DNA PACKAGING
1/18
Earn XP
Description and Tags
Exam 1
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
19 Terms
Heterochromatin
Top of the chromosome which is more condensed, has more silenced genes, gene poor (high AT content) and stains darker.
Euchromatin
Bottom of the chromosome which is less condensed, higher gene expression, gene rich (GC content) and stains lighter.
Telomeres
DNA protein structures at the ends of eukaryotic linear chromosomes. They prevent chromosome ends from being mistaken for broken DNA.
Centromeres
The site on each chromosome where the kinetochore (which attaches the chromosome to the spindle during mitosis) assembles.
Replication Origins
Specific DNA sequences where DNA replication begins.
Nucelosomes
A complex formed when linker DNA winds itself around core histones. DNA is wrapped around the histone approx. 2 times, forming the nucleosome.
Chromatin fibers
The result of nucleosomes interacting and contributes to higher order folding of chromatin threads.
Histone proteins
Positively charged proteins that package and organize the long strands of DNA into a chromatin. Each histone protein has an amino acid side chain which are hot spots for covalent modifications.
Histone octamers
A complex of 8 histones, specifically H2A, H2B, H3 and H4 (H2A and H2B form a dimer, H3 and H4 form a dimer).
Condensin I & II
Belong to a large family of chromosomal ATPases, structural maintenance of chromosomes (SMC) protein complexes.
Condensin forms loops within loops by making zip tie like structures on the chromatin fibers.
Condensin II is a ring shaped protein complex that initiates the process of chromosome condensation.
Why are chromosomes highly condensed during M phase and less condensed during interphase?
During interphase parts of the chromosome need to be decondensed and looser to allow for gene expression, whereas in M phase the genes no longer need to be expressed and are more tightly packaged and condensed.
What are histone protein post-translational modifications (PTMs)? How do they affect chromatin structure and function? How do these changes in chromatin structure affect gene expression?
They are covalent amino acid side-chain modifications and can change the charge and structure of the side chain. \
For example, acetylation makes the side chain more negative and loosens chromatin structure while methylation keeps a positive charge and keeps chromatin condense.
These modifications have specific enzymes, methyl groups are added by histone methyl transferases, and removed by histone demethylases. Acetyl groups are added by histone acetyl transferases and removed by histone deacetylase complexes.
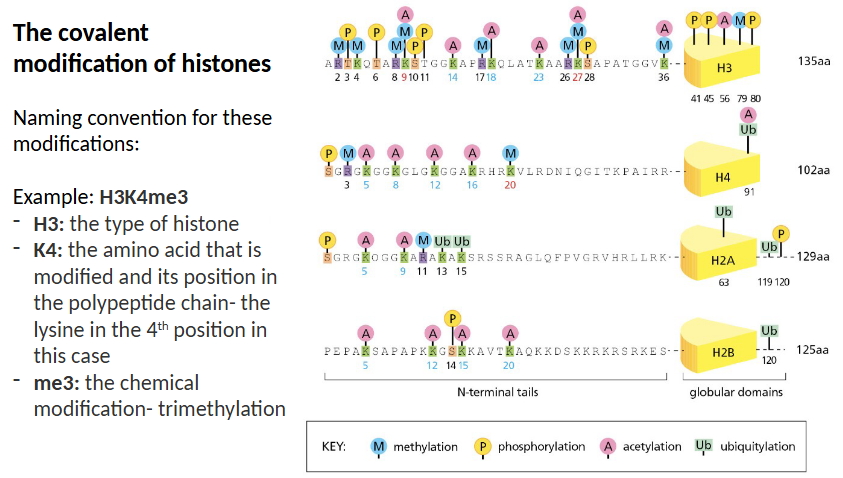
What is the naming convention for covalent histone modifications?
The type of histone
The amino acid that is modified and its place in the polypeptide chain
The chemical modification
How do cohesins (another type of SMC protein complex) work to organize interphase chromosomes?
Because of their ring shaped complex, they can form loops where cohesin loads onto DNA at certain sites and then makes the loop larger by pulling DNA in until it reaches a barrier.
How do ATP-dependent chromatin remodeling complexes reorganize nucleosome structure to affect the “readability” of certain DNA segments?
These complexes can shift the nucleosomes along the DNA molecule, so that polymerase can find the promoter. They can also completely remove histones, or exchange the core histones out with histone variants which allow for greater access to DNA because it is less condensed.
How are histone PTMs spread to other regions of the chromosome? How are these changes inherited across cell generations?
PTMs are spread through different regions of the chromosome as the reader-writer complex spreads heterochromatin along the chromosome until barrier DNA is reached. These changes are inherited across cell generations because during chromosome duplication, the DNA is split as is the histones and nucleosomes that these two structures form, so the tags that were added from the reader-writer complex are also split among the daughter chromosome, where the process repeats itself.
What is barrier DNA?
A protein that tells the reader-writer complex to stop tagging.
What are reader-writer complexes?
A complex which binds tightly to a region of chromatin that contains different histone marks that they recognize. This complex spreads heterochromatin along a chromosome by tagging heterochromatin specific histone tail modifications for the heterochromatin proteins to bind to. The heterochromatin spreads until it reaches a barrier DNA sequence.
How do eukaryotes replicate DNA sequences and chromosome structures (what role do ATP-dependent chromatin remodeling complexes and histone chaperones play)?
Step 1: DNA polymerases replicate the nucleotide sequence from replication origins. Access to the template is restricted by nucleosomes so chromatin os disrupted ahead of the replication fork.
Step 2: Nucleosomes are disrupted as the replication fork continues to open, and the H3-H4 tetramers are retained and transferred onto the daughter strands while the H2A-H2B dimers are often exchanged out for new ones. Chaperones ensure that old and new histones are incorporated into the newly replicated DNA for partial inheritance of epigenetic marks.
Step 3: At the replication fork ATP-dependent chromatin remodeling complexes loosen nucleosomes so helicase and polymerase can continue, reposition nucleosomes after replication so spacing and higher-order structure are restored, and overall work together with histone chaperones to make sure that nucleosomes are evenly distributed, not clumped together.
Step 4: After replication DNA needs to be packaged back into chromatin with correct spacing and higher-order folding. Old histones are recycled and new histones are added and then modified to match the parental chromatin pattern.