D103 Cell Surface Receptors with Intrinsic or Associated Enzyme Activity (ALS 18 and Video 32)
1/44
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
45 Terms
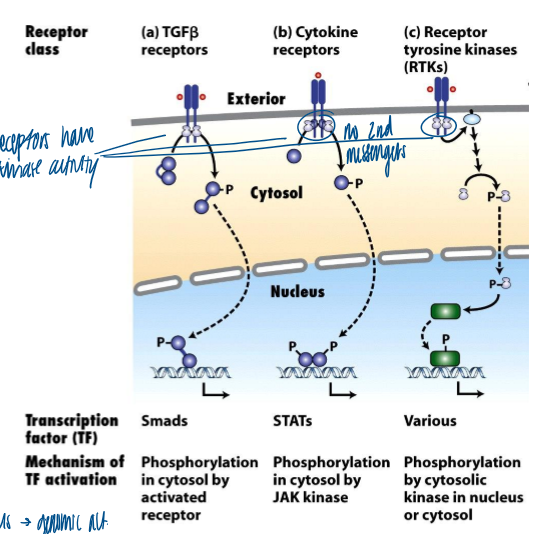
overview of kinase activation
ligand binding → conformational change → kinase activity → change in protein phosphorylation → nucleus → genomic activation
receptors have kinase activity
no second messengers
intrinsic
proteins in which the kinase domain is encoded by the same gene encoding the transmembrane receptor protein
associated
the protein with the kinase activity is encoded by a separate gene that does not encode the transmembrane receptor
cell surface receptors with enzyme activity and GPCR similarities
both are TM proteins with ligand-binding domain on the outside of the cell that transmit a signal across the PM via a conf change
cell surface receptors with enzyme activity and GPCR differences
enzyme-linked cell surface receptors are usually single-pass TM proteins, unlike GPCRs
cytoplasmic domain of enzymatic cell surface receptors
have either intrinsic enzyme activity or may be physically associated with an enzyme
enzyme in the cell surafce receptors
kinase (or sometimes a phosphatase) with specificity for tyrosine or serine/threonine amino acids
how were enzyme-linked cell surface receptors identified
by their ability to stimulate growth, proliferation, differentiation, or survival of cells exposed to extracellular proteins (growth factors)
result of growth factor exposure
cells can respond both rapidly (ex - changes in cytoskeleton within mins) and slowly (ex - changes in gene expression that take hours)
disorders in signaling
cause of developmental defects and cancer
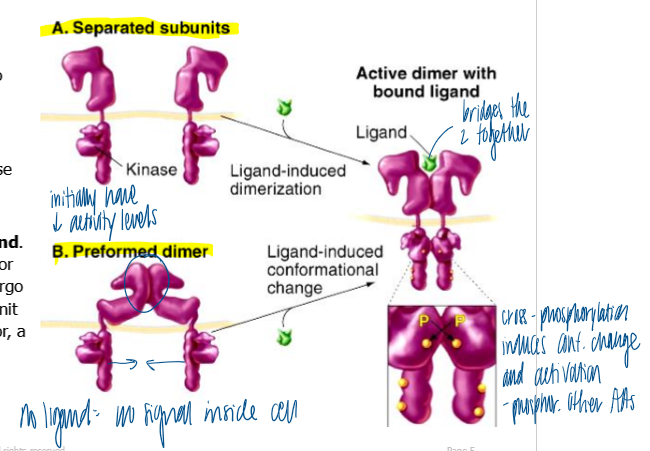
how are single-pass TM receptors usually activated
dimerization
ligands cause monomeric receptors to dimerize
separated: dimerized receptor subunits phosphorylate each other → kinase domains become fully active
preformed: preformed dimer in the absence of ligand; receptor still needs to be bound by ligand and undergo conf change before each subunit becomes fully active
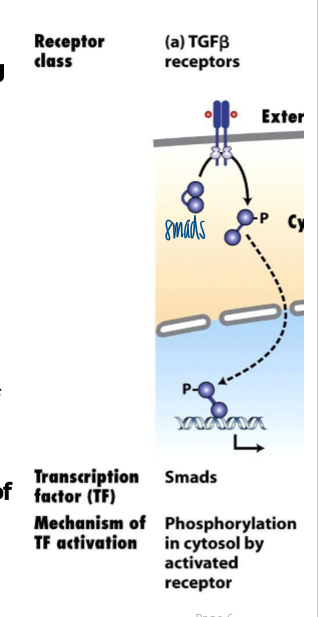
TGF-B receptor and transducing signaling proteins
receptor: TBFB-family receptors
transforming growth factor-B (TGFB) family members (includes TGFB and BMP ligands/ receptors) regulate embryonic development and differentiation by inhibiting cell proliferation and stimulating differentiation
transducing signaling proteins: Smads
ligands for TGFB/BMP receptors are secreted dimeric proteins
tgfb family receptors
hetero-tetramers (2 of each type I and II subunits)
each subunit has intrinsic serine/threonin kinase activity
tgfb family signals
signals from the cytosolic side of the receptor are conveyed to the nucleus by 3 functional classes of Smad proteins: receptor regulated (R-) Smad, Co mediator Smad, and inhibitory smad
combos of smads allow for large number of responses to different ligands
R and Co smads
proteins that form hetero-trimers that bind to DNA and recruit other factors required for regulation of target gene expression
I smads
can negatively regulate signaling by causing expression of inhibitors of transcription or by blocking interaction of R-Smad with the receptor
defects in tgfb family signaling
causes a set of developmental disorders and diseases including cancer, fibrosis, autoimmune, and CV disease
signaling thru TGFB receptor pathway
ligand binds to type II receptor (RII)
RII’s kinase activity is always on
Ligand binding to RII stimulates RII to interact with and change the conf of RI → Phosphorylated RI has increased affinity for R-Smad, which it subsequently phosphorylates
phosphoryation of the R-Smad on S at the C term changes its conf, which exposes a NLS
phosphorylated Smad3’s can associate via interaction on their MH2 domains and the phosphorylated S
importin B forms a complex with one or 2 molecules of phosphorylated R-smads, plus 1 molecule of non-phosphorylated co-mediator Smad
after nuc import, importin B undocks and the Smad trimer associates with different proteins (ex - transcription factor to form a transcriptional activation complex at target sites in the genome
r smads are continuously dephos and exported from the nuc, thus the conc of smads in the nuc is directly proportional to amount of TGFB receptor signaling occuring at the cell surface
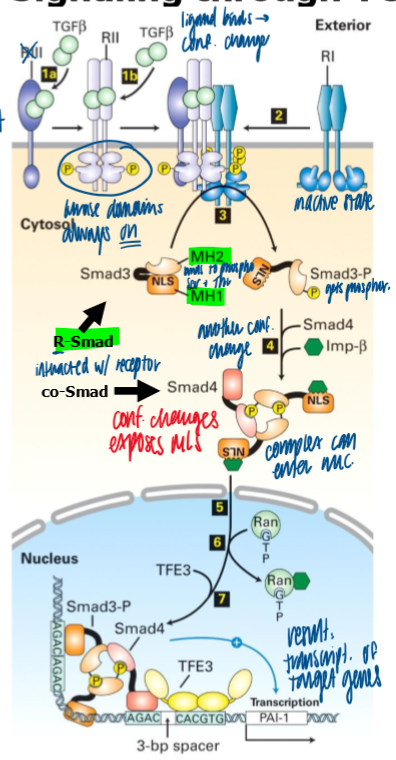
negative feedback of TGFB signaling
via i-smad dependent polyubiquination of TGFB Type I receptor and proteosomal degradation
I smad
activation of TGFB signaling can induce Smad7 (inhibitory) and competes with R smad for binding to T1 receptor
cannot be phosphorylated by the T1 receptor, preventing interaction of R smadds with the receptor (neg feedback)
recruits Smurf 1 and 2 to the TGF B receotor, which results in poly ubiquination of the T1 receptor and its degradation by the proteosome (negative feedback)
fibrodyplasia ossificans progressiva (FOP)
a dom mutation in a TGFB receptor pathway affects skeletal homeostasis
rare genetic disease; progressive physical impairment due to ectopic ossification
accel ossification following muscle injury (formation of secondary skeleton)
susceptible to infection
sensitizes mesenchymal stem cells to form bone
cytokine receptors and transduction signaling proteins
receptors: cytokine receptors
transduction signaling proteins: JAKs, STATs
combos of receptors and signals allows the cell to respond in a sophisticated manner to different ligands
cytokines
family of polypeptide hormones and growth factors that regulate many cellular processes
ex - production of RBC, milk, body growth, stimulation of body’s defenses against infection
cytokine receptors
usually a homodimer or heterodimer
some receptor subunits are shared by many different types of receptors
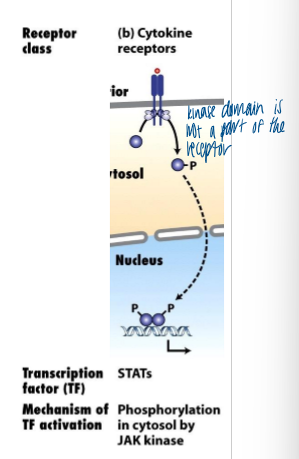
no intrinsic kinase activity, but instead associate with cytosolic tyrosine kinases (JAKs) = extrinsic
cytokine signals
signals from the receptor-JAK complex are conveyed to the nucleus by STAT proteins (similar to Smads)
JAK-STAT signaling can be negatively regulated by dephosphorylation of JAKs, destruction of JAKs via SOCS-mediated polyubiquination
STAT = signal transducers and activators of transcription
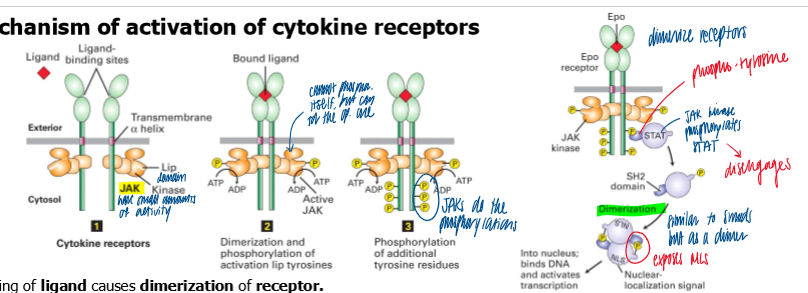
mechanism of activation of cytokine receptors
binding of a ligand causes dimerization of receptor
initially weak kinase activity of each associated JAK phosphorylates a tyrosine on the opposite JAK → conf change to activate both JAK
active JAK → phosphorylate tyrosine AAs on cytosolic tails of their receptors → serve as binding sites for STATs via a SH2 domain
JAKs phosphorylate the receptor bound STATs
phos of stats = release and dimerization thru ints involving SH2 domains
dimer of STATs cause conf change that exposes a NLS
STAT dimer translocate to the nuc for transcription
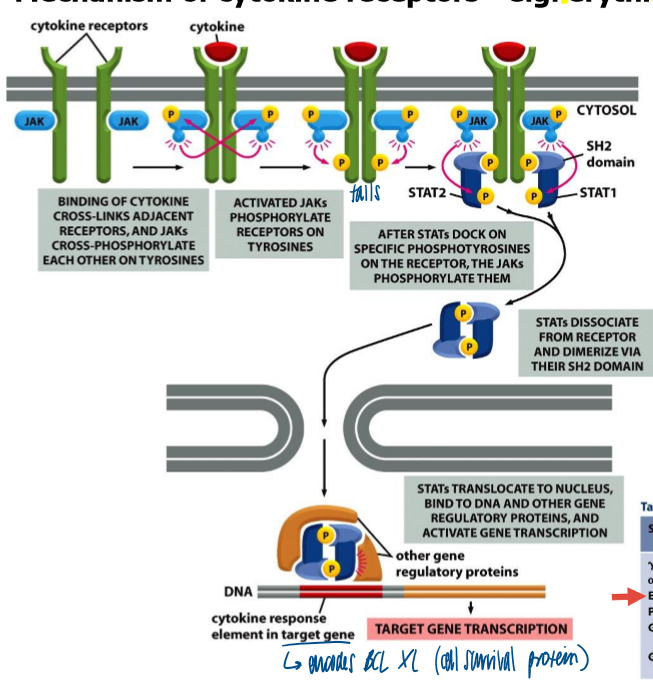
mechanism of cytokine receptors
binding of cytokine crosslinks adjacent receptors, and JAKs crossphorphorylate each other on tyrosines
activated JAKs phosphorylate receptors on tyrosine
after STATs dock in specific phosphotyrosines on the receptor, the JAKs phosphorylate them
STATs dissociated from receptor and dimerize via their SH2 domain
STATs translocate to nucleus, bind to DNA and other gene regulatory proteins, and activate gene transcription
inactivation of cytokine receptors
via dephos or protein degradation
protein phosphatases (SP1) can bind phosphotyrosines in cytoplasmic domain of receptor using a SH2 domain
SHP1 can then turn off cytokine receptor signaling by dephosphorylating JAKs and cytokine receptors (neg feedback)
suppressor of cytokine signaling (SOCS) proteins can also bind phosphotyrosines using a SH2 domain in SOCS
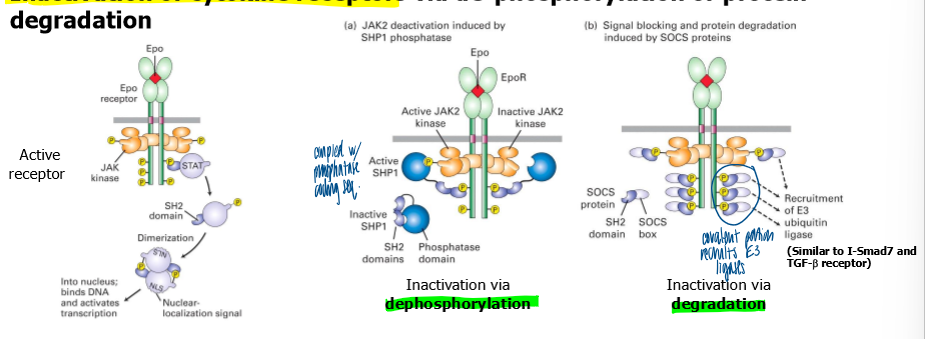
SOCS
have an amino acid sequence recognized by the ubiquitin E3 ligases, which targets the receptor and or JAK for degradation via the proteosome system
consequences of defects in cytokine receptor sig in mice
loss of function of erythropoietin receptor or JAK2 in gene knock-out mice cuases failure in fetal erythropoises
giantism in SOCS-2 gene knock-out mice
RTK receptors and signals
receptor: receptor tyrosine kinases (RTKs)
signals: Ras/MAP kinase
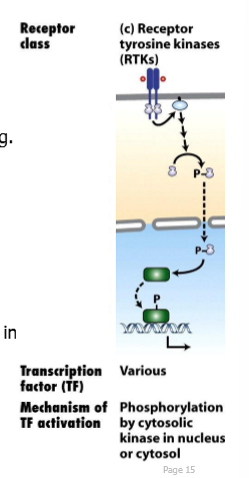
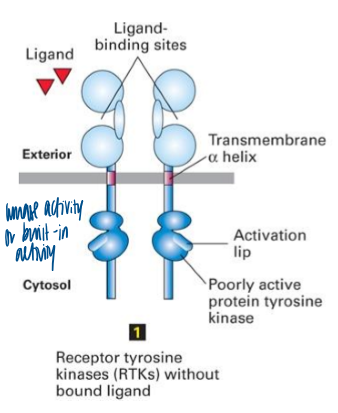
receptor tyrosine kinase
receptors have intrinsic tyrosine kinase activity in their cytoplasmic domain
Ras/ MAP kinase
ligands for RTKs include soluble or membrane-bound proteins (ex - nerve growth factor, fibroblast growth factor, epidermal growth factor)
ligand-induced activation of a receptor stimulates its tyrosine kinase activity
downstream signaling from RTKs often invovles Ras- MAPK pathway
role of RTK signaling pathway
prolifeation, differentiation, survival, and metabolism of cells
defects in this path are often associated with defects in human development and homeostasis (cancer)
RTK vs cytokine signaloing
RTK: kinase activity is intrinsic to the receptor on its cytoplasmic tail (encoded as part of the same gene)
same mech of activation of a lip domain in the kinase by low-level kinase activity following receptor dimerization
cytokine receptor: kinase (JAK) is extrinsic with the receptor on the cytosolic side of the PM (associates non-covalently)
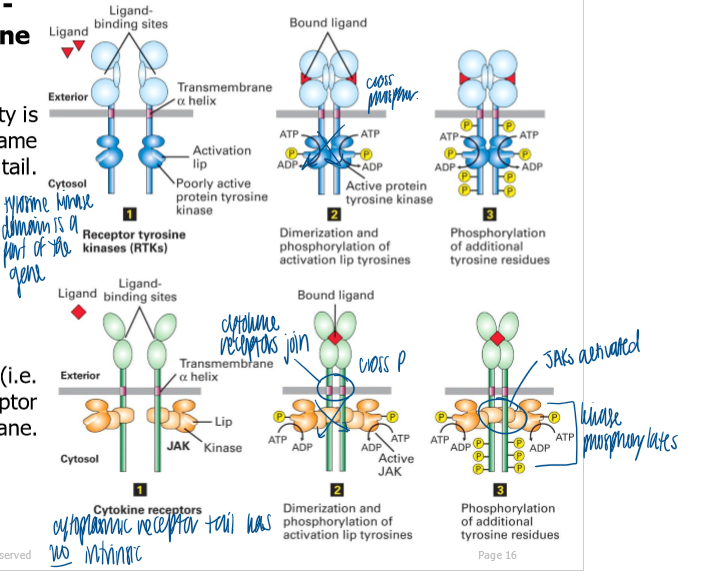
what does ligand binding cause with rtks
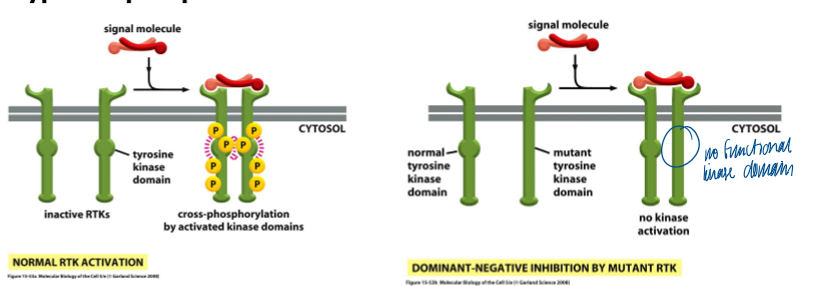
dimerization of RTKs followed by activation by cross/trans phosphorylation
kinase domains of monomeric RTKs are initially mostly inactive
binding of ligand causes dimerization of RTKs that initially have low kinase activity
each RTK’s kinase phosphorylates the kinase domain on the adjacent receptor, resulting in activation of full kinase activity in both kinases (ex - siblings fighting in the back seat)
kinases phosphorylate additional tyrosines on the cytoplasmic tails of the receptors that serve as binding sites for downstream signal proteins (scaffolding function)
signal transduction via ras by receptor tyrosine kinases
phospho-tyrosines are targets for SH2 domains in different proteins
binding of a protein to another often causes a change in each proteins’ conformation (SH3 domains on GRB2 bind to a proline-rich region on SOS, a GEF)
binding of SOS (GEF) to Ras (GTPase) changes the conf of Ras and cuases it to exchange GDP for GTP
Ras-GTP activates downstream signaling via several pathways including mitogen activated protein kinase pathway
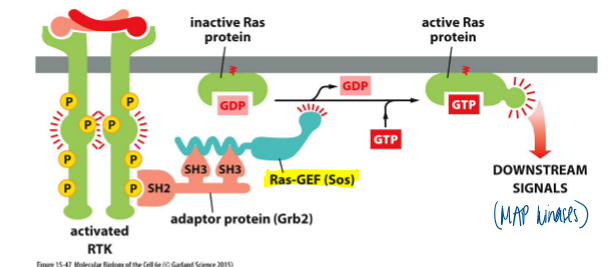
MAPK pathway
downstream mediators of ras action
activated Ras triggers a casacade of serine/threonine kinases
MAP3K = Raf
MAPK2 = Mek
MAPK = Erk
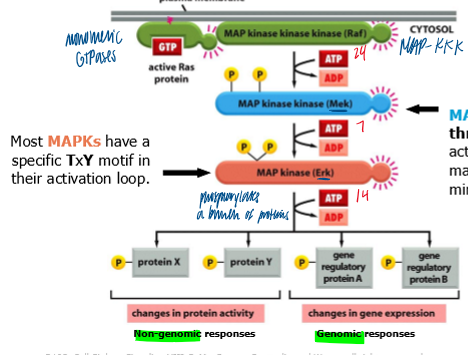
MAP2K
highly specialized
can phosphorylate both threonine and tyrosine simultaneously
only known target is MAPK; with so many kinases around, this is a useful safety mechanism to minimize accidential act of MAPK
MAPK phosphorylation
MAPK phosphorylates nuclear components that regulate cell cycle control
MAPK phosphorylation of MAP3K mediates negative feedback of signaling pathway
how do dominant mutant receptors can perturb the function of a WT receptor partner
a mutant receptor subunit can often dimerize with a normal receptor subunit and thereby perturb th efunction of its normal counterpart
some mutant-WT heterodimeric complexes cannot cross-phosphorylate (ex of dom neg inhibition)
other mutant-WT heterodimeric complexes constantly cross phosphorylate in absence of a ligand (dom act)
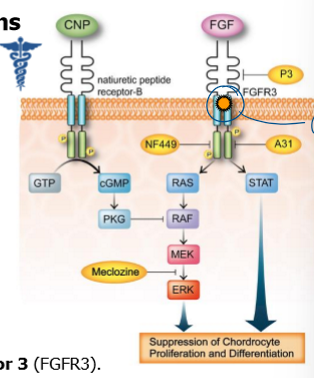
achondroplasia
short-limed dwarfism in humans
caused by G380R mutation within FGF receptor 3
mutation causes constitutive activation of receptor, some of which is ligand-independent
hyperactive FGFR3 inhibits chondrocyte proliferation → arrest in prolif
Which of the following best describes how an I-Smad (e.g. Smad7) participates in regulation of signaling via TGF-a family member receptors ?
A. The I-Smad co-associates with R-Smads and Co-Smads (e.g. Smad 4) and prevents activation of gene expression in the nucleus by interfering with binding to DNA.
B. The I-Smad binds with R-Smads only and prevents R-Smads from activating gene expression in the nucleus.
C. The I-Smad binds the the Type-I receptor and once phosphorylated interacts with and prevents R-Smads from being imported to the nucleus to mediate gene expression.
D. The I-Smad binds to the Type-I receptor but cannot be phosphorylated by it, thereby preventing interaction of R-Smads with the receptor.
D. The I-Smad binds to the Type-I receptor but cannot be phosphorylated by it, thereby preventing interaction of R-Smads with the receptor.
Based on your knowledge of cellular signaling you are asked to develop a strategy to boost the concentration of red blood cells (erythrocytes) in cancer patients. Which of the following combinations of changes in activity of targets might produce the best strategy?
A. Increase activity of SOCS proteins; decrease activity of Epo receptor; increase activity of SHP1 phosphatase.
B. Decrease activity of SOCS proteins; increase activity of Epo receptor; increase activity of SHP1 phosphatase.
C. Decrease activity of SOCS proteins; increase activity of Epo receptor; decrease activity of SHP1 phosphatase.
D. Decrease activity of SOCS proteins; decrease activity of Epo receptor; decrease activity of SHP1 phosphatase.
E. Increase activity of SOCS proteins; increase activity of Epo receptor; increase activity of SHP1 phosphatase.
C. Decrease activity of SOCS proteins; increase activity of Epo receptor; decrease activity of SHP1 phosphatase.
Ras GTPase was first discovered as being a gene that can change normal cells into cancer cells. Ras is normally activated by a receptor tyrosine kinase (RTK). However, in many cancers the Ras gene is mutated in a way that makes its signaling hyperactive. The hyperactive Ras conveys signals that drive cell division and tumor growth. You have learned that activated Ras can bind MAP3K, which in turn activates MAP2K, which activates MAPK that targets effector molecules leading to cell growth and division.
What types of mutations in a Ras gene could produce hyperactive signaling Ras ?
I - Missense mutations that stimulate Ras to bind Ras GTPase-activating proteins.
II - Missense mutations that decrease the ability of Ras to hydrolyze GTP.
III - Missense mutations that prevent Ras from binding to Ras-GEF.
II