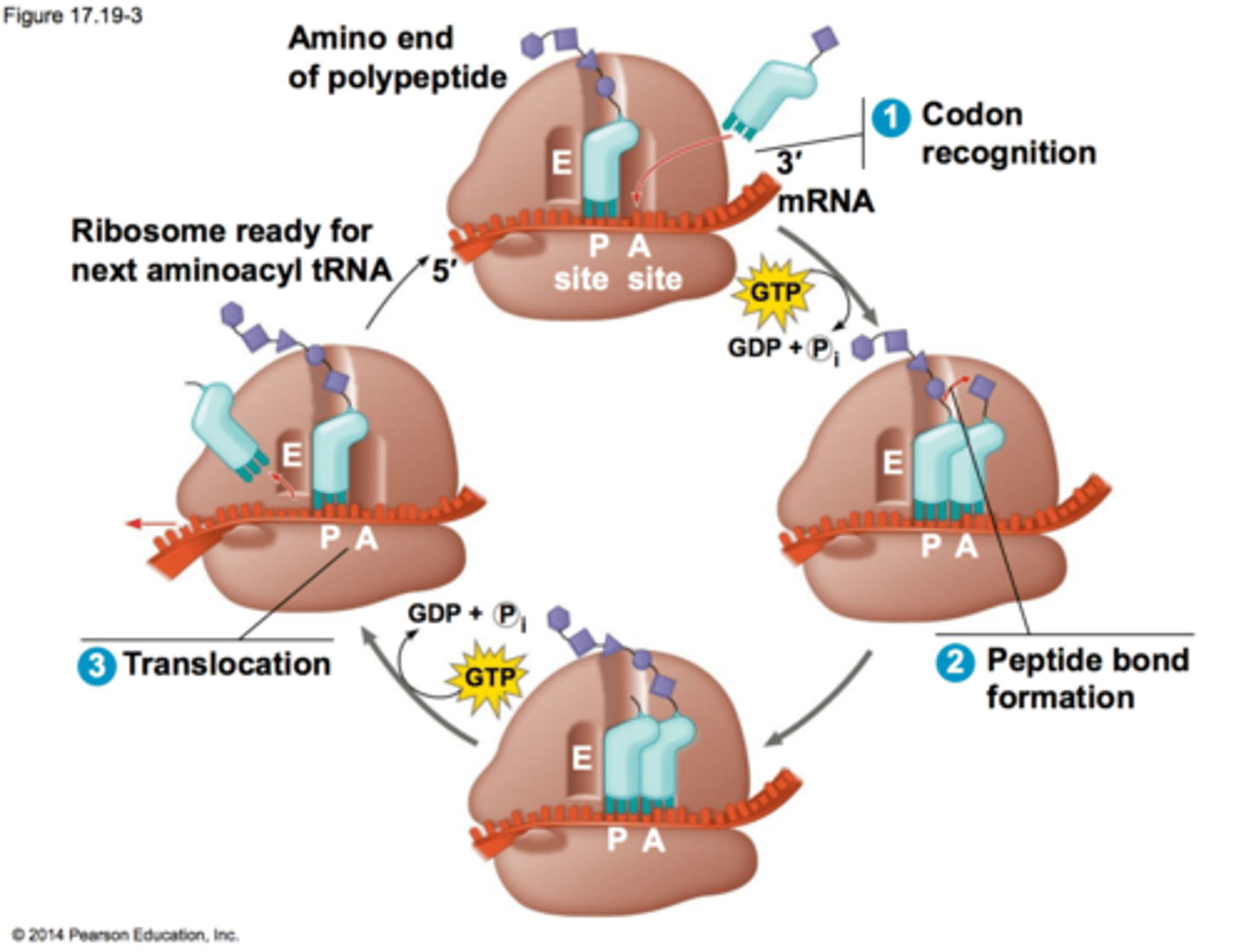
Peptide Transferase function?
an enzyme in the large subunit of the ribosome; forms the peptide bond using GTP
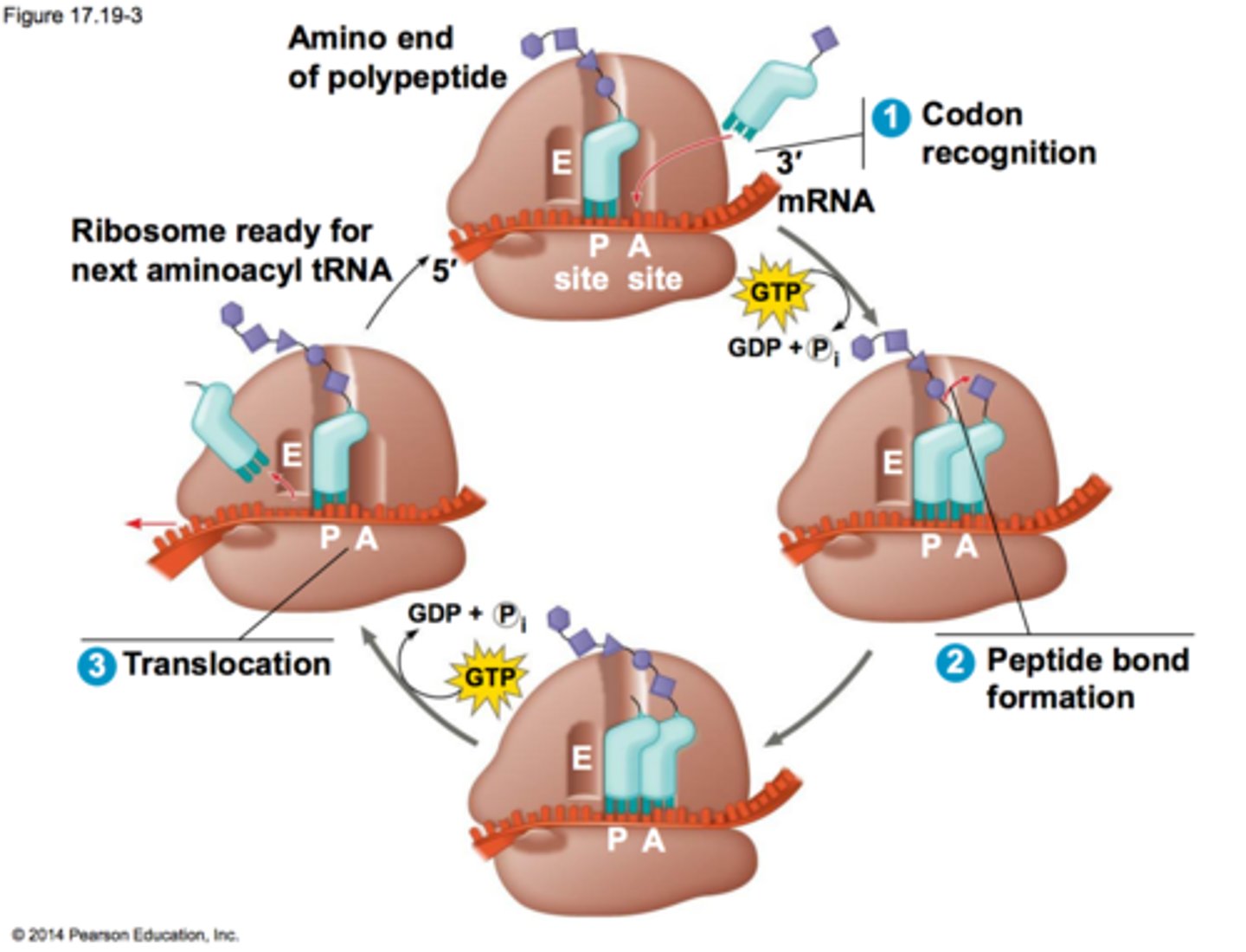
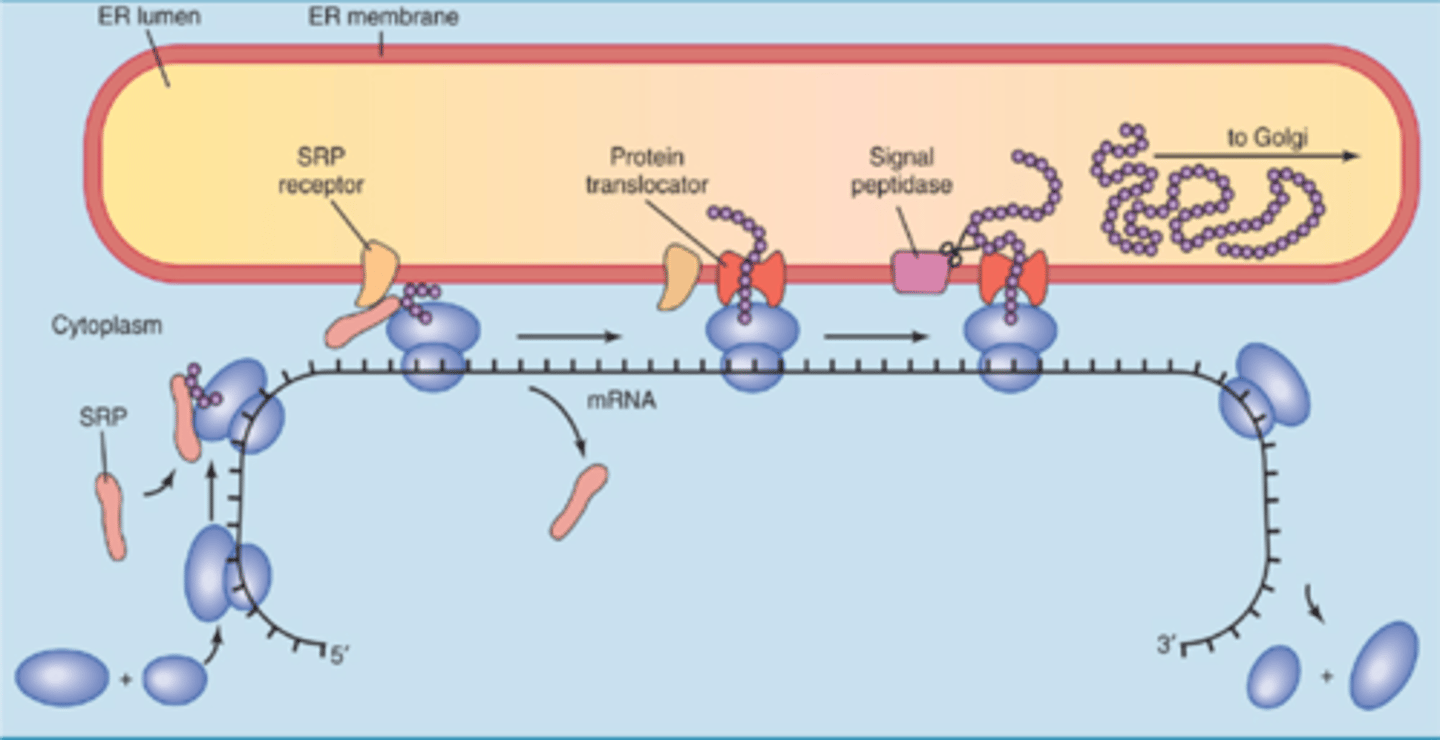
Signal Sequences (translation)
sequences on a protein that designates a destination for it: Secreted Proteins (hormones, enzymes) are directed to ER and translated into the lumen of rough ER, and then sent to golgi or secreted in a vesicle
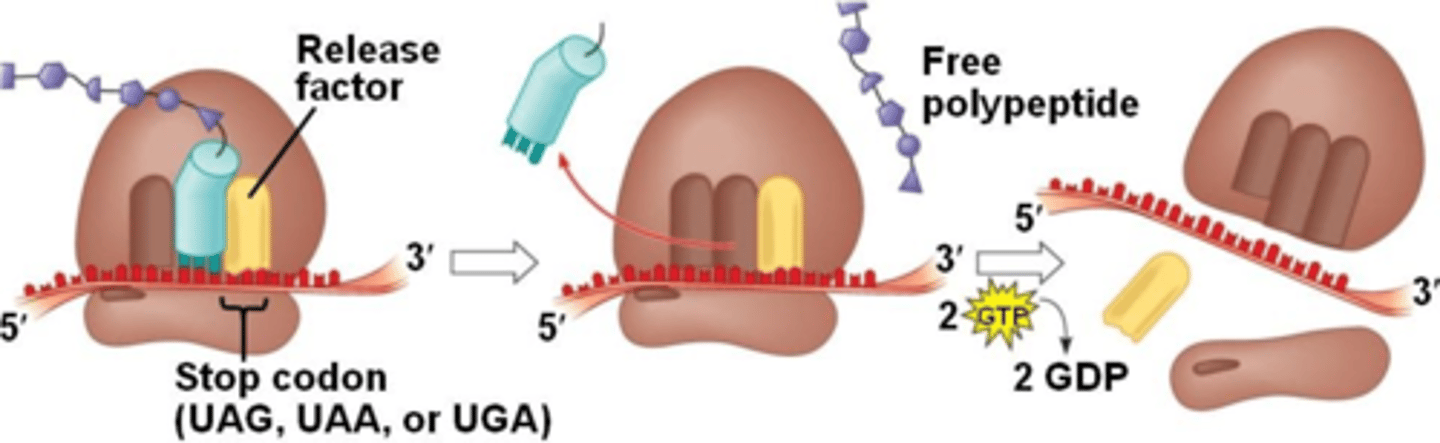
Translation: Termination
when any of the 3 stop codons move into the A site, Release Factor (RF) protein binds to the termination codon adding a water to the peptide, which signals peptidyl transferase & termination factors to hydrolyze off the peptide chain from the last tRNA, releasing the tRNA from the P site and releasing the 2 ribosome subunits
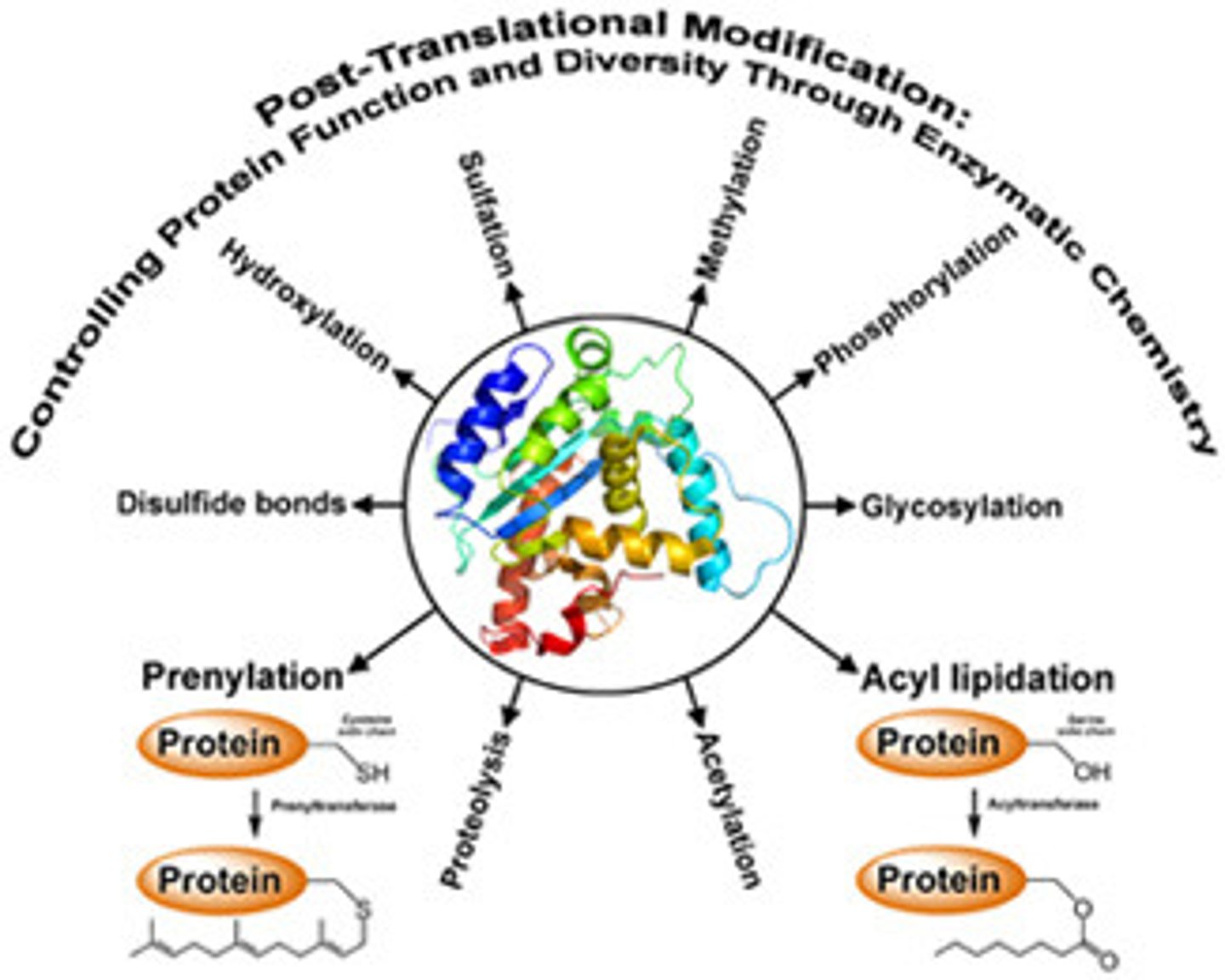
Post-translational processing (4)
after translation, proteins are modified before functioning; commonly folded via chaperones; commonly cleaved to the active protein; commonly form 4˚ structure; groups can also be added
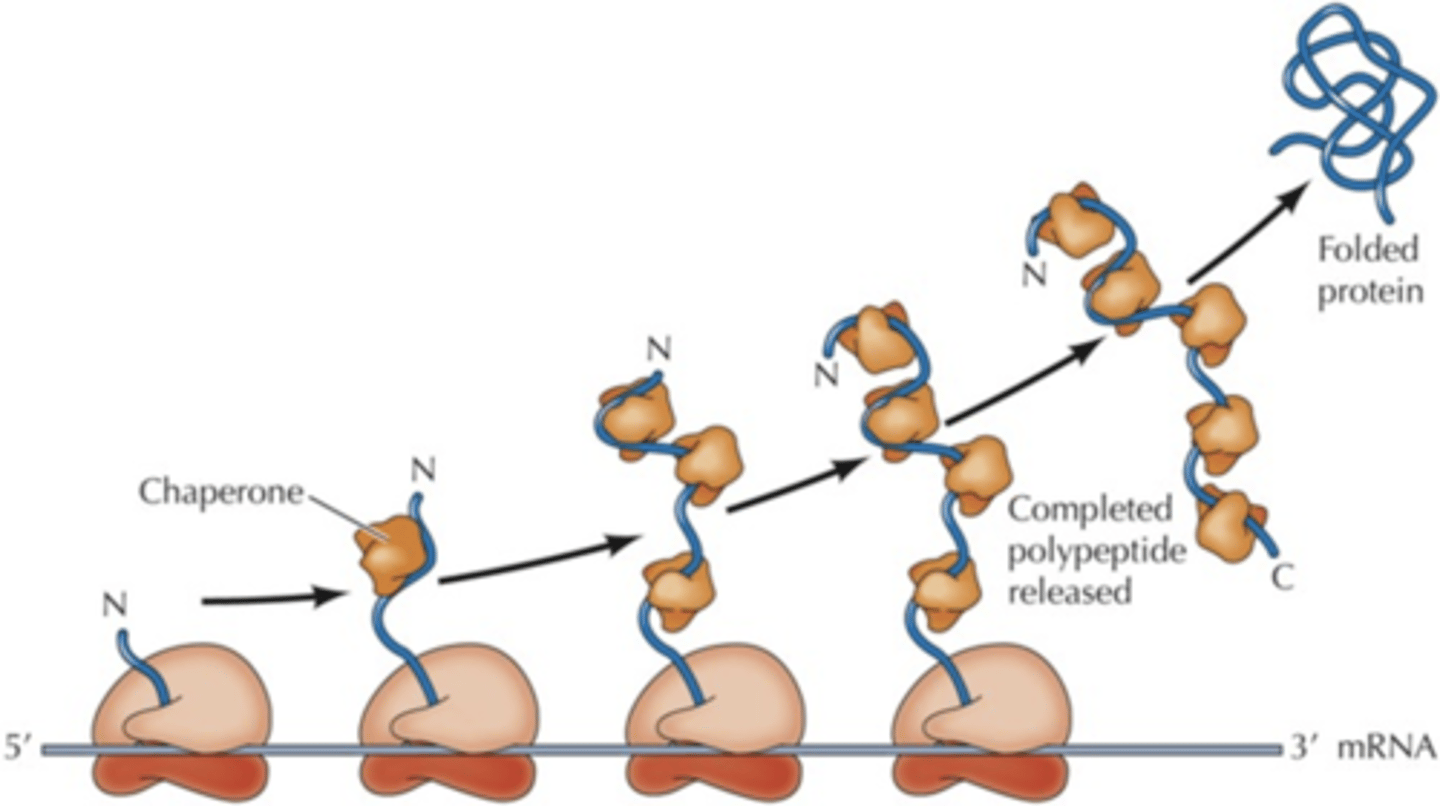
Chaperones (translation)
special proteins that post-translationally fold proteins properly
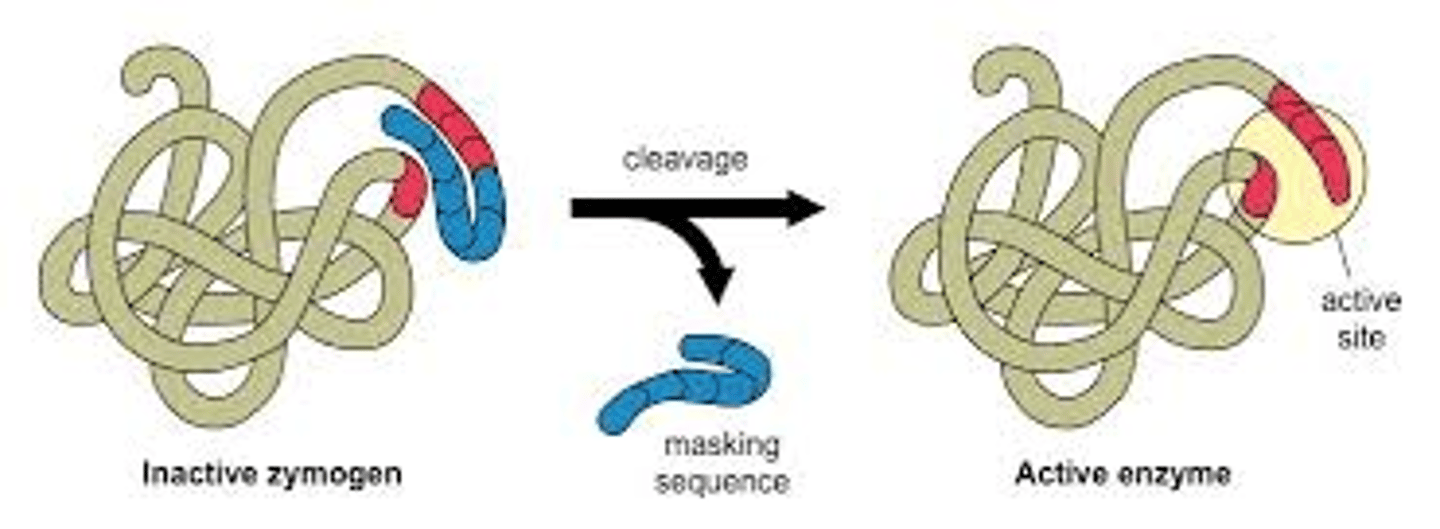
Cleavage Proteins (translation)
proteins that post-translationally cleave parts off proteins to active them
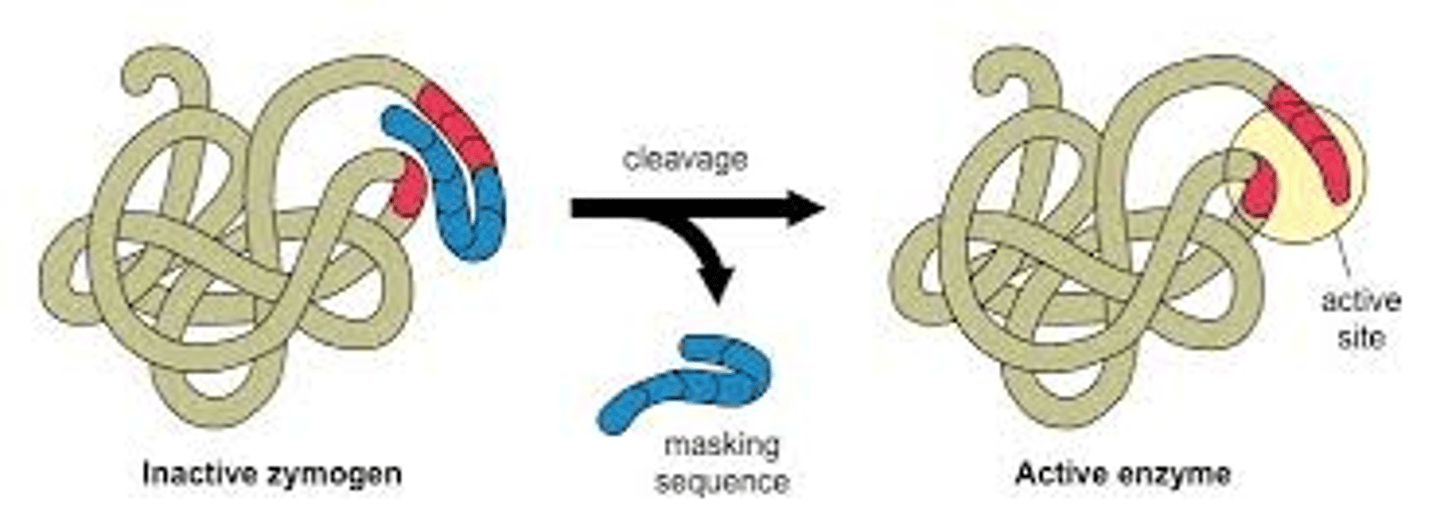
For proteins with _____________ _____________, this must be cleaved off when at the correct location before it can enter and perform its function
signal sequences
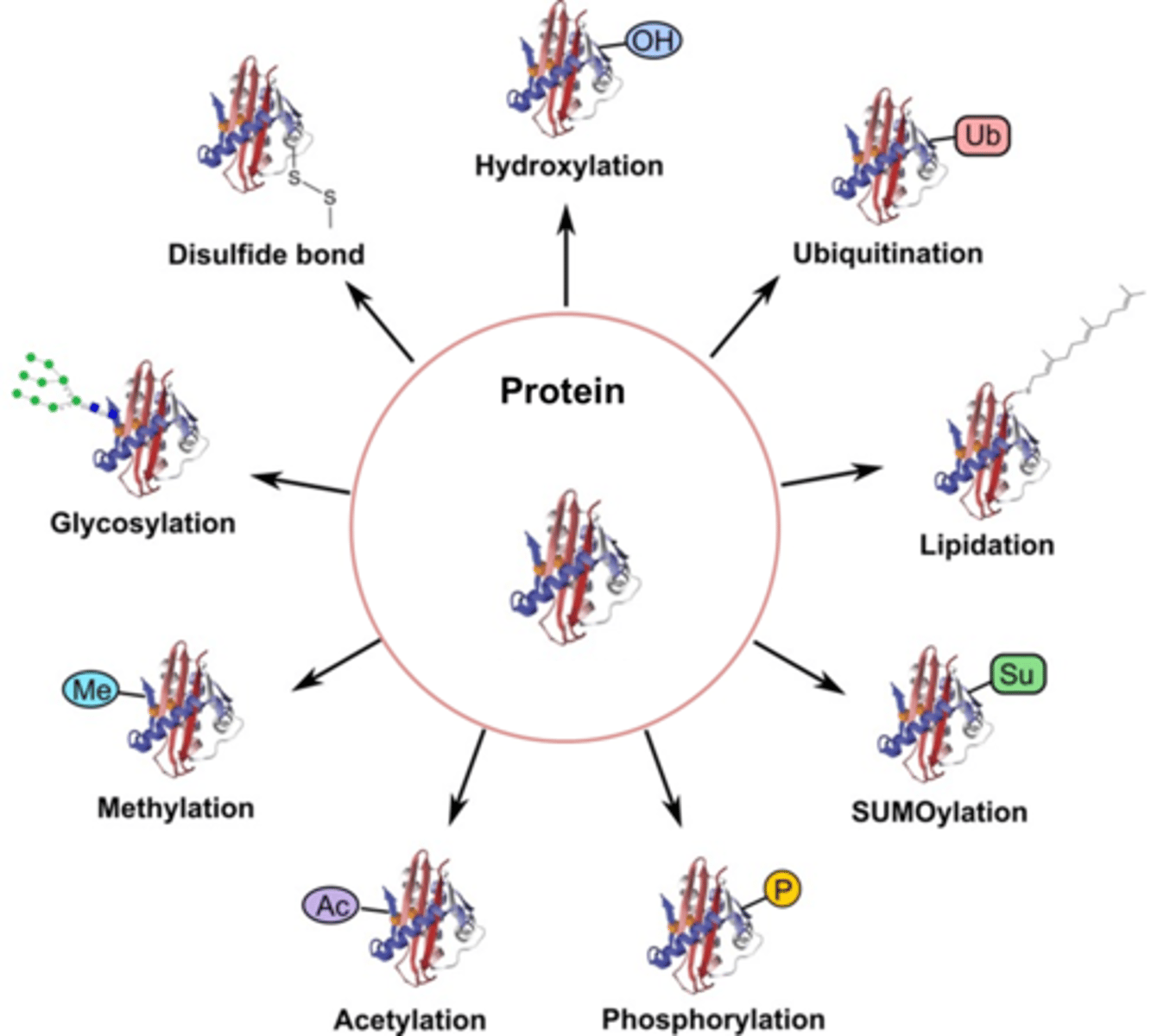
4 groups that can be added to proteins post-translationally
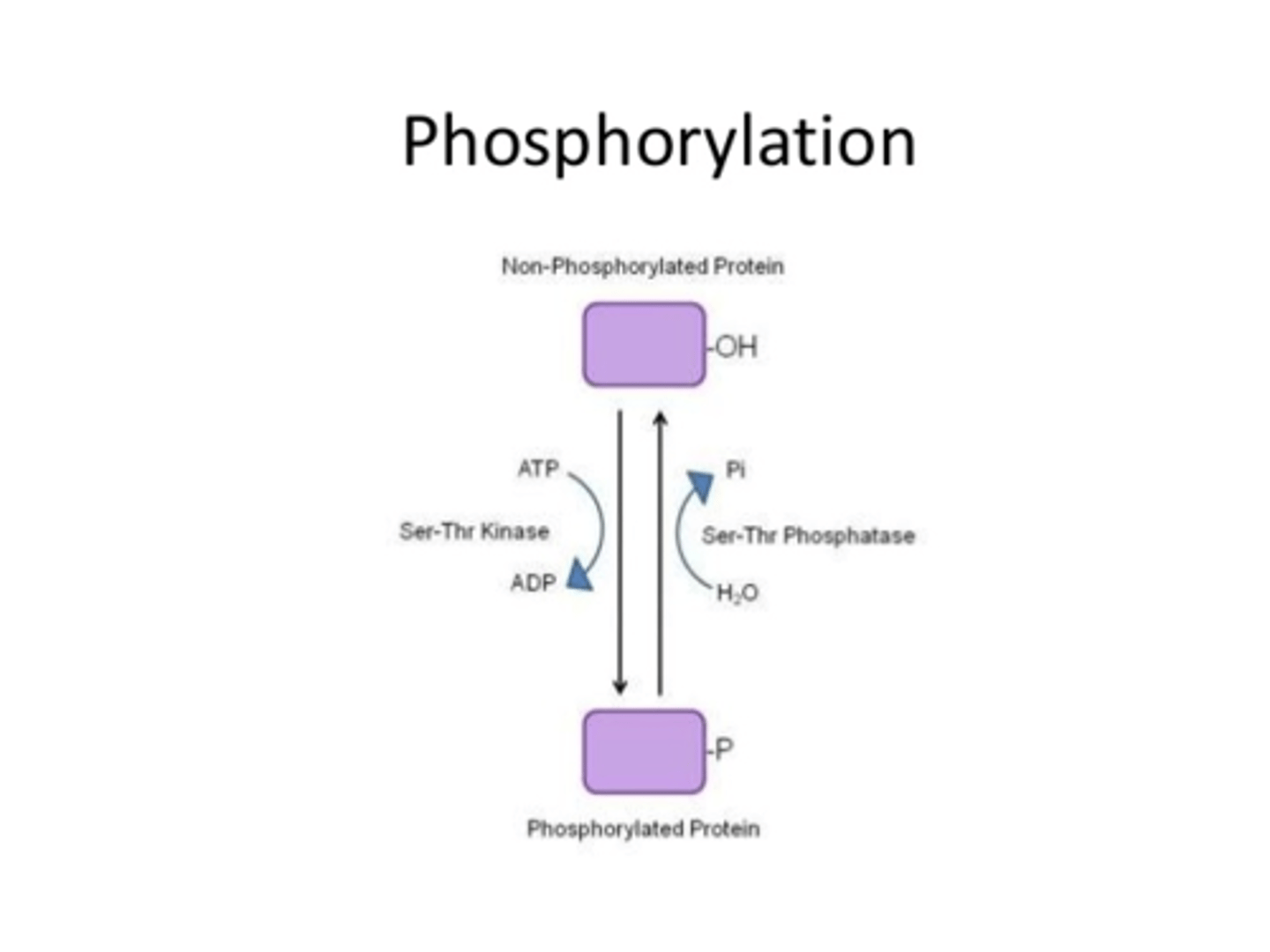
1) Phosphorylation (phosphate added via kinase to activate/deactivate; ONTO serine, threonine, or tyrosine with their OH),
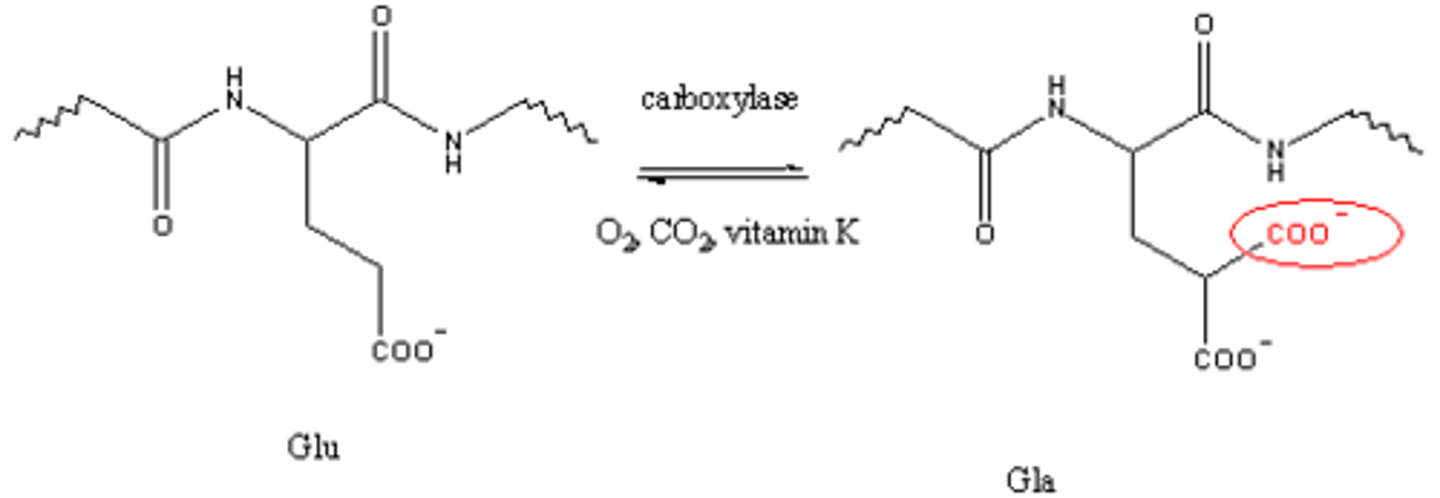
2) Carboxylation (adding carboxylic acid to a protein to bind Ca2+),
4) Glycosylation (adding oligoosaccharides to proteins as they pass through the ER and golgi that directs them to a location),
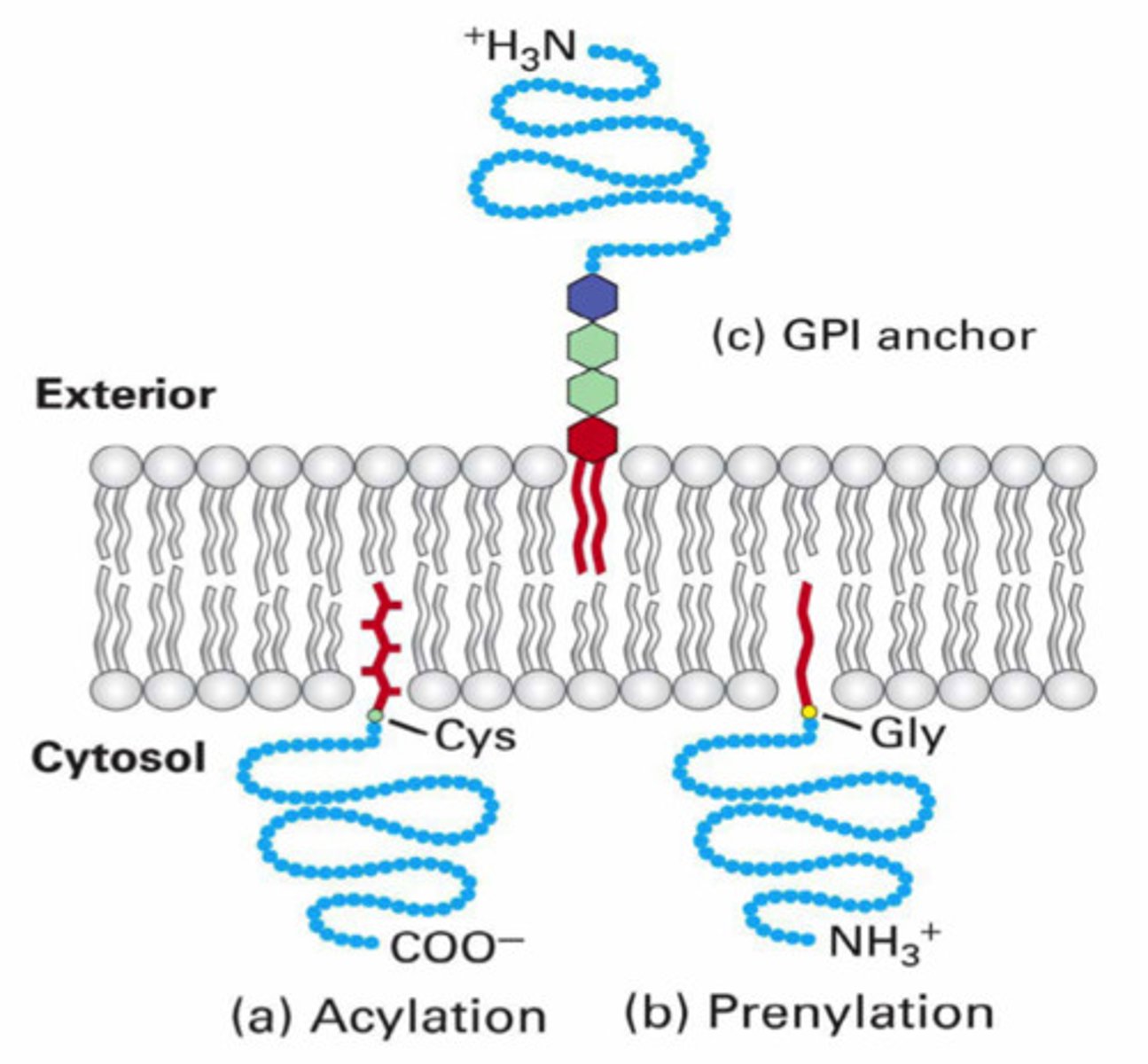
4) Prenylation (adding lipid to membrane-bound proteins/enzymes)
Onto which amino acids in a protein does phosphorylation usually occur?
1) Phosphorylation (phosphate added via kinase to activate/deactivate; ONTO serine, threonine, or tyrosine with their OH),
2) Carboxylation (adding carboxylic acid to a protein to bind Ca2+),
4) Glycosylation (adding oligoosaccharides to proteins as they pass through the ER and golgi that directs them to a location),
4) Prenylation (adding lipid to membrane-bound proteins/enzymes)
Carboxylation post-translational modification
adding carboxylic acid to a protein to bind Ca2+
Glycosylation post-translational modification
adding oligosaccharides to proteins as they pass through the ER and golgi that directs them to a location
Prenylation post-translational modification
adding lipid to membrane-bound proteins/enzymes
Operons
clusters of genes with 1 promoter transcribed as 1 mRNA in prokaryotes; on-off switch for genes
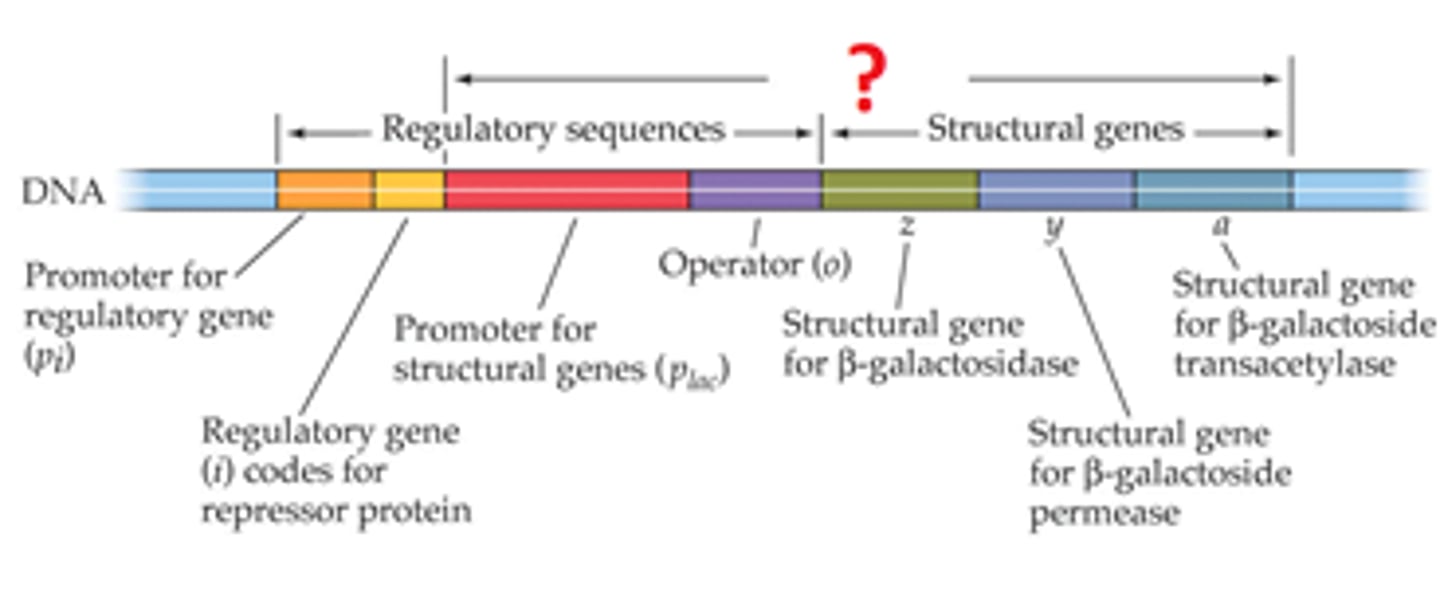
Jacob-Monod Model of Operons? 4 parts?
states that operons contain structural genes, an operator site, a promoter site, and a regulator gene:
Structural Gene codes for the protein,
upstream Operator Site is un-transcribed DNA which binds a repressor,
further upstream is Promoter Site which binds RNA pol,
further upstream is the Regulator Gene which codes for the repressor protein
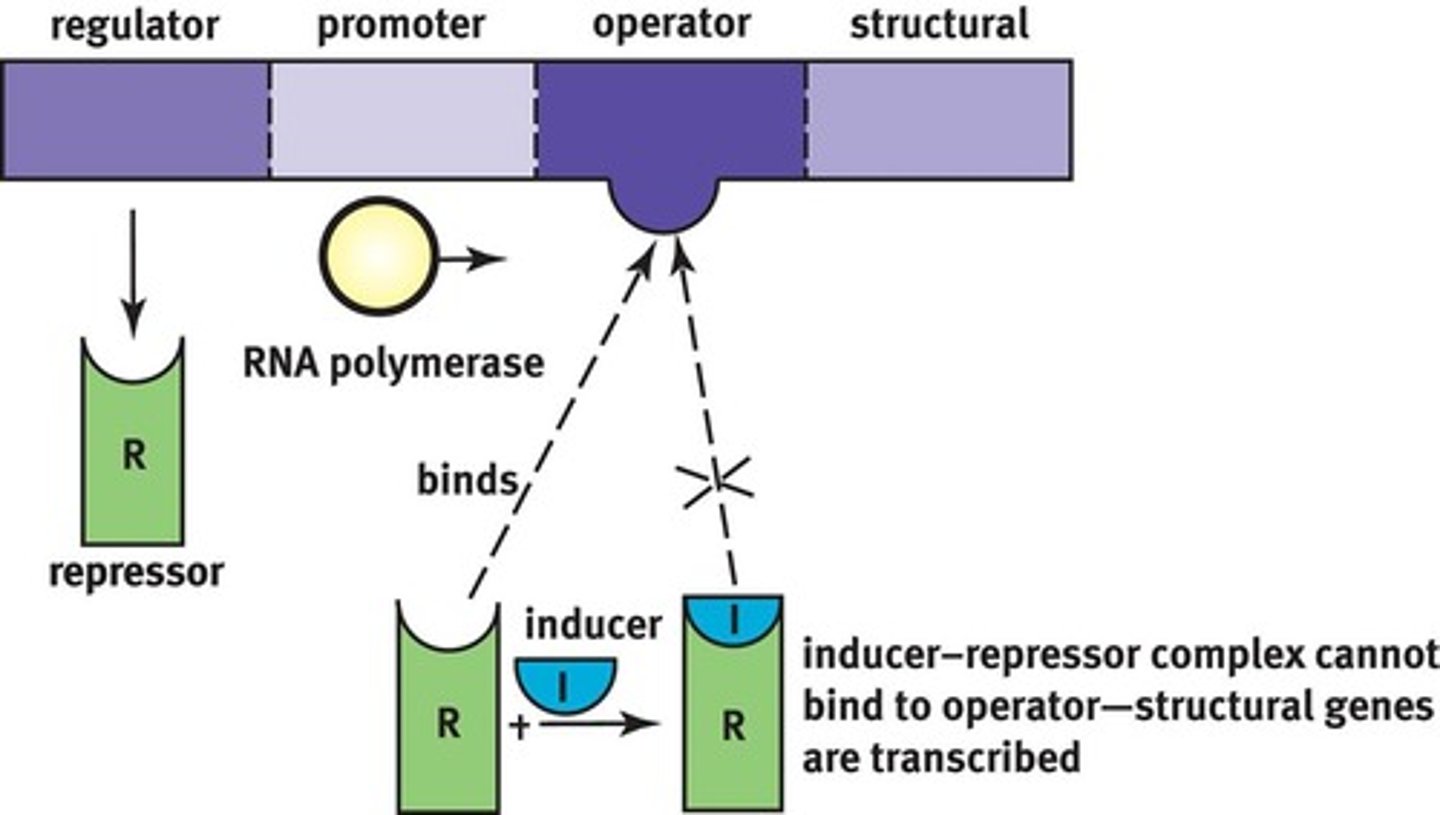
Order of genes in the Jacob-Monod model of operon from 5' to 3'
Regulator Gene ➝ Promoter Site ➝ Operator Site ➝ Structural Gene
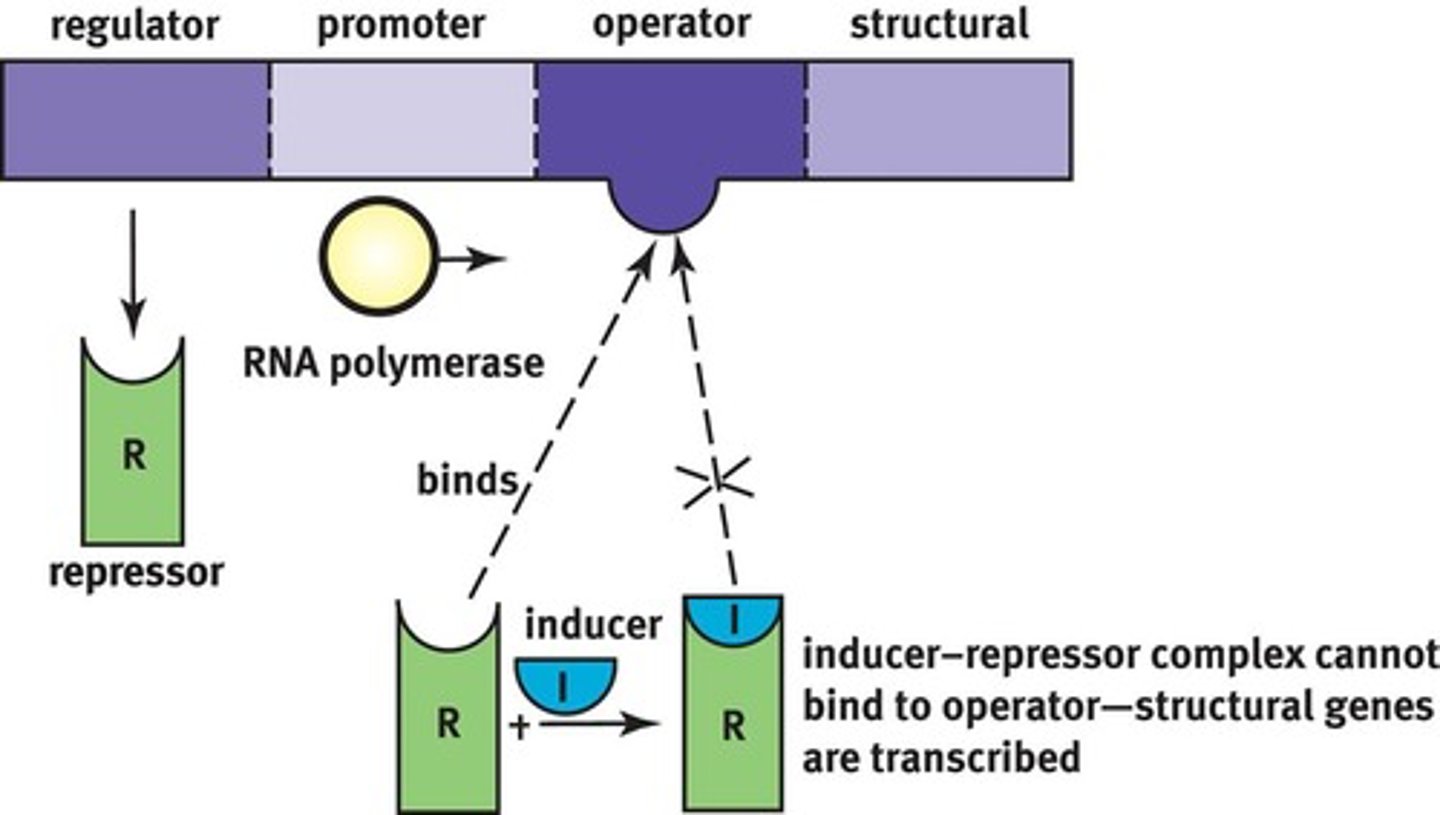
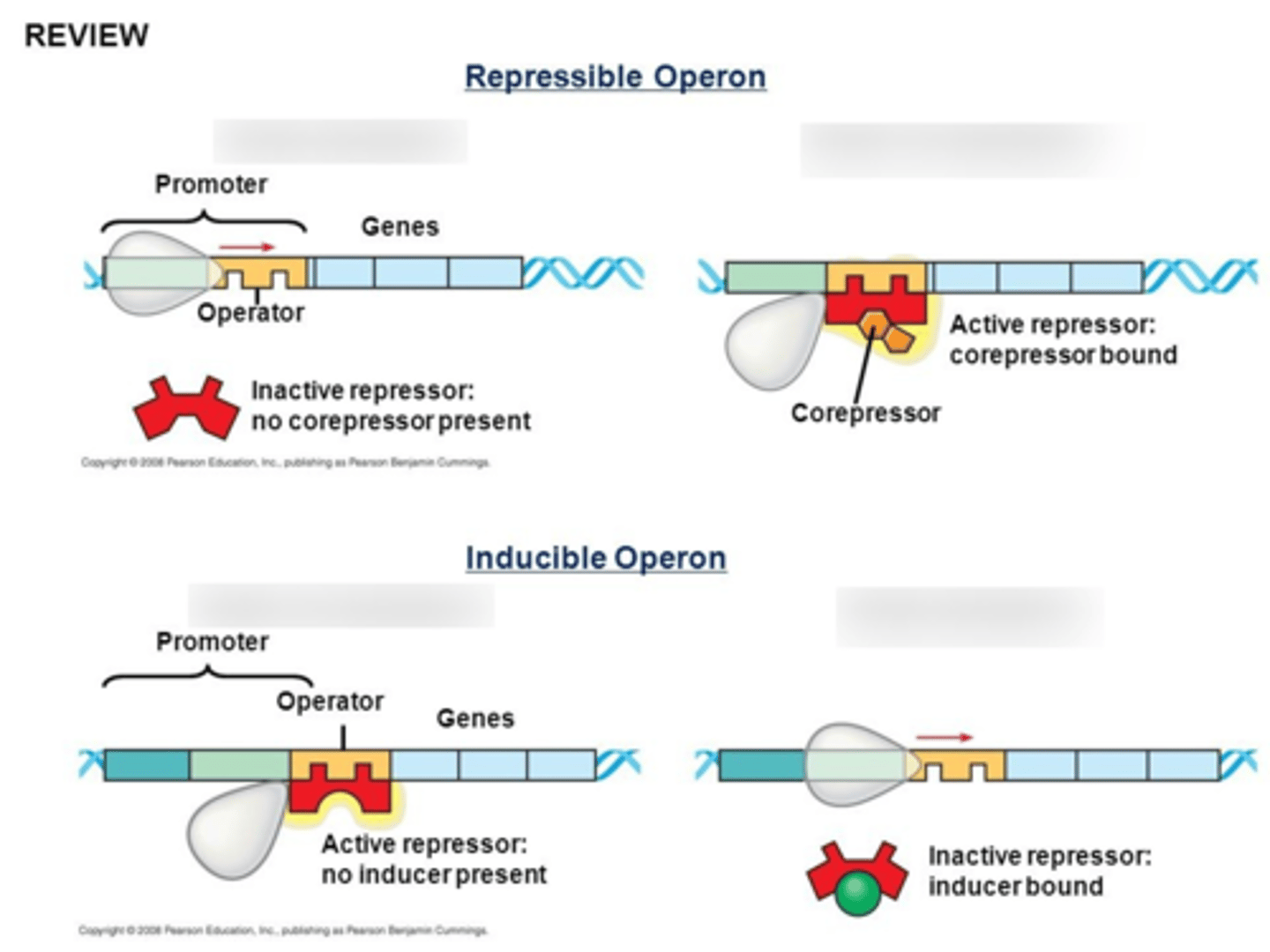
2 types of Operons:
1) Inducible (activated)
2) Repressible (inactivated)
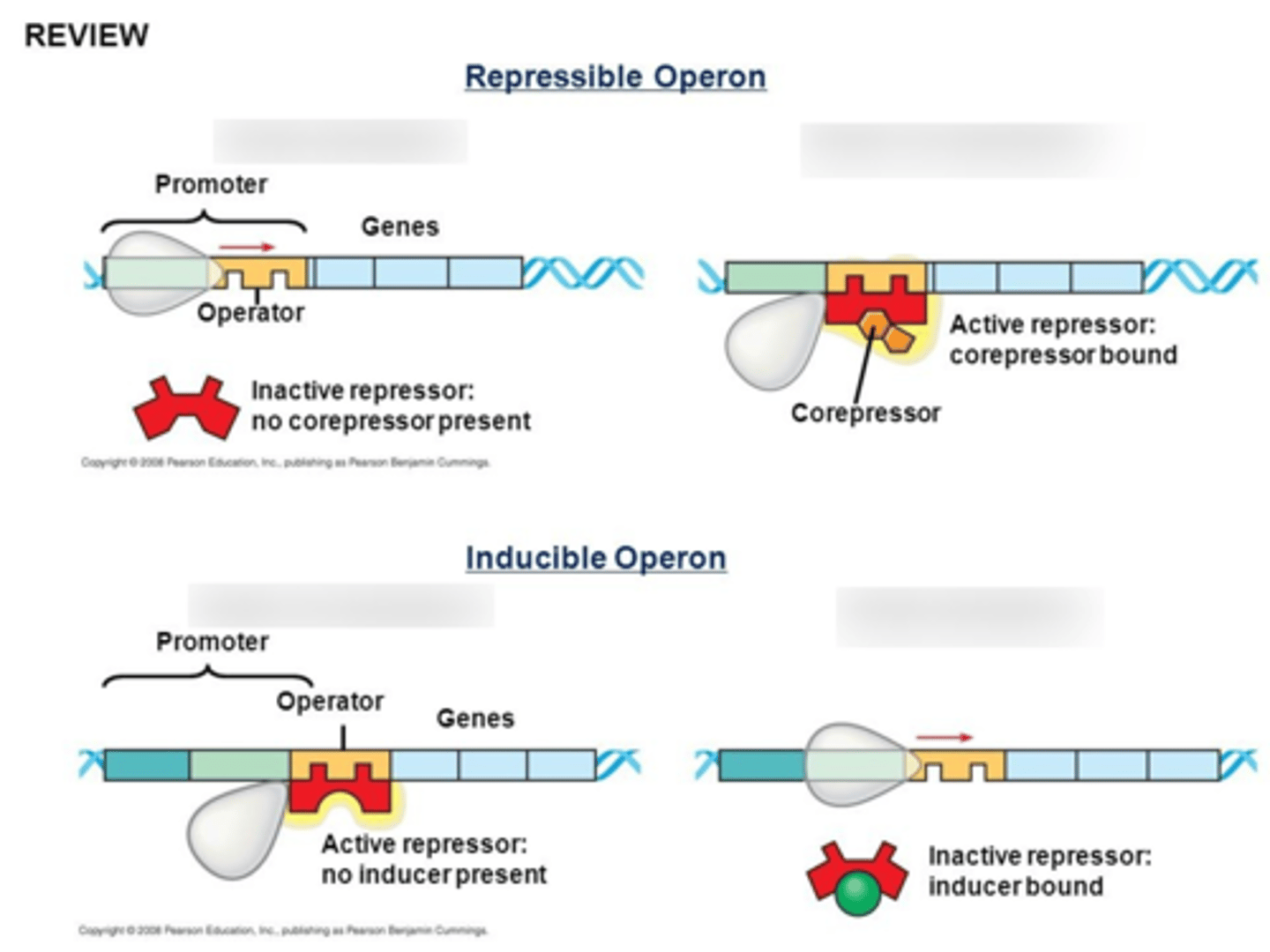
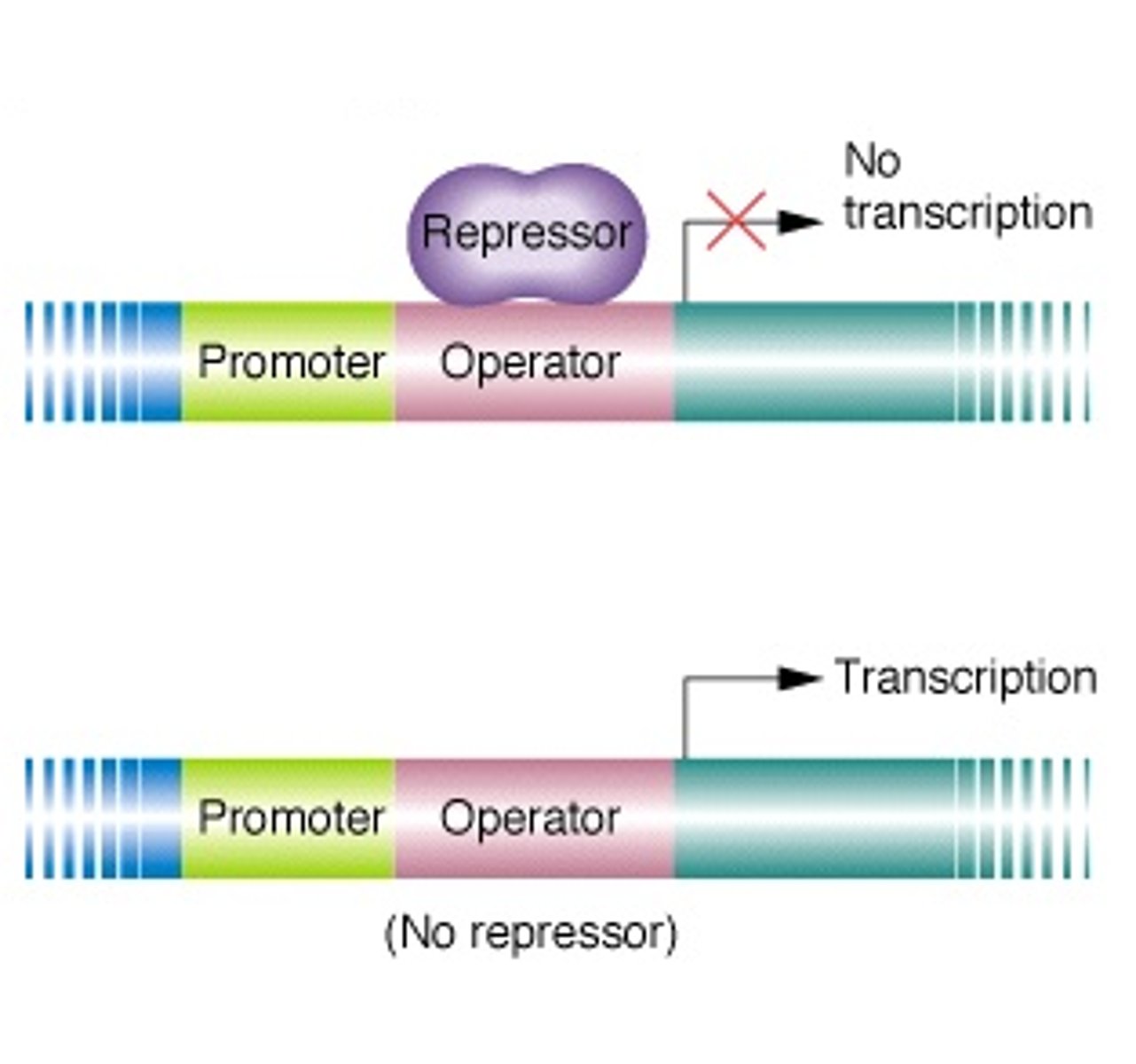
How does an inducible operon work?
repressor is usually bound to operator; RNA pol cannot pass it to transcribe the structural gene
Negative Control
to activate it, an inducer must bind to the repressor so the complex falls of and RNA pol can bypass; inc conc'n inducer causes competition to activate it
In general, inducible operons allow genes to be activated only when ____________________. Why are they usually off?
only when needed! They are off usually to not waste energy
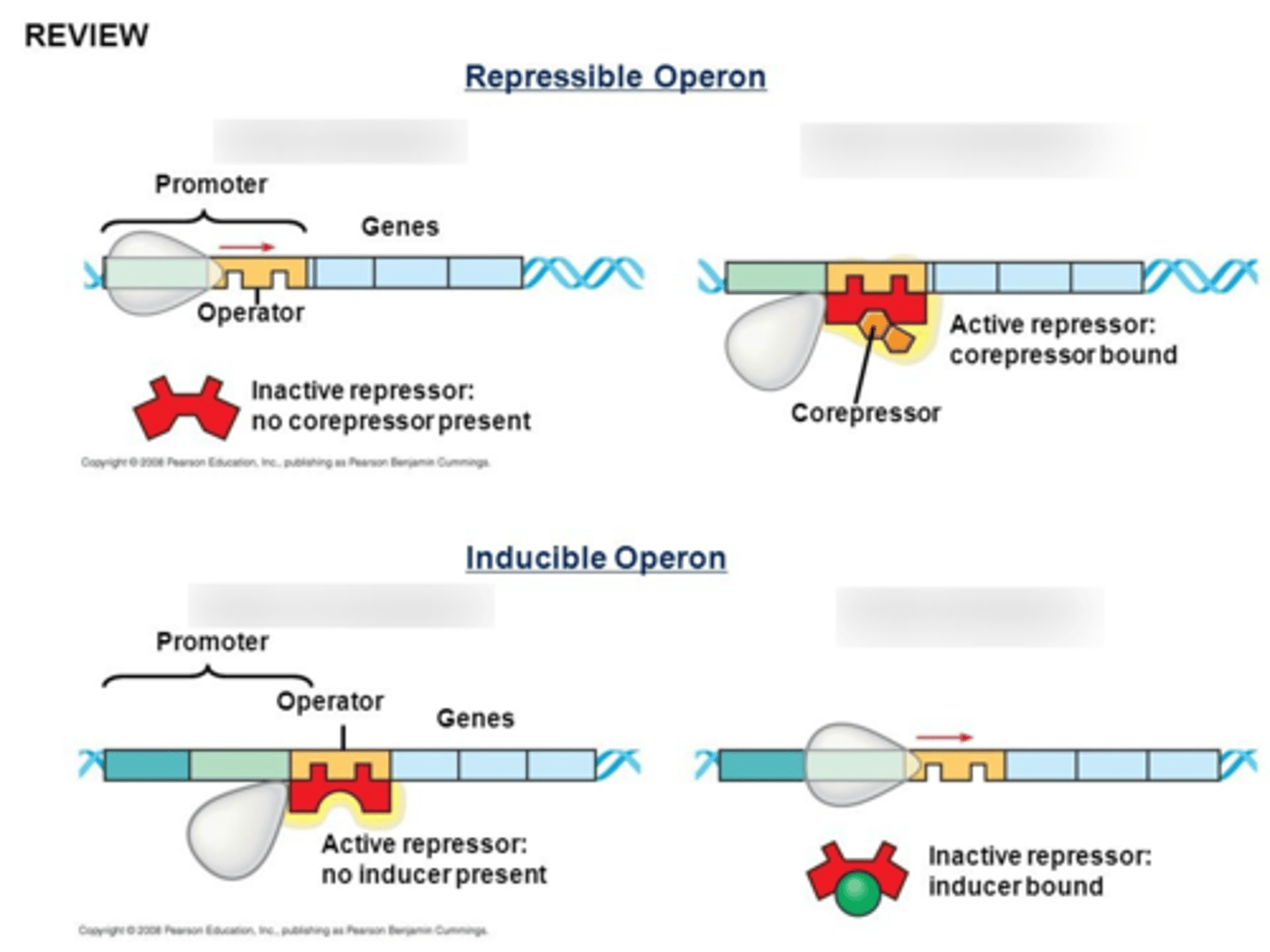
Lac Operon? when is it on? when is it off? CAP and cAMP levels?
inducible operon that codes for lactase
it is energetically more expensive to digest lactose than glucose, so the operon is only active when glucose is absent and lactose is present: repressor falls off when bound to lactose, and Catabolite Activator Protein (CAP) activates the promoter in low glucose once cAMP (increased in low glucose) binds to it and it conformationally changes
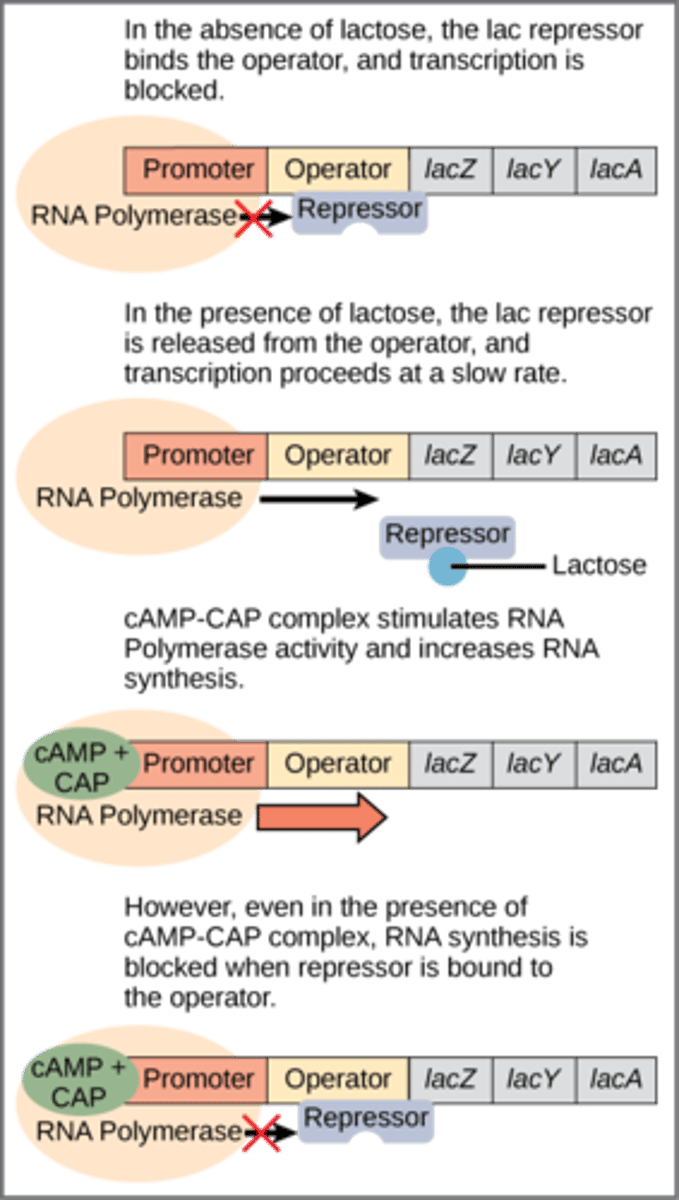
Positive Control vs negative control (genetics/operons)
POSITIVE = a gene system where a protein bound turns ON the gene (ex: trp)
NEGATIVE = a gene system where a protein bound turns OFF the gene (ex: lac)
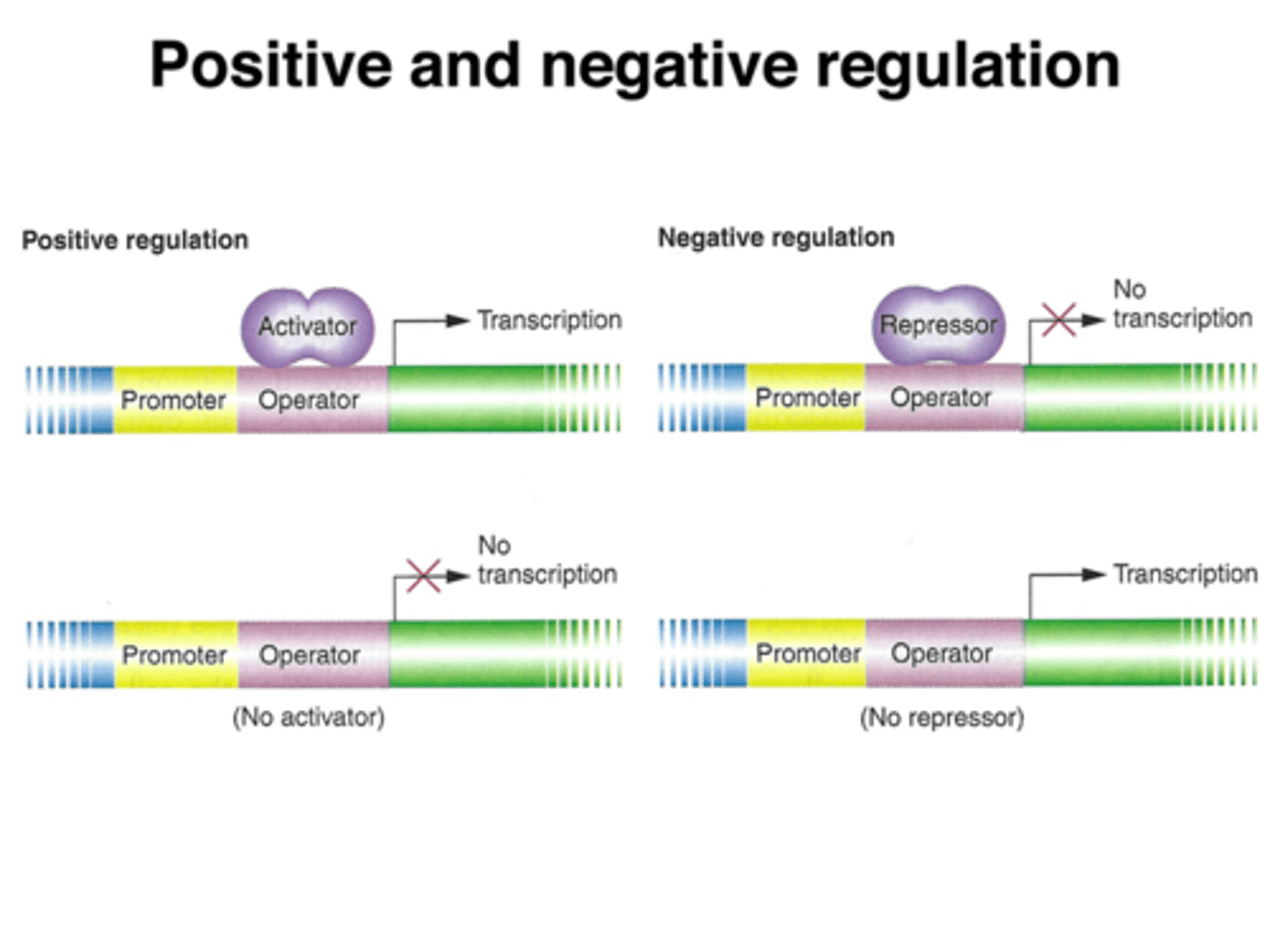
how repressible operons work?
usually constant production of the protein occurs; the repressor must bind to a corepressor to bind to the operator site in a negative feedback way (usually the gene product is a corepressor and inactivates itself when levels are too high)
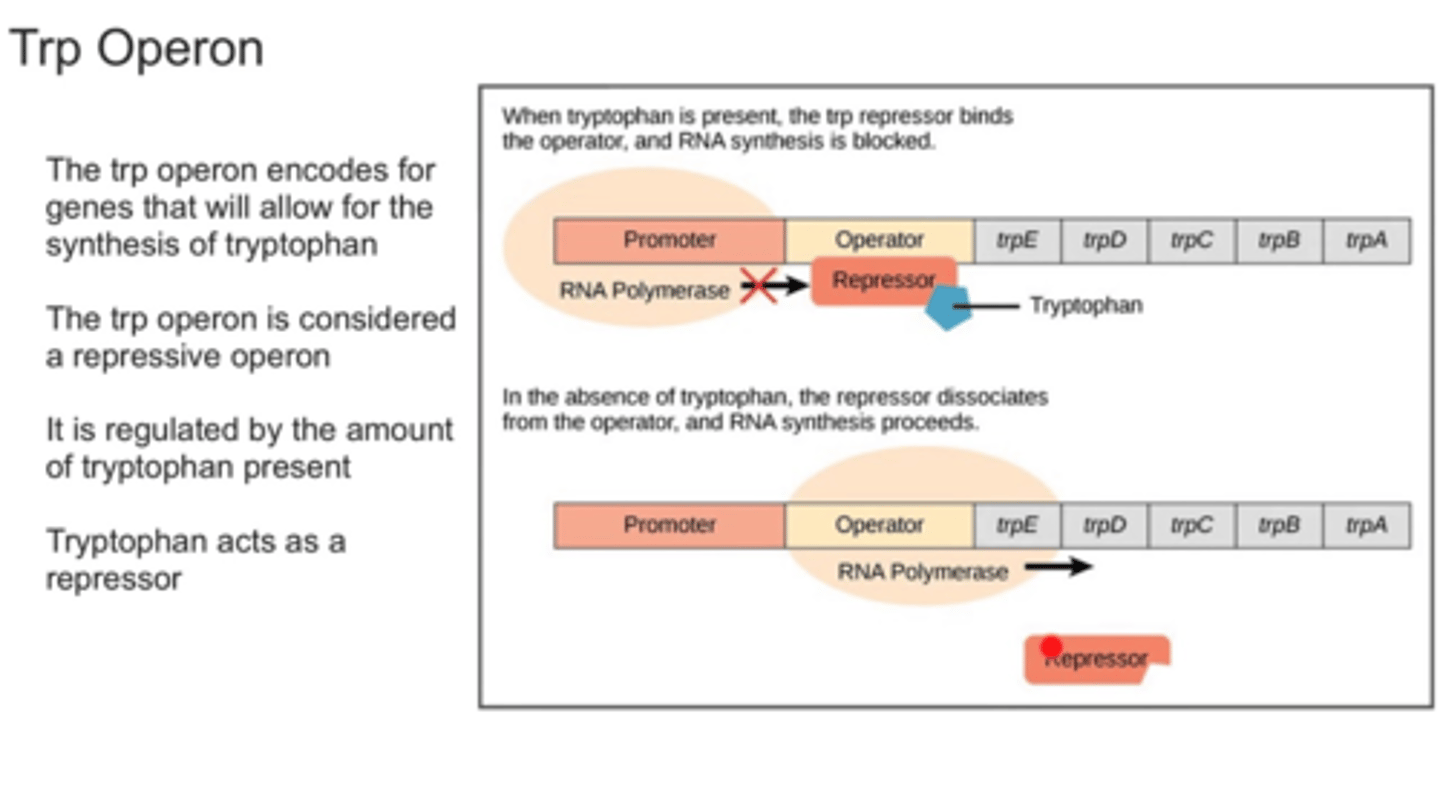
Trp Operon? what does it code for? inducible or repressible?
repressible operson that codes for tryptophan
Trp is a corepressor that activates a repressor for the operator site when x2 Trp bind, turning off synthesis of the energetically-expensive Trp
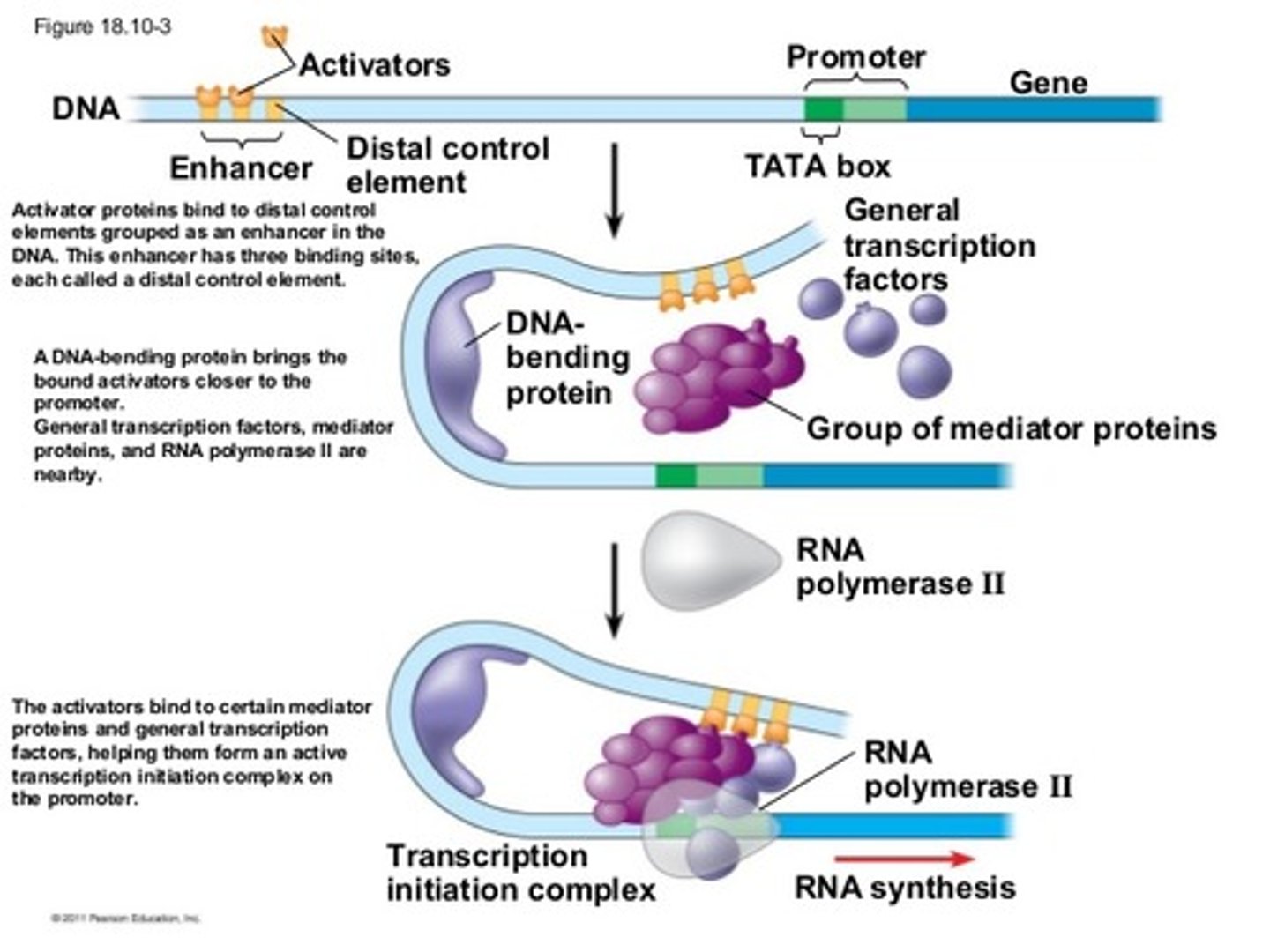
Eukaryotic Transcription Factors & Gene Regulation
they activate transcription by looking for specific DNA motifs; 2 domains: DNA-binding Domain (that binds to a DNA-response element that is specific to the TFs) & Activation Domain that binds TFs and regulatory proteins (RNA polymerase, histone acetylases)
Gene Amplification
when expression is increased due to signalling (hormones, growth factors, conditions)
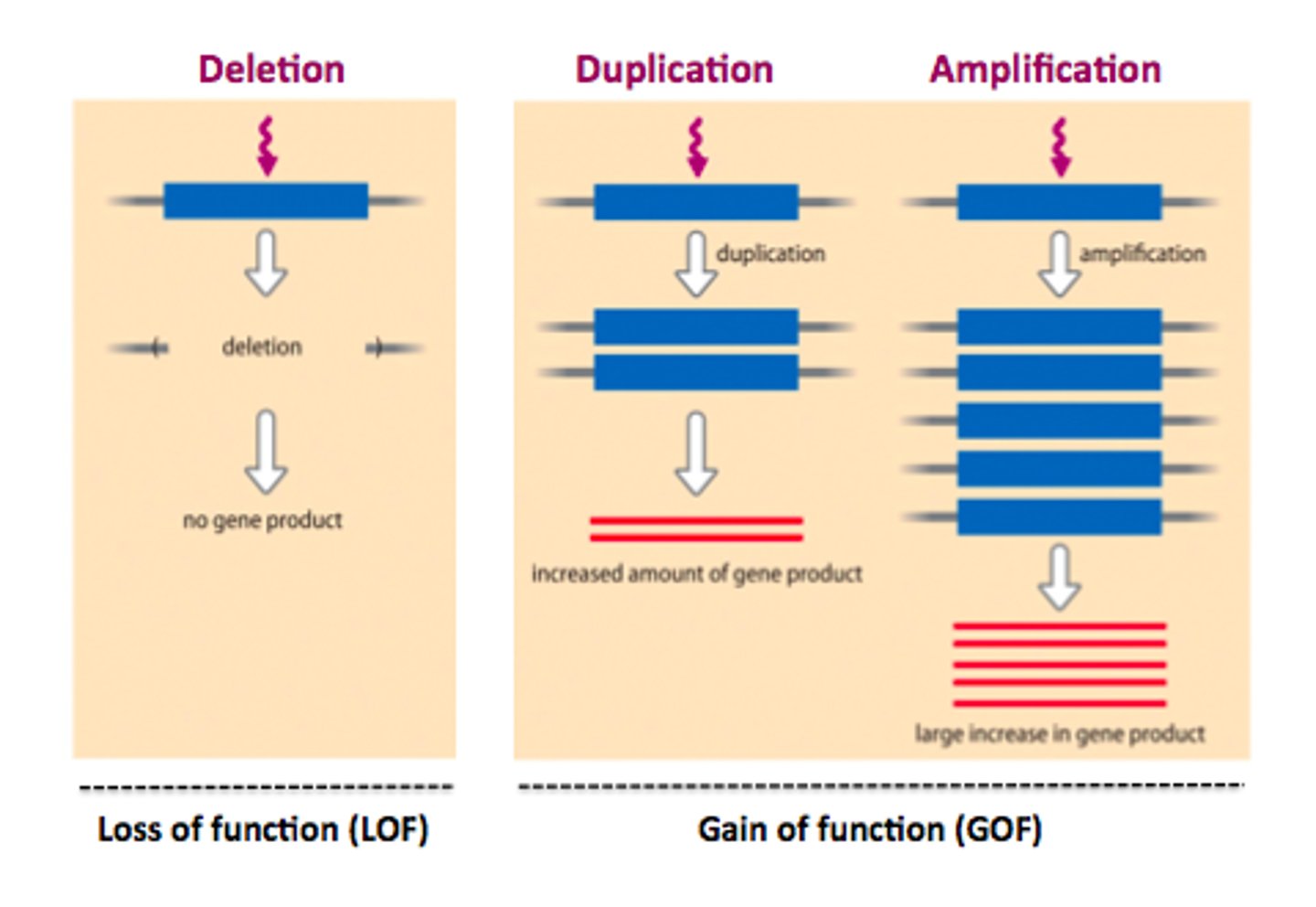
2 things that can amplify eukaryotic genes
1) enhancers,
2) duplicating the gene
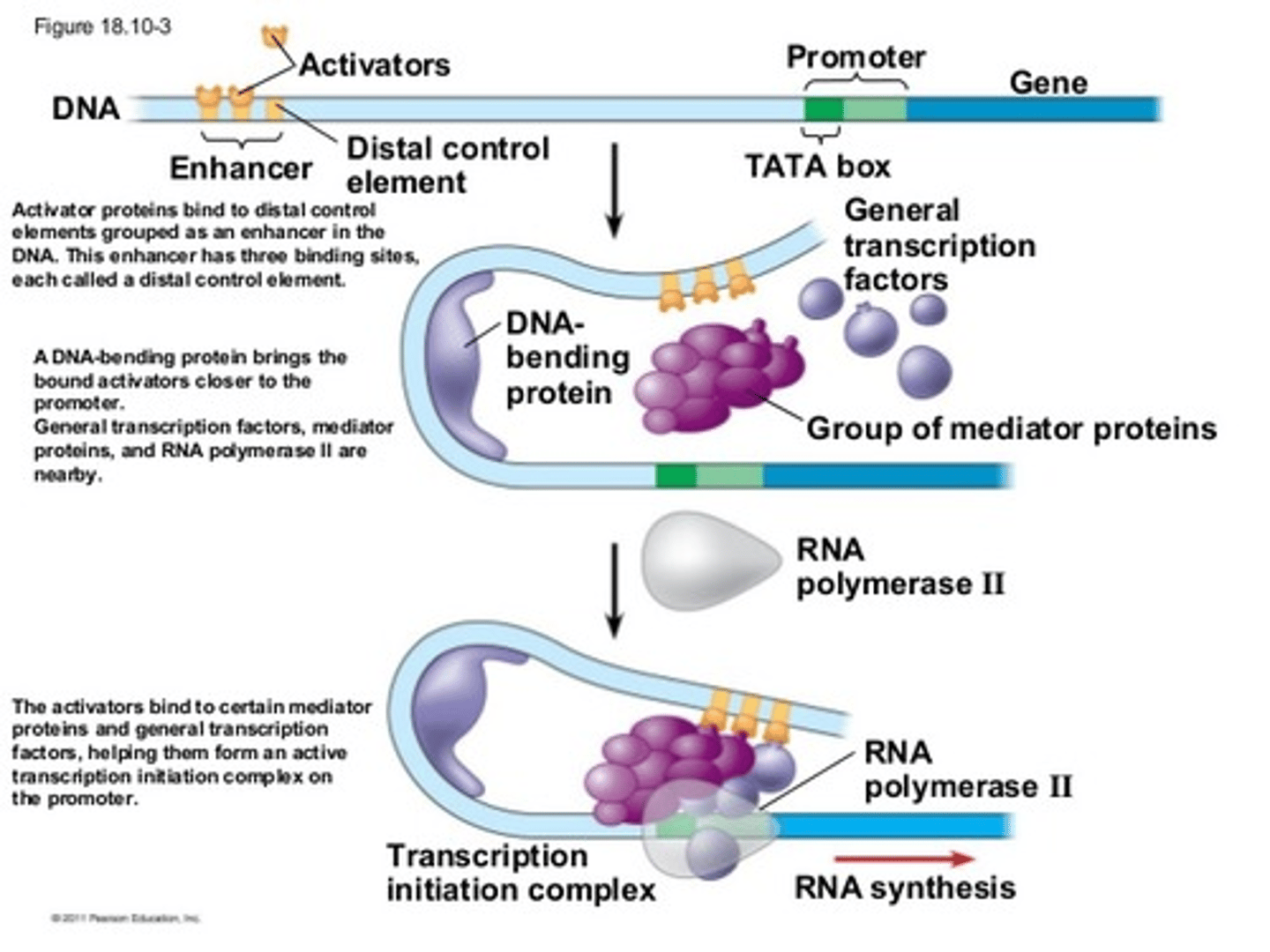
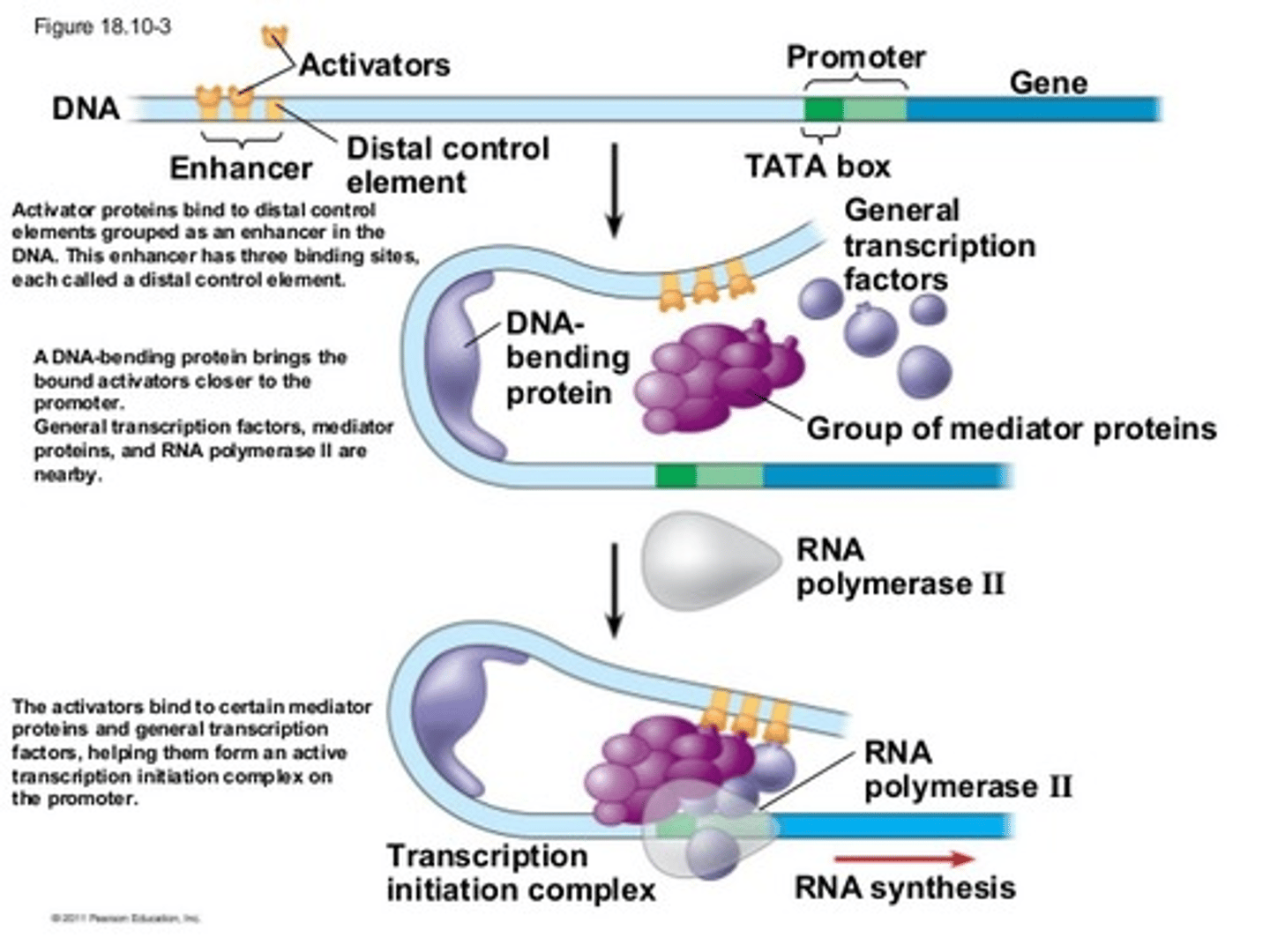
Gene Enhancers
Response Elements (promoters outside of the regular promoter region) group together into Enhancers on DNA, which bind specific TFs, which themselves bind molecules like cAMP, cortisol, estrogen, etc. bind to amplify gene expression: MANY OF THEM USUALLY ACTIVATED AT ONCE TO GROUP TOGETHER AS AN ENHANCER
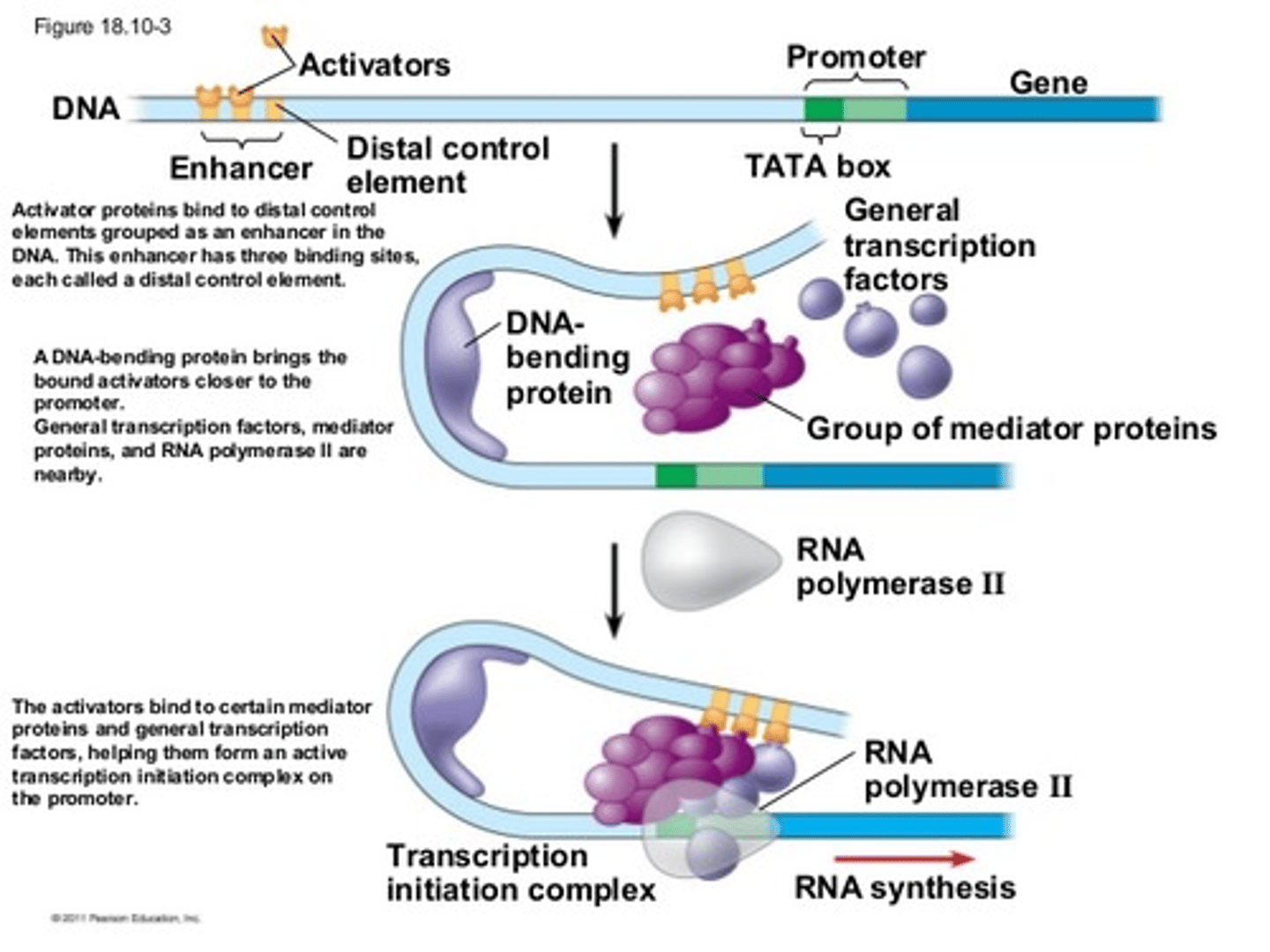
cAMP Response Element Binding Protein (CREB)
A protein activated by cyclic AMP that binds to specific regions of DNA, thereby increasing the transcription rates of nearby genes.
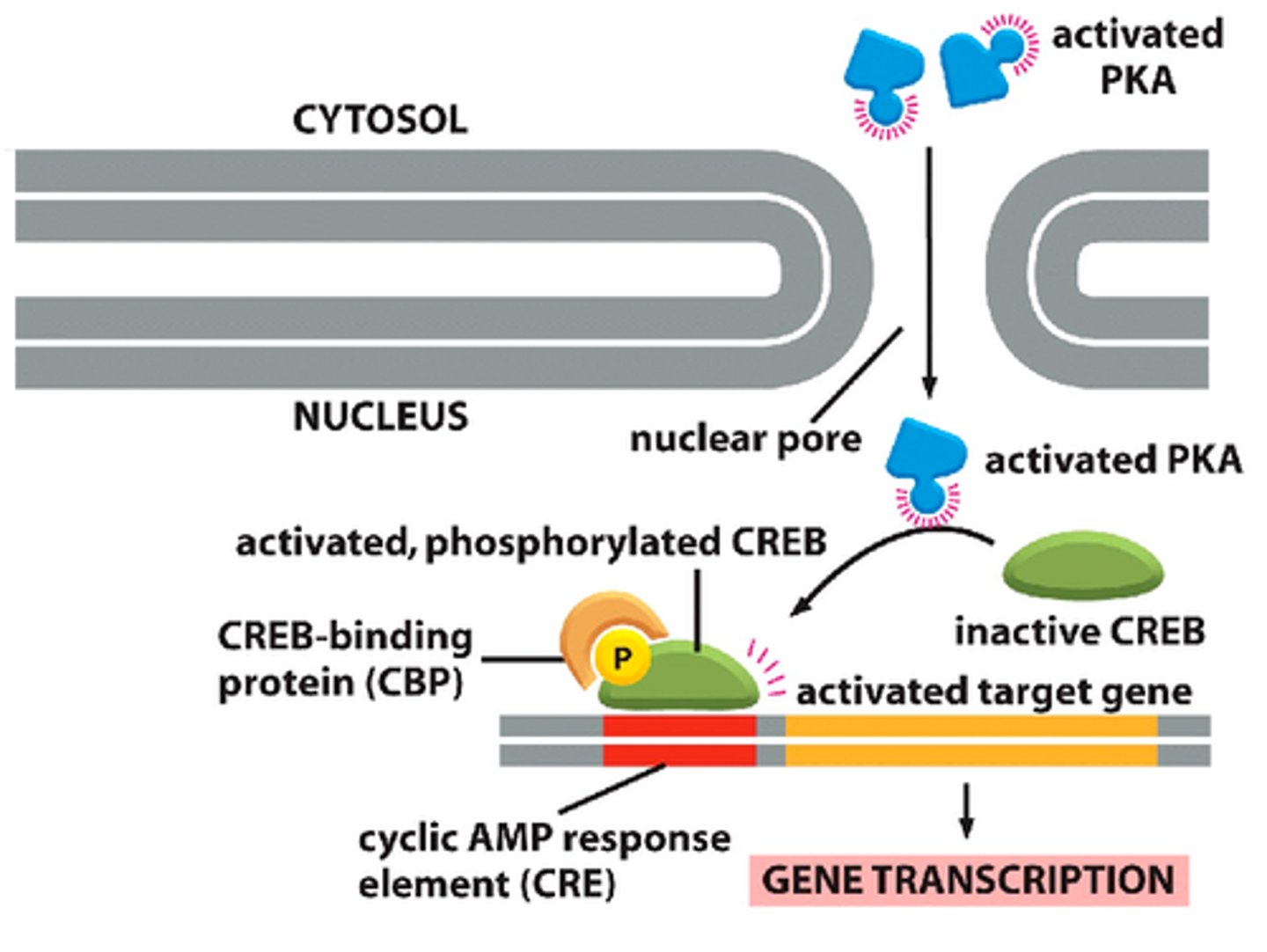
DNA must often bend into a ________ shape due to the distance (up to 1000bp) between the ________ and ________ regions when amplifying a gene
hairpin; enhancer; promoter
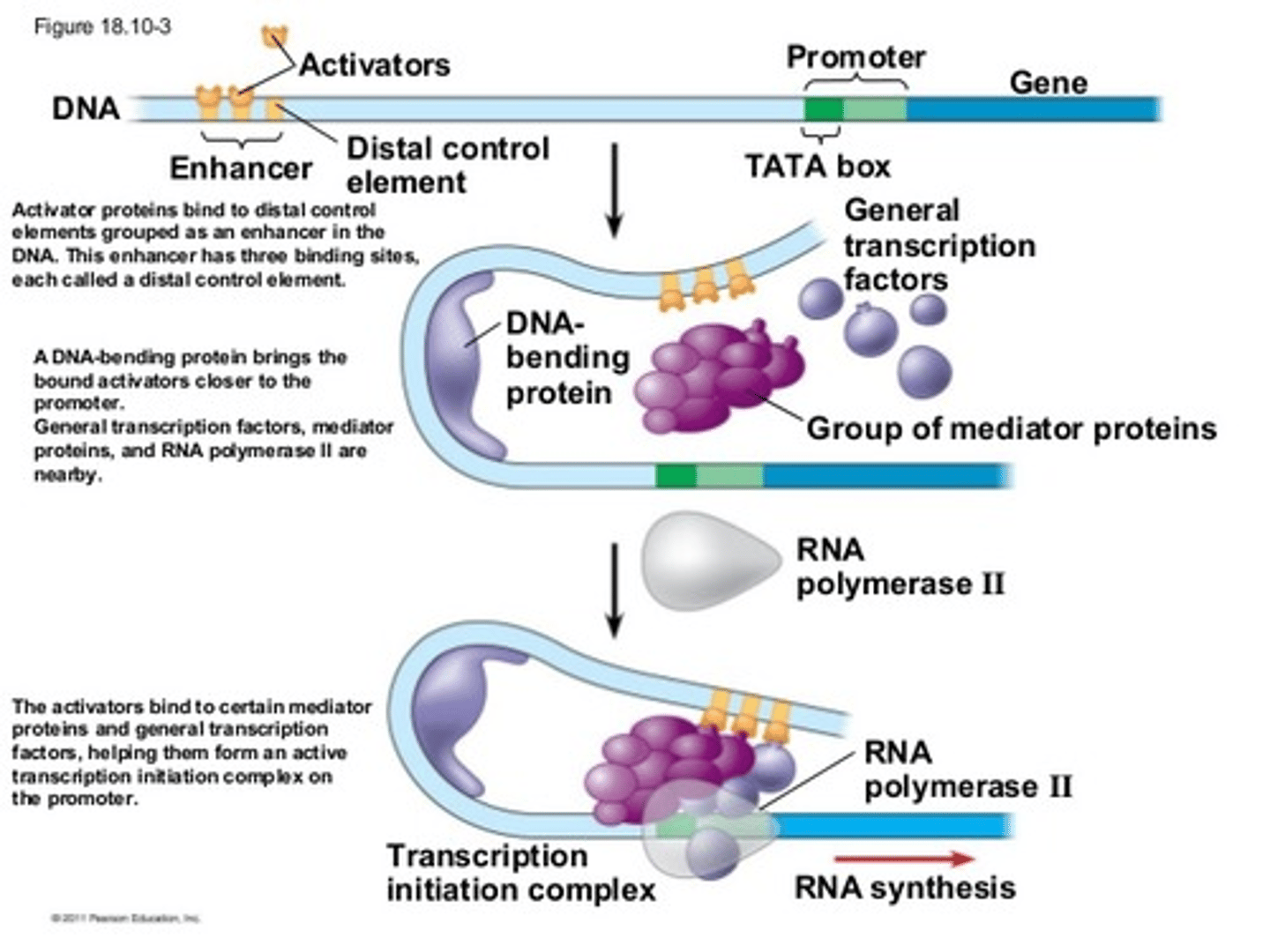
Why are enhancers useful if multiple signals can activate them?
the multiple signals increases the likelihood that SOMETHING will activate the enhancer
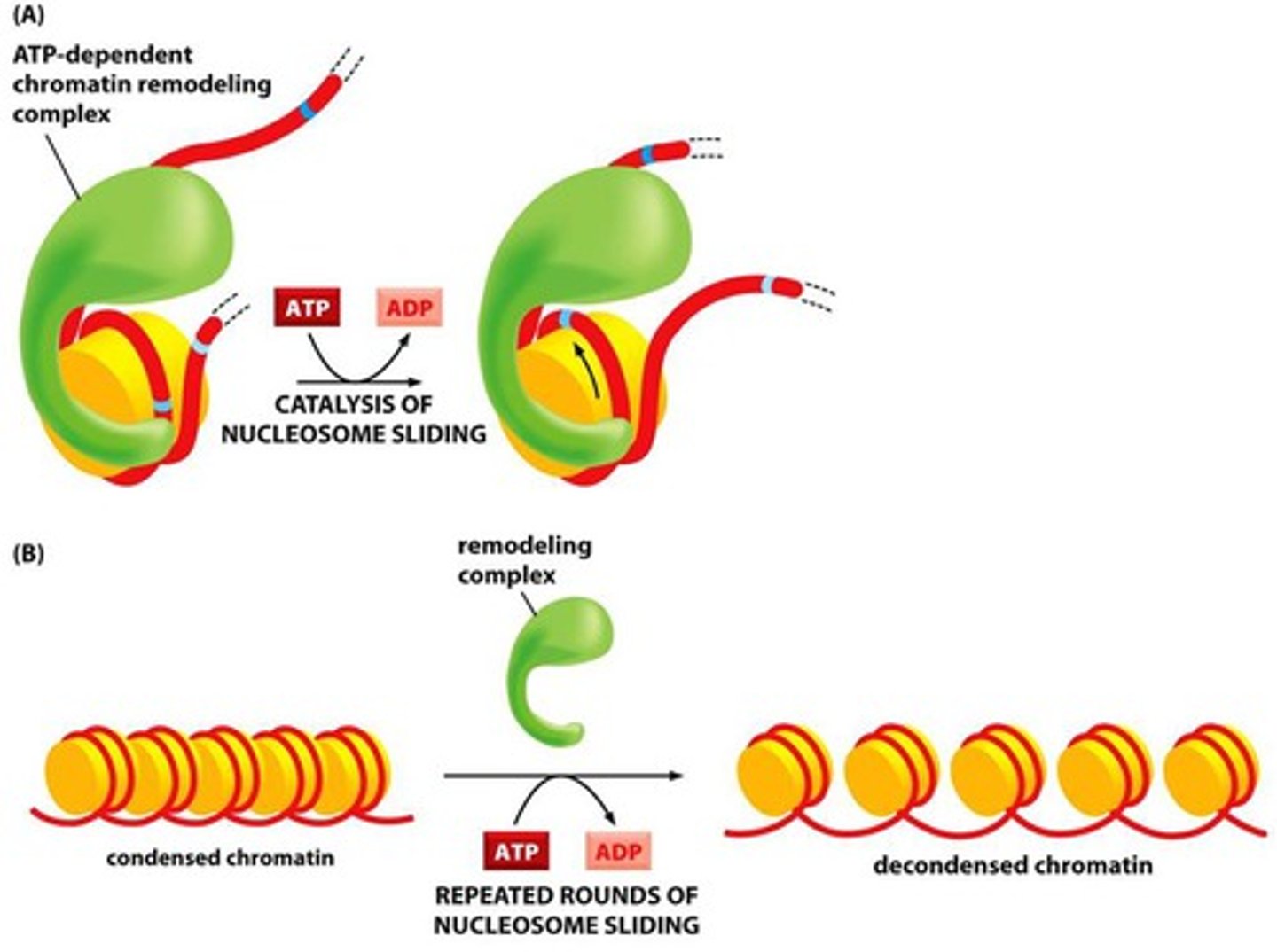
Why is chromatin remodelling/decondensing necessary for transcription?
this opens/closes DNA so transcription factors and such can be transcribed (heterochromatin to euchromatin, loosening it)