Exam 3
1/82
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
83 Terms
What is the physiology of double muscling? Specifically in cattle?
High number of muscle fibers
Less marbling (less fat)
Reduced fertility, high embryo mortality rate
Cattle: DM in cattle contains less connective tissue, show sings of fatigue faster (reduced blood circulation)
What are the genetics of double muscling?
Caused by the myostatin (MSTN) gene mapped on chr. 2 (highly conserved)
Myostatin is responsible for skeletal muscle growth → mutations lead to muscle hypertrophy
Six mutations: deletions, insertions, point mutations
How did the backcross design in mapping myostatin work?
Microsatellites
Naturally existing polymorphisms. Short sequences of DNA repeated (tandem arrays → TGTGTG = TG3)
Comparative mapping approach
Method for establishing the relative genomic positions of orthologous (genes from common ancestor that retained similar function in different species) gene pairs in two species (human/mouse)
Positional Candidate Gene Approach
Combines genetic mapping techniques with targeted selection/function analysis of candidate genes to identify genetic factors linked to a specific trait or disease.
MSTN mutation affect on protein function within dogs
A 2-bp deletion leads to a premature stop codon instead of producing regular cysteine. The loss of cysteine results in lack of formation of disulfide dimers required for protein function (lack thereof causes double muscling).
Myostatin is a negative regulator of muscle growth
Controls total number of muscle fibers by regulating myoblast proliferation
Absence of functional protein (mutations) leads to more muscle fibers made
What factors affect genetic diversity?
Founder effect
Genetic drift
Population Sire effect
Population bottleneck
Founder Effect
Most breeds were established using a few dogs that possessed certain desirable traits
Genetic Drift
The use of small number of founders caused fluctuations in allele frequencies and the loss of rare alleles
Population Sire effect
A dog with desirable traits is used more frequently for breeding than other males in population
Population bottleneck
A sudden reduction in numbers. Recovery of the breed depends on a small number of individuals, resulting in genetic drift and increased homogeneity
Transposable Elements (TE)
DNA Transposons → move using cut-and-paste mechanism
Retrotransposons → move using copy-and-paste mechanism (duplicates the element into a new genomic location via RNA intermediate)
SINE and LINES, use copy n paste to invade genomic regions
DNA methylation mechanism represses activation of TE
SINEs and LINEs
SINEs have an internal promotor for transcription but depend upon enzymes produced by LINEs for reverse transcription (RNA → cDNA) and integration into genome.
Once integrated, SINEs/LINEs passed from generation to generation.
What attributes have been linked to an insertion of a SINE element?
Merle Coat pattern: caused by insertion at the intron 10/11
Extreme piebald: insertion upstream of MITF
Saddle Tan: insertion in intron 1 of ASIP (agouti signaling protein)
Genome-wide Association Studies (GWAS)
Used for identification of genetic variants underlying both simple and complex traits.
Utilizes unrelated cases/controls to compare between cases for identification of associations
GWAS for a neurological disorder in Cavalier King Charles Spaniels
Strong association with the phenotype on chromosome 7 led to identifying the causative mutation. Extent of linkage disequilibrium (LD) in dogs is 100x longer than humans bc of long stretches of homozygosity.
What makes dogs a good animal model for studying human diseases?
Large litter sizes
a short gestational period
accelerated aging/disease progression
Large population have dogs making data widely available
Share similar environments
Diseases are commonly shared between dogs and humans
similar organ size
Fewer regulatory guidelines than human medicine
Importance of Merle Phenotypes
Cryptic Merle → dog with merle genotype but does not have merle phenotype (very rare) and produces merle offspring
All breeds → double merle genotype can be sublethal and associated with multiple skeletal, cardiac, and repro abnormalities
Linkage Disequilibrium Analysis
1. Two loci in close proximity may be in high LD
2. Two loci in high LD typically are in close proximity
LD = the nonrandom association of alleles at two or more loci
Whole Genome Association Study
Sample drawn from general population
Phenotypes with discrete classes (disease)
case (affected) vs control (unaffected)
Phenotypes for continuous traits
sample from tails of the distribution
Linkage Disequilibrium Analysis
Genotype LARGE number of SNPs
Test association between phenotype and marker genotype
Association between causative polymorphism and linked markers diminished for all but most closely linked
Assumption: disease predisposition due to common allelic variants at multiple loci (compared to rare variants with HUGE effects)
Meta-analyses (combining data across studies)
Identified associated regions that failed to pass significance thresholds in original studies
Follow-up by identifying causal variant associated with SNP-associated (can be difficult)
Lessons Learned with conducting a GWAS
Need large sample
Must account for multiple stat tests
Replication of results is essential
accurate assessment of phenotype, narrowly defined
Lessons learned - results of conducting a GWAS
Complex traits are highly polygenic
Pleiotropy is common (a single gene affects multiple, seemingly unrelated traits or characteristics)
How do you identify causative variants?
Is there a candidate gene coding sequence change? If not..
Combine independent sources of info (multi-omics)
Results from GWAS where: phenotype is trait, gene expression, or protein level.
Results from genomic analysis of epigenetic mods
Results from identification of regions with open chromatin
Applications for causative variants (how is GWAS relevant to human or veterinary medicine)
Drug choice/dosage
Identify personal disease risk (polygenic risk score)
Drug targets
Genomic selection for livestock
Ovine Progressive Pneumonia (OPP)
Caused by ovine lentivirus infection. Slow progression (2+ yrs).
Loss of body condition (but normal appetite)
Increased breathing effort at rest
Common for second bacterial infection → fever, cough, lethargy, nasal discharge
Hard bag (enlarged, firm udder, no milk)
How is OPP spread?
Respiratory in adults → consumption of infected colostrum or milk
Integrates into host genome
Infected never clear virus → no treatment
Manage through reducing prevalence (testing, remove infected, increase genetic resistance)
Sensitivity vs Specificity
Sensitivity- proportion of patients with disease that test positive
Specificity- proportion of patients without disease that test negative
Porcine Reproductive and respiratory syndrome (PRRS)
Causes repro, respiratory, and reduced growth problems
Caused by single-stranded RNA virus (Arterivirus)
Different breed susceptibilities suggest genetic component
GWAS study for PRRS
Pigs donated from companies challenged with virus
Eliminate exposure questions
Dedicated facility for pig housing
Study conducted in replicates of 200 pigs
Pigs tested for viral growth and load
PRRS results
Strongest signals for viremia found on SSC4 and SSCX
What was the characterization of the positional candidate gene (SSC4)
Several GBP family members in region (all candidates) → mediate proinflammatory immune response
GBP5 → thought to function in uncovering virus in endosome (splice site mutation, frameshift mutation, low expression of wild-type)
How does understanding PRRS help us?
Wild-type less common → greater opportunity to improve population by selection
Wild-type is dominant → no need for homozygous genotyping
Bovine Respiratory Disease (BRD)
Caused by variety of pathogens, viral and bacterial
Symptoms: fever, loss of appetite, shallow breathing, coughing, salvation, watery/pus/bloody nasal/eye discharge
How was BRD in a GWAS studied?
Scored values visually
Samples cultured to determine pathogen
genotyped cases and matching controls (herd, gender, pen, location)
Results from BRD study
no single genomic region of large effect
Multiple regions with small to modest effect
Microphthalmia
Abnormal eye development, prominent third eyelids, small eyes, blind, partial deafness
Melanin Synthesis
Extension Locus (E)
E → normal extension. Allows black pigment to be expressed
e → recessive red. Coat is red or yellow instead of black
E^M → Black mask around muzzle (boxer)
E^m → larger black mask (pug)
E^G → grizzle (pale face with a widow’s peak above the eyes i.e. afghan hounds)
Dominance: E^M > E^G > E > e
MC1R
Important Concepts
Genes and their functions are conserved across species
Specific mutations differ btwn species
phenotypic effect of mutations may be similar
Important Concepts #2
One gene may affect multiple phenotypes
pleiotropy
MC1R allelic variants in humans effect: skin/hair color, freckling, risk for skin cancer, pain sensitivity
Important concept #3
Multiple genes can have complex interactions in determining phenotype
Epistasis → phenotype dependent on combination of genotypes at multiple loci
Tyrosinase Function (involved with B locus)
If tyrosinase low and cysteine high → Phaeo Melanogenesis (reddish yellow color)
If tyrosinase high and cysteine low → Eumelanogenesis (Brownish/black color)
Brown (B) Locus
B → dominant black. Dogs with at least 1 B with express black
b → brown. Dogs with two bb will only have brown coat with no black
TYR1P
Agouti (A) locus
ay → fawn or sable. Dominant to other alleles
aw → Wild-type agouti. Produces banded hairs, resulting in a wolf-grey or sable appearance
at → tan point pattern. Causes tan or cream pattern specific to areas of the body.
a → recessive black. Causes solid black coat color
OTHER: A^DY > A^SY > A^AG > A^BS > A^BB > A^a
DY (dom yellow)
SY (shaded yellow)
AG (agouti)
BS (black saddle)
BB (black black)
A (recessive black)
ASIP
K Locus
KB → dominant black. Dogs with at least 1 KB will express black
ky → non-solid black. causes brindle, tan points, or other patterns. Requires 1 E to express
kbr → brindle (needs two kbr)
k → allows expression of other alleles at other loci, red or dilute
Dominance: k^B > k^br > k^y
CBD103
Dilution (D) locus
D → Full pigmentation. No dilution of color
d → dilution. Causes dilution of black pigment, resulting in blue (from black) or isabella (from brown)
Causes from mutations in melanophilin (MLPH gene)
Spotting (S) locus
S → no white spotting. Solid coat
sp → piebald. causes white spotting on coat in specific patterns
si → irish spotting. Results in white markings on the face, chest, belly, and feet.
sw → extreme white (deaf and blind)
MITF
Interaction of E and K loci
If Ee or EE and k^br/k^br or k^br/k^y → will be brindle with the color of the phaeomelanin part of the brindling in turn affect by agouti alleles
If Ee or EE and k^y/k^y → the distribution of eumelanin and phaeomelanin will be determined solely by the agouti alleles
Becker’s Muscular Dystrophy (BMD)
In-frame deletions in the DMD gene that allow synthesis of an internally truncated, but functional protein
Duchenne Muscular Dystrophy (DMD)
Mutations that cause premature translation termination (nonsense or frameshift) → loss of functional DMD gene product
Dystrophin gene responsible
recessive/fatal (X-linked) → affects males
What is the path of different types of muscular dystrophy in dogs?
Symptoms of MD in dogs, what causes different variations?
Symptoms: chewing but dropping food, muscle stiffness, pneumonia, cardiac failure, enlargement of the tongue (hypertrophy)
Why variation? Different mutations with different effects, gene x environment interactions.
Dystrophin
Maps to chrm. X → needed for mechanical stability of muscle cells
RNA splicing
Original mRNA copy of all exons and introns → before RNA moves from nucleus to cytoplasm → introns are removed and exons are spliced together
GT-AG rule
Introns begin with a GT and end with a AG. Mutations near either end of an intron may cause a change in splicing exons and produce abnormal protein
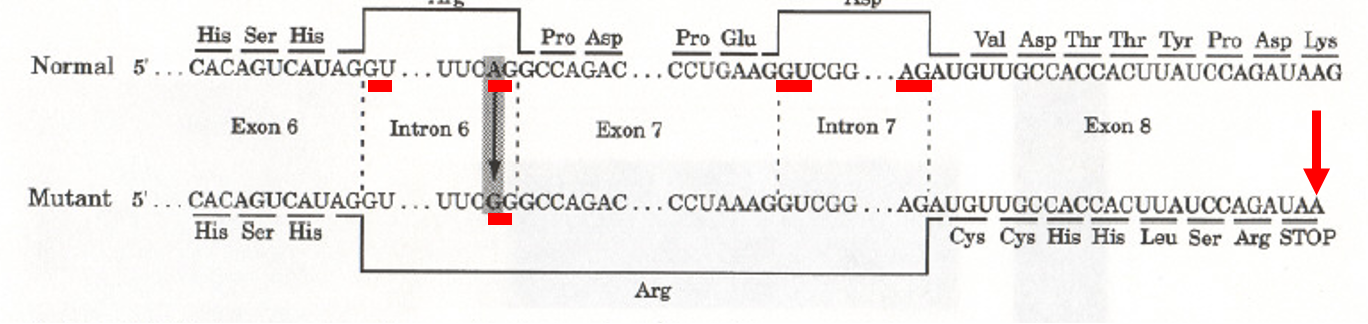
List the process for a frameshift mutation creating a stop codon in exon * of dystrophin in dogs
Base substitution in the splice-acceptor site of intron 6 of the dystrophin gene (AG to GG) → exon 7 is spliced along with intron 6 and intron 7 → deletion of exon 7 → stop codon in 8 → result: truncated transcript of dystrophin
“Treatments for MD”
Molecular: gene therapy/gene correction, gene editing using CRISP/cas9, Cellular stem cell transplants into muscle, pharmacologic compounds that reduce inflammation.
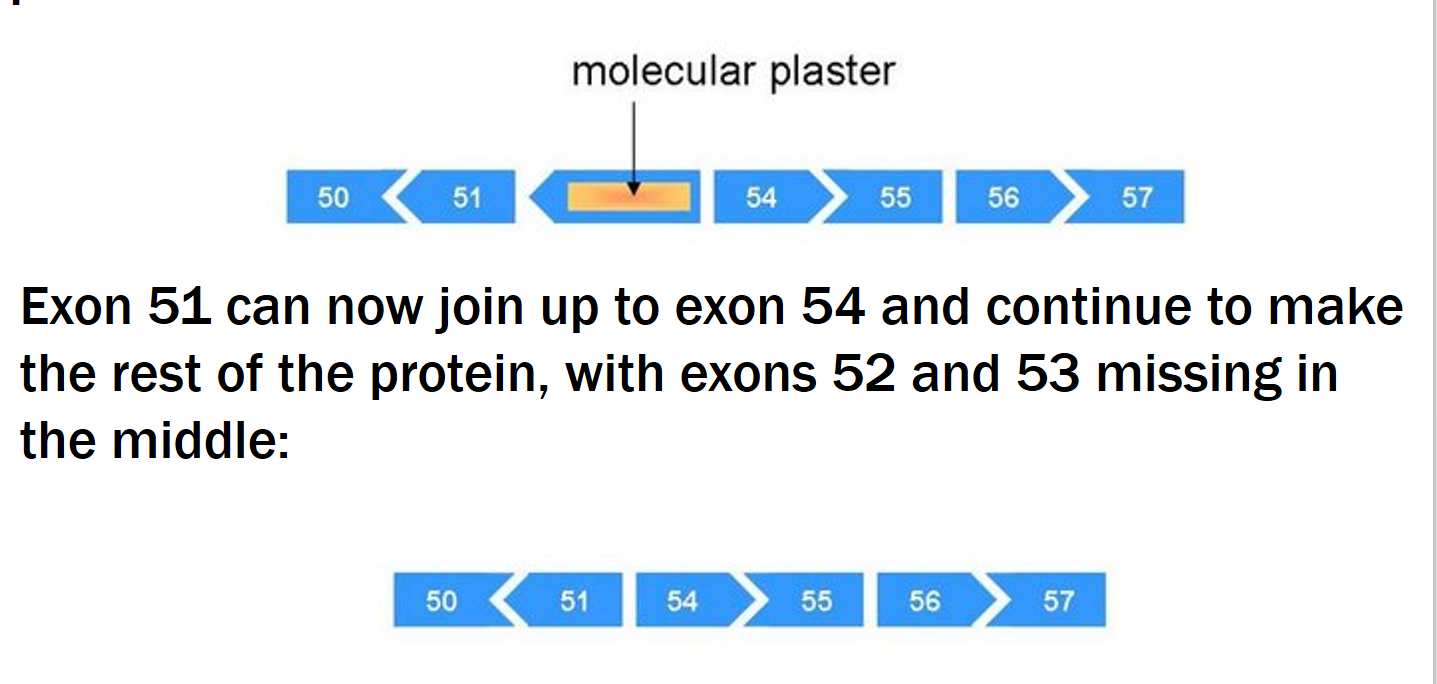
Exon Skipping therapy
Small pieces of DNA called antisense oligonucleotides (AOs) (molecular patches) → used to mask the exon that you want to skip → ignored in protein production.
Gene Editing types and challenges?
Non-homologous end joining (NHEJ) or homology directed recombination (HDR)
Germline editing not feasible → ethical issues, many mutations make it impractical to use for everyone, components must be delivered efficiently in vivo to skeletal muscles and heart
Cas9 Gene editing
Cas9 can cut DMD gene to have deletion or insertion of a few nucleotides to restore correct alignment of triplet codons
Ringo!
Born with another mutation that protected him from MD!
Jagged1 plays important role in muscle regeneration
Mutation in the promoter region caused upregulation of the gene in ringo
Overexpression of Jagged1: normal muscle phenotype
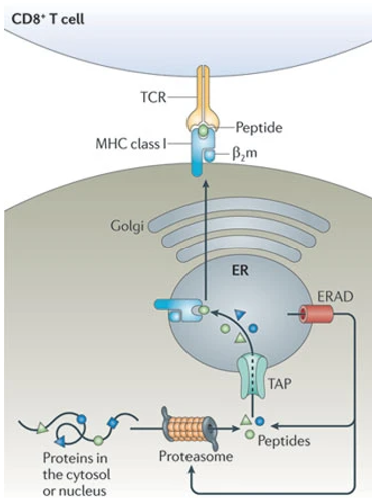
MHC Class 1 Formation Process
degradation of cell proteins by proteasomes into peptide fragments → fragments transport into lumen of ER where they are complexed with MHC 1 receptors → peptide-receptor complex is then transported to plasma membrane for antigen presentation to CD8+ T cells
MHC class 2 Formation process
Foreign peptides taken up by antigen presenting cells and degraded into small fragments → fragments complexed with MCH class 2 receptors and presented to t-helper cells → T-helper recognize the MHC-peptide complex which stimulates adaptive immune response.
MHC class 1: in infected vs uninfected cells
Uninfected: peptides only derived from host cell proteins
Infected: foreign peptide-MHC complex activates t-cytotoxic cells → T-cyto produce cytokines and other proteins to trigger apoptosis of the infected host cell
Peptides are not all equal which means…
Different shapes and amino acid sequences determine peptide binding → 2 different MHC alleles may bind to different peptide fragments → different individuals respond differently to same pathogen
Dendritic cells
Antigen-presenting cells of mammalian immune system. Function is to process antigen material and present it on the surface to T cells
MHC Class 1 vs MHC class 2
Class 1:
present on cell surface of all nucleated cells
consists of 2 non-identical chains: alpha (chrm 6) and beta (chr 15).
Responsible for presentation of peptides to T-cells
Class 2:
Present on the surface of antigen-presenting cells (B cells, macrophages, dendritic cells)
Consists of 2 identical chains (both on chr. 6)
Responsible for presentation of peptides to T-cells
Role of MHC in immune response process
MHC Present T cells a small fragment of pathogen → fragment held within a groove
Characteristics of the immune system
Specificity: each antibody is specific for one antigenic determinant (epitope)
Diversity: immune system can recognize >10 million different epitopes
Memory: once exposed to antigen, the immune system remembers that antigen
Distinguishing self from non-self
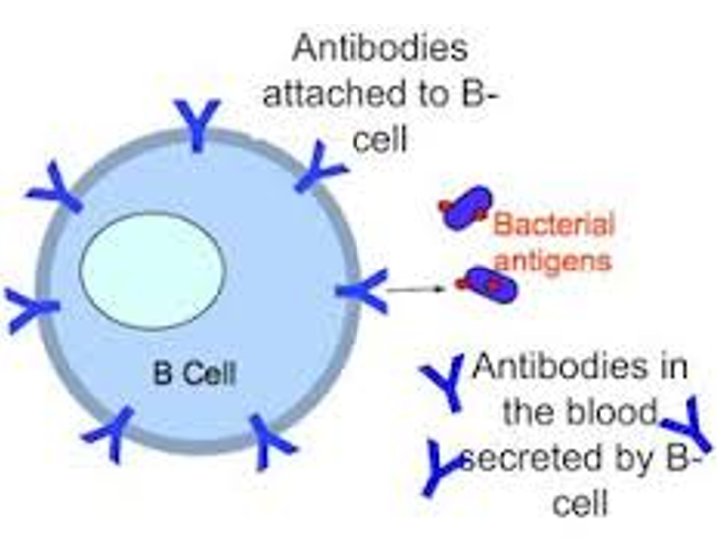
Immunoglobulins
Produced by B-cells: either found on the surface of B-cells (b-cell receptor or antigen receptor) OR secreted into extracellular fluid (antibodies)
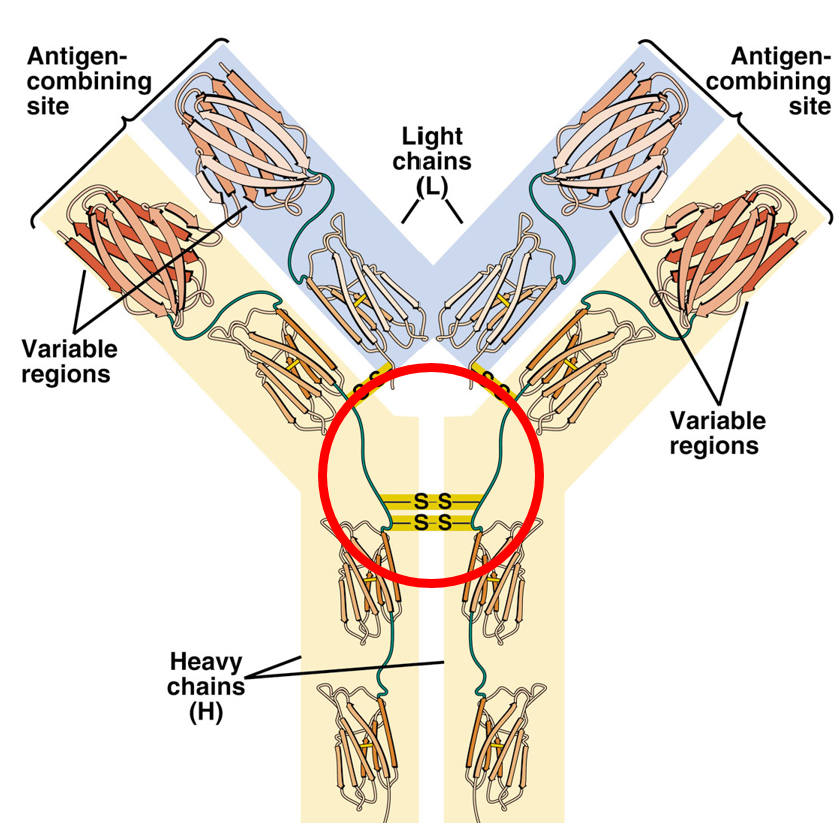
Structure of Immunoglobulin G (IgG) molecule
The light chains have three domains: V (variable), J (joining), and C (constant). Heavy chains have 4 domains: V, D, J, C
Sources of immunoglobulin diversity
Gene rearrangements, class switching, nucleotide insertion/deletion, receptor editing, gene conversion, somatic hypermutation
Gene Conversion
Involves the transfer of genetic (sequence) info from one gene to another (basically copy and paste)
Myxomatosis info
Infects only rabbits. Caused by myxoma virus. Introduced into Australia to control rabbit infestation. Innocent rabbits are imported → CSIRO releases virus → almost all rabbits died due to high susceptibility and virulent pathogen → rabbits become resistant and virulence decreases → CSIRO releases new strains.
Trypanosomiasis
“Sleeping sickness.” caused by a parasite named Trypanosome. Become infected from bite of infected tsetse fly. Effects humans and animals and decreases production and fertility.
Trypanosomiasis early stages vs late stages
Early: Onset occurs 1-3 weeks after bite. Fever, headache, general weakness, and weight loss occur. Multiple organs (liver, spleen, skin, eyes, etc) affected.
Late: start to psychotic behaviors (violence, mania, suicide, etc.), motor, sensory, sleep abnormalities (wanting to sleep all the time and changes to sleep cycle).
Trypanosomiasis late stages
Psychiatric, motor, sensory abnormalities, sleep disturbances.
Psychiatric: personability changes, headache, violence, hallucinations, suicidal tendencies, mania
Sleep disturbances: tiredness, distractibility, spontaneous urges to sleep, reversal of the normal sleep-wake cycle
Trypanosome Biology
Animals are reservoir for parasite. Flies become carrier after feeding on effected host, parasites undergo morphological and biochemical changes in fly’s anterior midgut.
Antigenic Variation
Parasite encapsulated by glycoprotein coat. Antigenic variation: the parasite changes its variant surface glycoprotein (VSG) coat to evade the immune system. Keeps parasite one step ahead. →Unexpressed VSGs are scattered among the different chromosomes.
→All expressed are located near telomeres.
→ telomeres contain expression sites (ES) under control of promoter, but only one ES can be active at a time
→ trypanosomes are preprogrammed to produce sequence of antigens (spontaneous)