Lecture 7: Protein Structure and folding
1/28
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
29 Terms
Primary structure: amino acid and genetic code
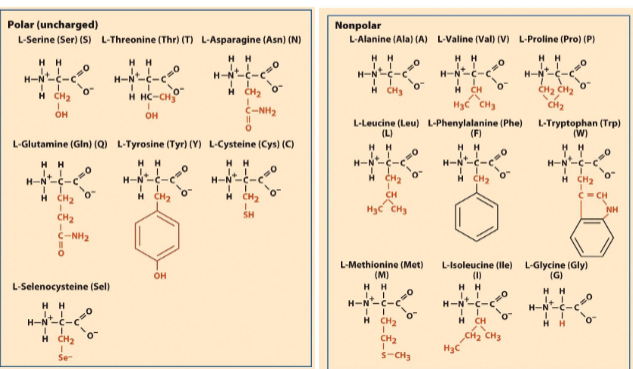
The 22 amino acids found in proteins
proteins are chain-like polymers of AA
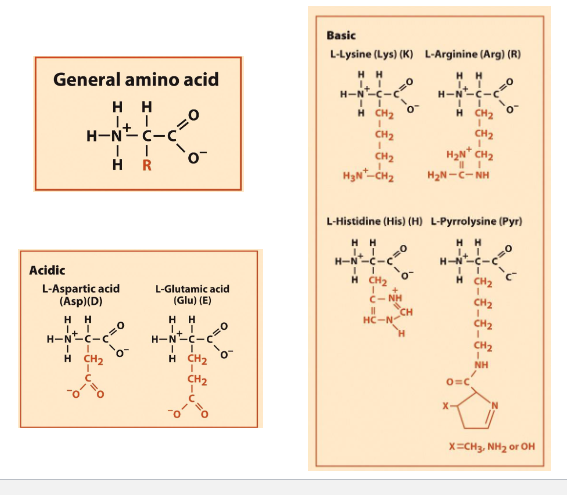
Each AA has amino/ carboxyl group
diff R groups
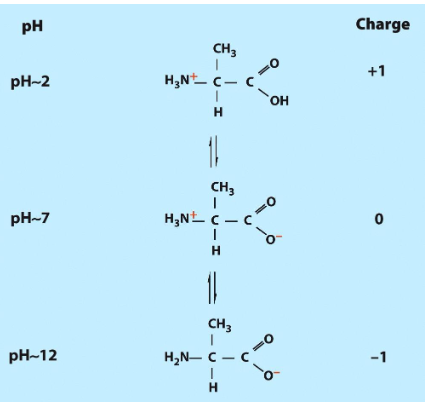
pH 7 = charges on backbone
PH2= +1 change
PH7= 0change
Ph12= -1 charge

AA to remember
Lysine, Arginine, Histidine, Pyrrolisine (Basics)
N terminal has lots of Lysine, adn arginine, that are modified and are epigenetic modifications
Histidine is fron histone- interact with DNA
Pyrrolisine (modified lysine)
Aspartic acid (asparagous), glutamic acid (acid)
Tyrosine- phosphorylated adn dephosphorylated
Cysteine (created disulfide bonds) for protein protein interactions (P)
Methionine (s-group) (NP)- first AA in eukaryotic and codon is AUG
selenocysteine (modified cysteins)
AA
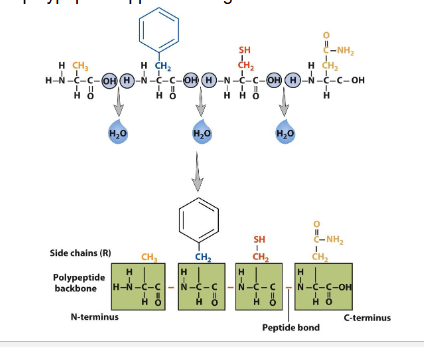
AA help by petide bonds due to condensation reaction
peptide= sequence of AA
has partial double bond character due to resonance
free rotation occurs between a-carbon and peptide unit
trans and cis config possible due to rigid peptide bond
AA cont
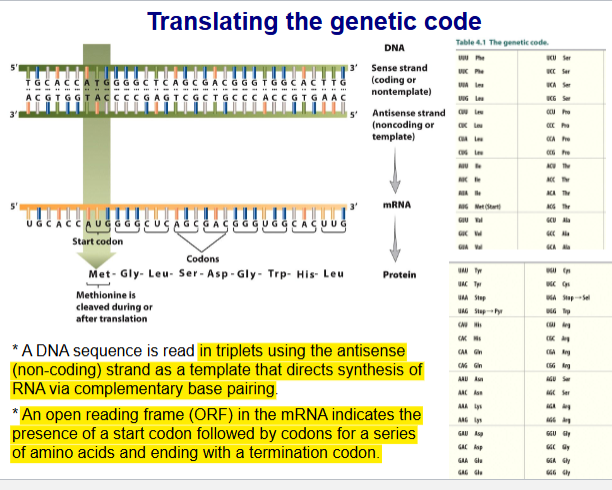
Translating genetic code
DNA seq read in triplets using antisense (non-coding) strand as a templet that directs synthesis of RNA via complementary base pairing
ORF (open reading frame) in mRNA indicates presence of start codon followed by codons for a series of AA and ending with termination codon
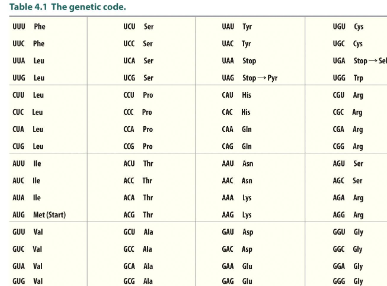
The genetic code
codon box
made of 4 three letter codes (64 in all)
61 codons recognized by tRNA for incorporation for the 20 common AA
3 codon signal termination or code for selenocysteine and pyrrolysine
The genetic code is degenerate
tRNA is specific to specific AA recognize multiple codon triplets that differ in 3rd letter only
Pairing for first 2 codon position are the same for codon and anticodon using complementary base pairing rule
wobbles” (non-Wat/crick base pairing) occue at third position
look at graph
3rd of codon and 1st on anitocodon has flexibility

Genetic code is not universal
In certain organisms and organelles the meaning of select codons has been changed.
The 21st and 22nd genetically encoded amino acids
UGA code for selenocysteine found in (UGA used as codon to expresses selenocysteine)
15 genes in prokryotes involved in redox reactions
40 genes in euk code for various antioxidants and type 1 iodothyronine deiodinase
UAG code for pyrrolysine found in(UAG used as codon to expresses pyrrolysine)
some archaebacteria and eubacteria
Codon bias
freq at which different codon are used vary between diff organisms and between proteins expressed at high or low levels within same organism
codon bias can have major impact on effeciency of expression of proteins if they contain codons that are rarely used in desired host
D- and L-amino acids in nature (na)
enantiomers
living org maininly have L-AA
ribosomes use L-AA to make proteins
D-AA made in pathways that do not involve ribosomes
can be made by L-AA by post translation process
The three-dimensional structure of proteins
Dalton (Da)= describes molecular wieght of proteins
most polypeptide chain habe molecular weight of 20-70kDa
avg mol/weight of AA is 110
2nd structure
stabilized by H-bonds (amino is donor, and carb is acceptor) that ceate a peptide bond
structures
a-helix
Prolein (helix breaker) cannot act has a donor in H-bonding
B-pleated sheet
stab by H0bind
Parallel
same directions
Antiparallel (more stable)
opp directions
unstructured turns
turns connect a and b in proteins
short loops that do no exhibit a defined secondary structure
tertiary structure
mostly stabilized by noncovalent bonds:
hydrophobic interactions
H-bonds
Primary covalent bond within/between polypeptide are disulfide bridges between cysteins (S-S)
structures
globular proteins
most proteins are spherical
fibrous proteins
long filamentous or rod-like structures
membrane proteins
differ from soluble proteins in relative distribution of hydrophobic AA residues
7 transmembrane helix structure is a common motif in membrane proteins
Quarternary structure
1 or more polypeptide subunits creates functional protein
great versatility of function
Protein function adn regulation of activity (na)
proteins made of domain with specific function
each domain made of continous AA sequence
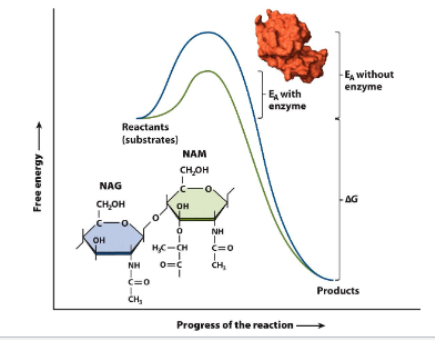
Ezymes are biological catalysts
lower activation energy → speed up reaction
substrante bind complex with enyme by binding to active site
induced-fit mechanism
Regulation of protein activity by post-translational modifications
post translation mod
ex: phosphorylation adn allosteric effectors
After translation- proteins joind cov& noncov to other molecules
exL lipoproteins, phycoproteins
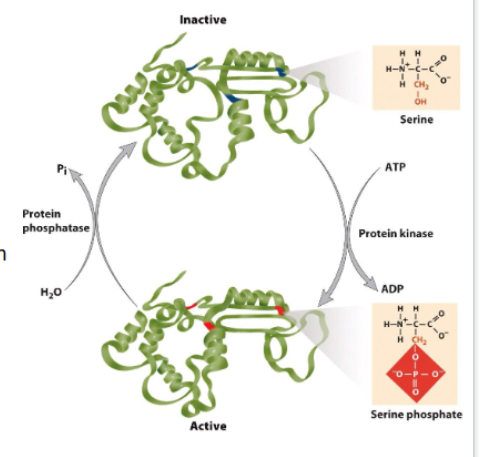
reversible phosphorylation of a AA side chain is most common regulatory mechanism
Protein phosphorylation (NON remember)
can cause protein to change shape
phosphorylates side chain can be part of motif to facilitate formation of a multiprotein complex
phosphorylated side chain can promote dissociation of a multiprotein complex
AA: Ser, Thr, Tyr
Kinase catalyze addition of phosphate group
2 protein kinase groups
phosphorylate serine/threonine side chains
phosphorylate tyrosine side chain
phosphatase remove phosphate
less specif
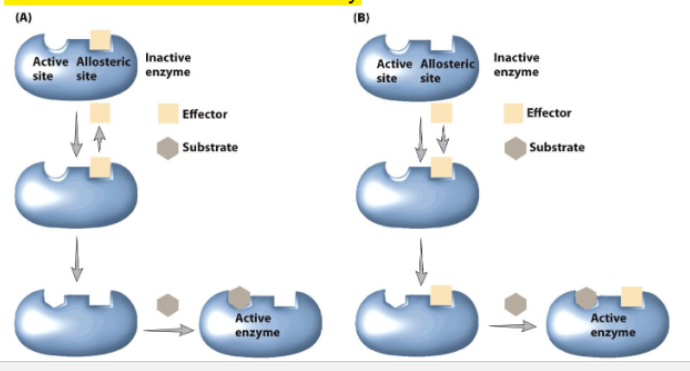
Allosteric regulation of protein activity
ligand induces conformational change
an active site/ another binding side is altered in a way that increases or decreases activity
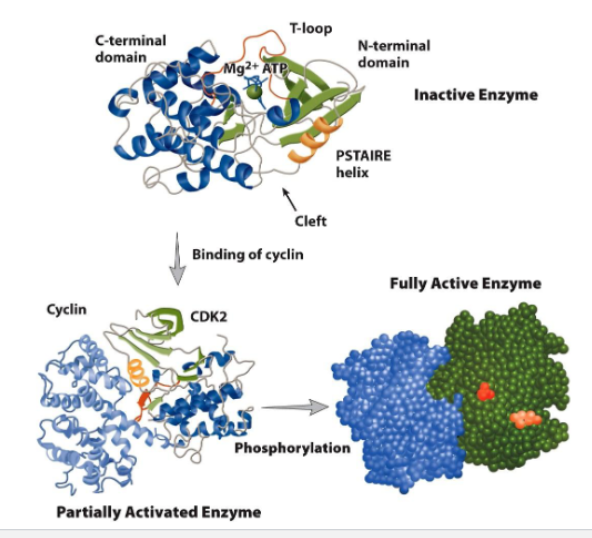
Cyclin-dependent kinase (CDK) activity is regulated by both allosteric modification and phosphorylation
Partial activation of CDK
binding cyclin to CDK causes conformational change
T loop moves away from entrance of active side
Fully activation of CDK
phosphorylation of THr160 in T loop by CDK-activating kinase (CAK)
stabilizes active site (catalytic cleft)
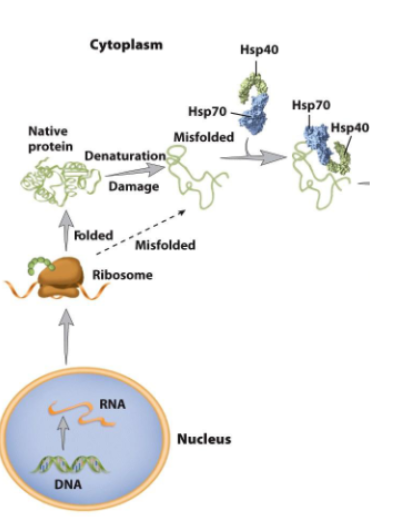
Protein folding and misfolding (No remember)
protein folding is innitiated before completion of protein synthesis
other protein go major olding after reease into cytoplasm or organelle
Molecular chaperones
Increase efficiency of protein folding.
Reduce the probability of competing reactions such as
aggregation.Heat-shock proteins promote protein folding and aid in the destruction of misfolded protein.
e.g. Hsp40, Hsp70, Hsp90
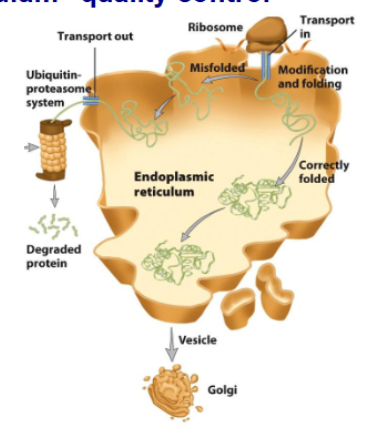
Endoplasmic Reticulum (ER) quality control
secreated protein are translocated into ER
folding happens before secretions go into Golgi apparaturs
incorrectly folded proteins are directed by unfolded protein response and targeted for degradation
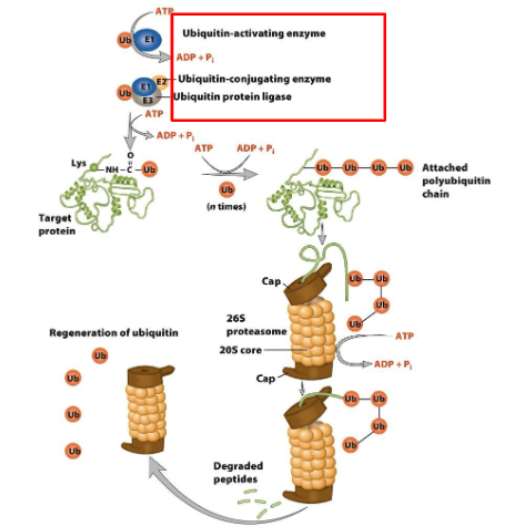
Ubiquitin-mediated protein degradation
Ubiquity (a 76 amino acid polypeptide) is attached to a protein by a series of enzyme-mediated reactions.
The ubiquitin-conjugated protein is then targeted to the 26S proteasome.
Ubiquitin is released and the target protein is degraded
by proteases
ubquity activated by atp (E1) activating enzyme
Ubiquitin-conjugating enzyme(E2) congregate enzyme
Ubiquitin Protien ligase (E3) ligase enzyme
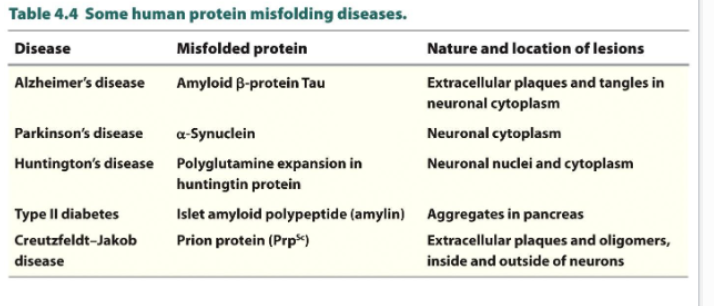
Protein misfolding diseases
Formation of protein aggregates is linked to at least 20 different human diseases
Amyloid or amyloid like fibrils: normal soluble proteins accumulate as insoluble deposits
these proteins in amyloid-like fibrils fold into cross B-spine
Prions
Primary cause of transmissible spongiform encephalopathies (TSEs)
progressive neurodegenation
dementia
loss of muscle control of voluntary movements\
No cure, death 6monts 1 year
Animal forms
scrapie (sheep)
BSE(mad cow disease) bovine spongiform encephalopathy
chronic wasting disease (elk/deer)
The “prion only” hypothesis of infection
Stanley Prusiner: Nobel Prize in 1997.
Lack of immune response characteristic of infectious diseases.
Long incubation time (up to 40 years for kuru).
Resistance of the infectious agent to radiation that destroys living microorganisms
(e.g. viruses, bacteria)
radiation destroryes DNA/RNA
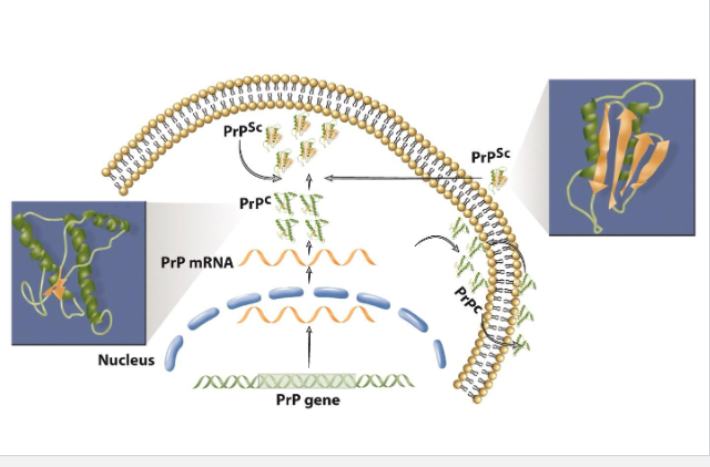
infectious agent is a protein called PrP^Sc that can replicate self within the body
normal cells have normal PrP^c , the PrP^Sc has same AA sequence but prion is misfolded into diff 3D structures
in infected cell, host protein PrP^c misfolds to form new prions called PrP6Sc
can survive sterilization techniques