Chapter 17a: Transcription
1/29
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
30 Terms
transcription
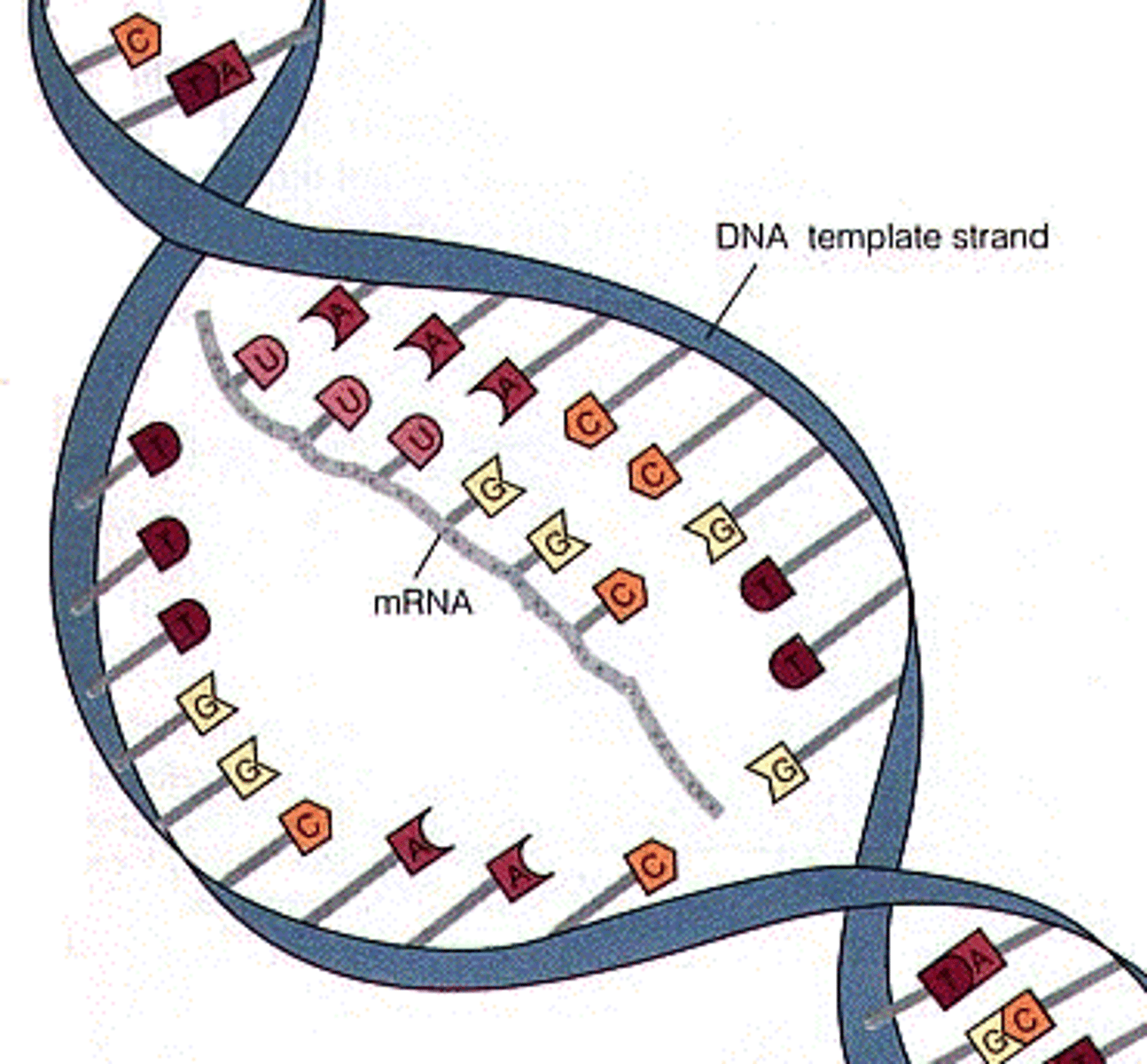
the synthesis of RNA from a DNA template
products of transcription
mRNA, tRNA, rRNA, and other small RNAs (used in expression of genes)
mRNA
will be translated for protein synthesis
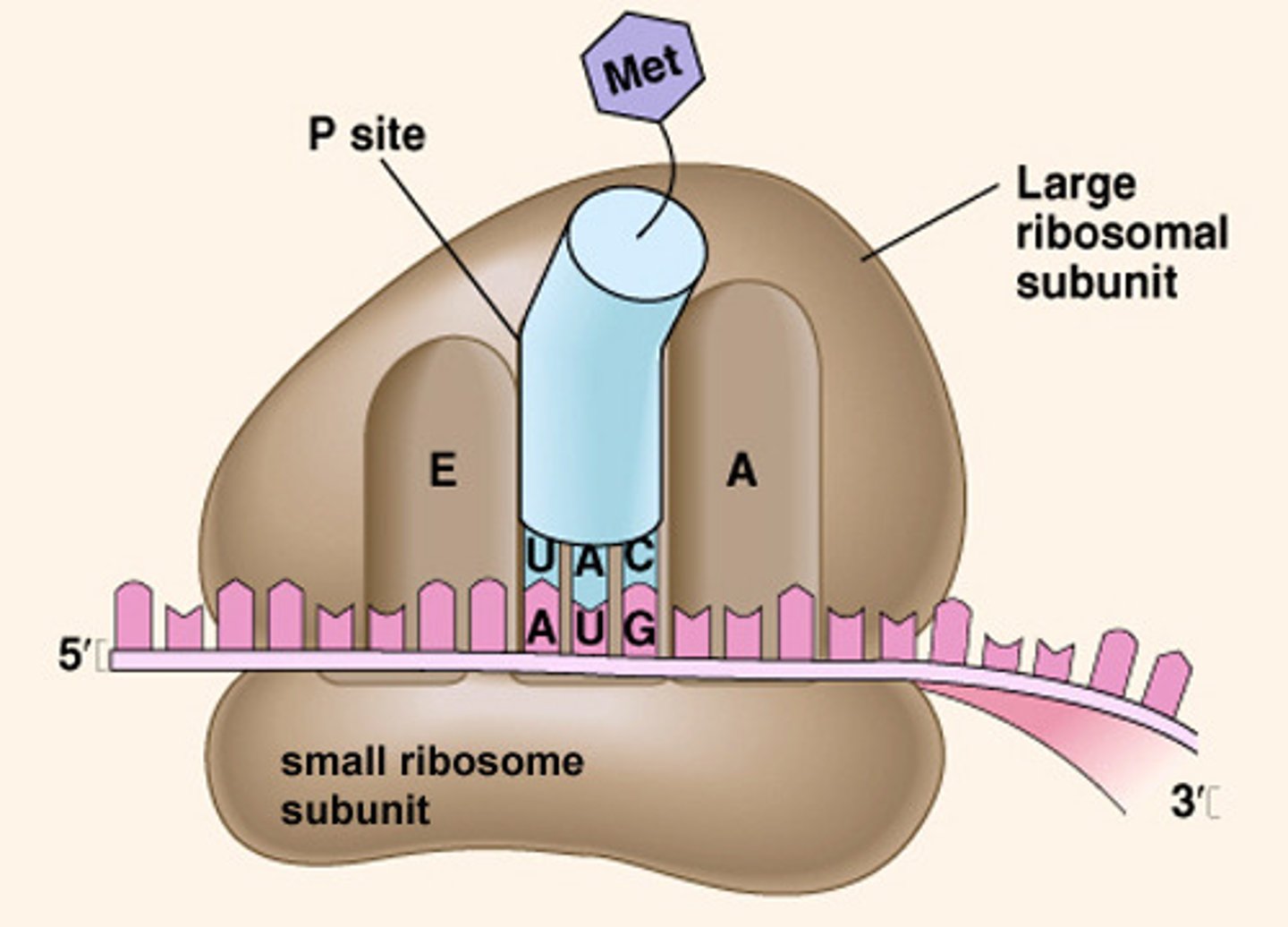
rRNA
structural and functional part of the ribosomes
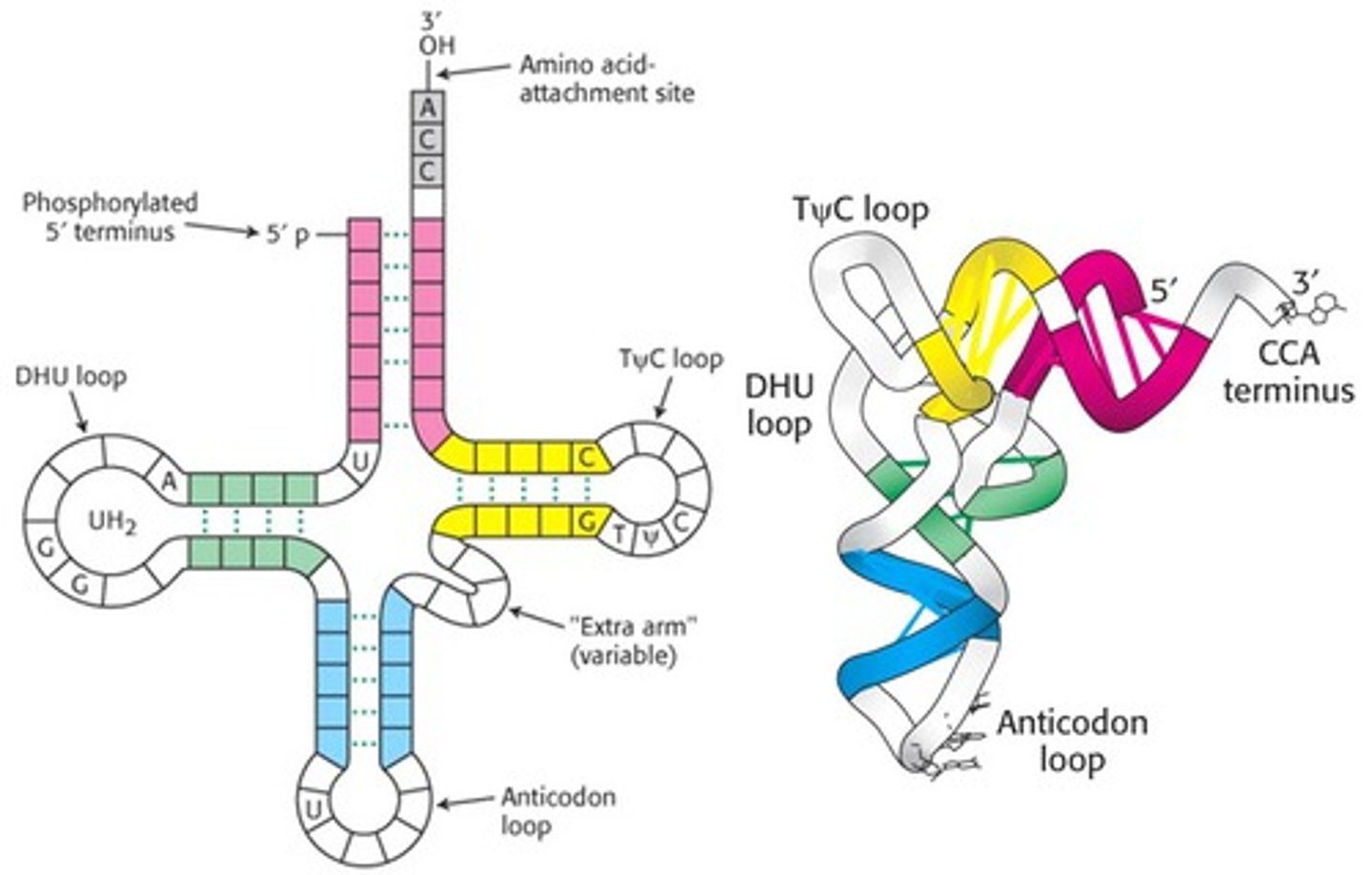
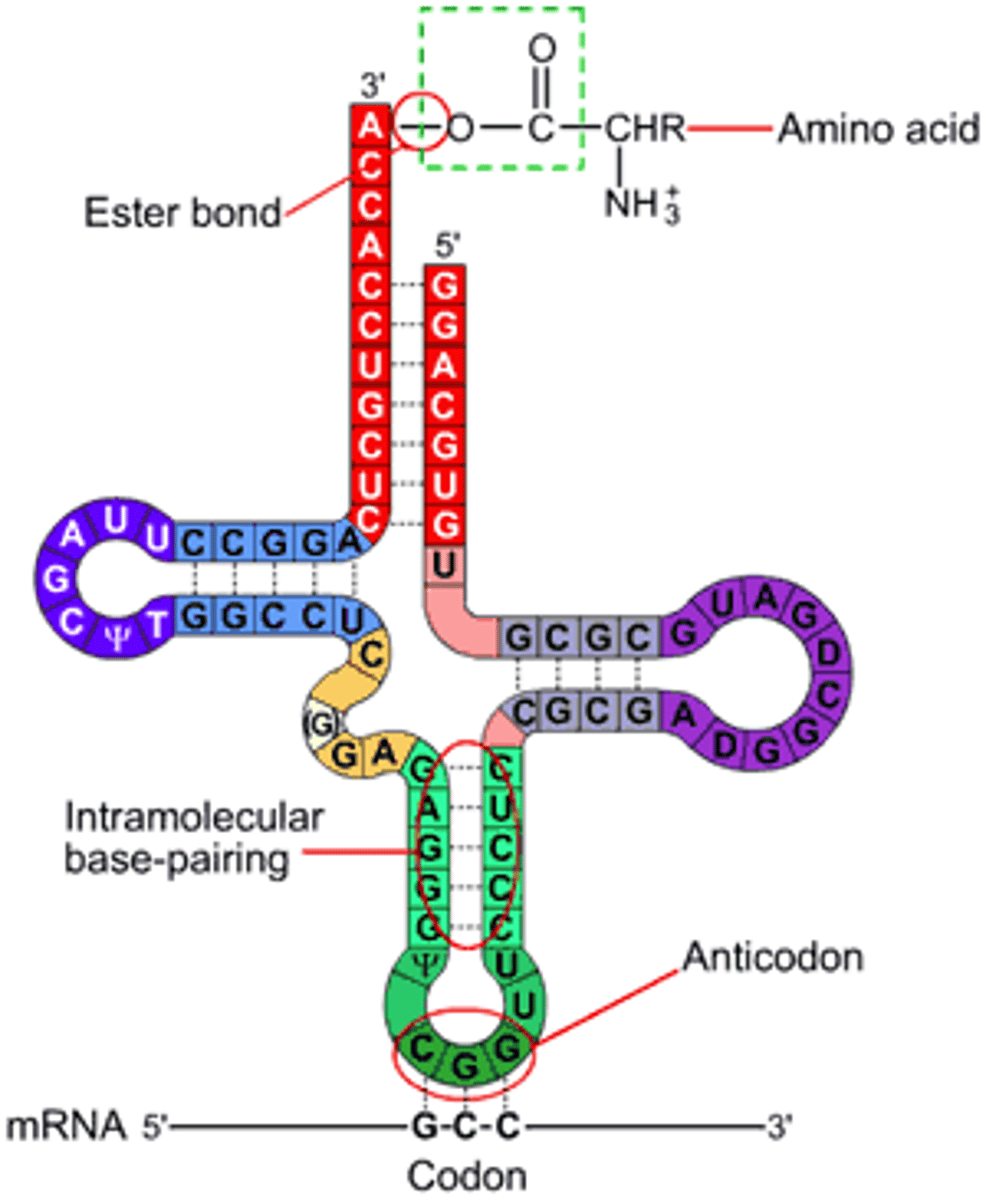
tRNA
anticodon binds to the mRNA codon in translation and attaches to an amino acid
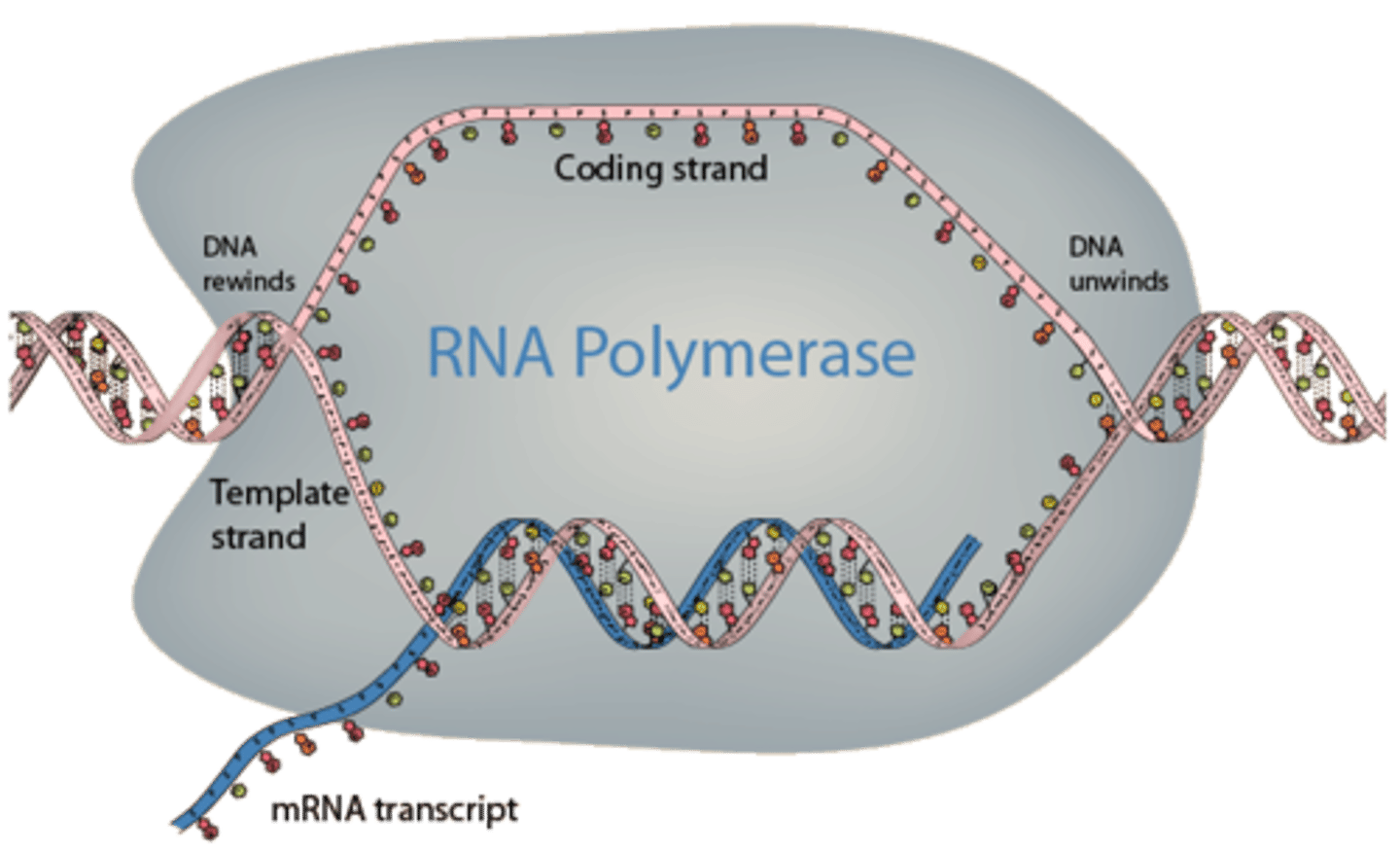
RNA polymerase
-perform template directed synthesis in the 5'->3' direction
-do not require a primer to begin transcription
-bacteria have one type of RNA polymerase
-eukaryotes have at least 3 distinct types: RNA polymerase I, II, and III
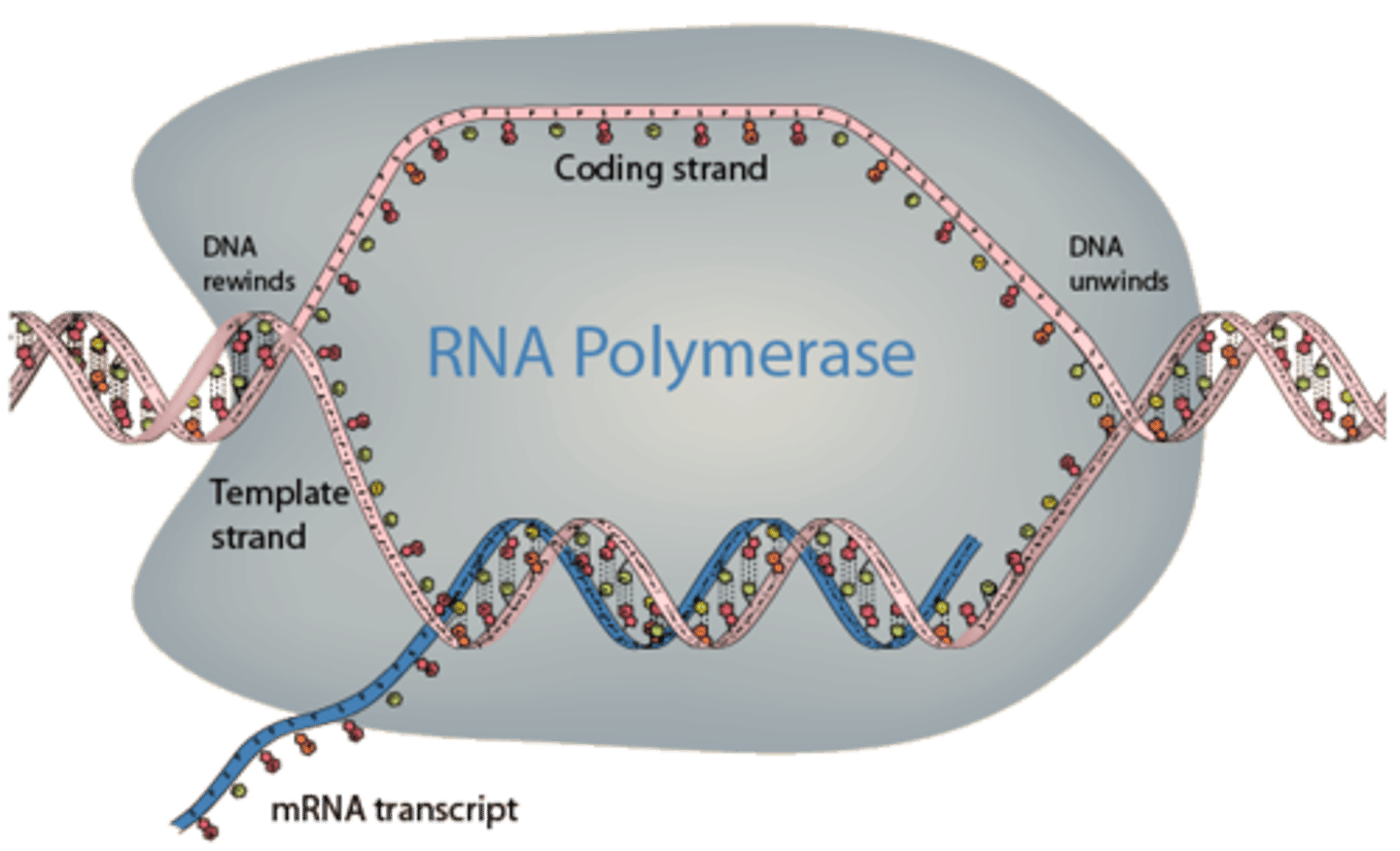
polymerization step 1: initiation
-where it starts
-promoter sequence needs to be transcribed into RNA
polymerization step 2: elongation
puts ribosomal subunits on and forms phosphodiester bond
polymerization step 3: termination
terminator sequence tells RNA polymerase to stop
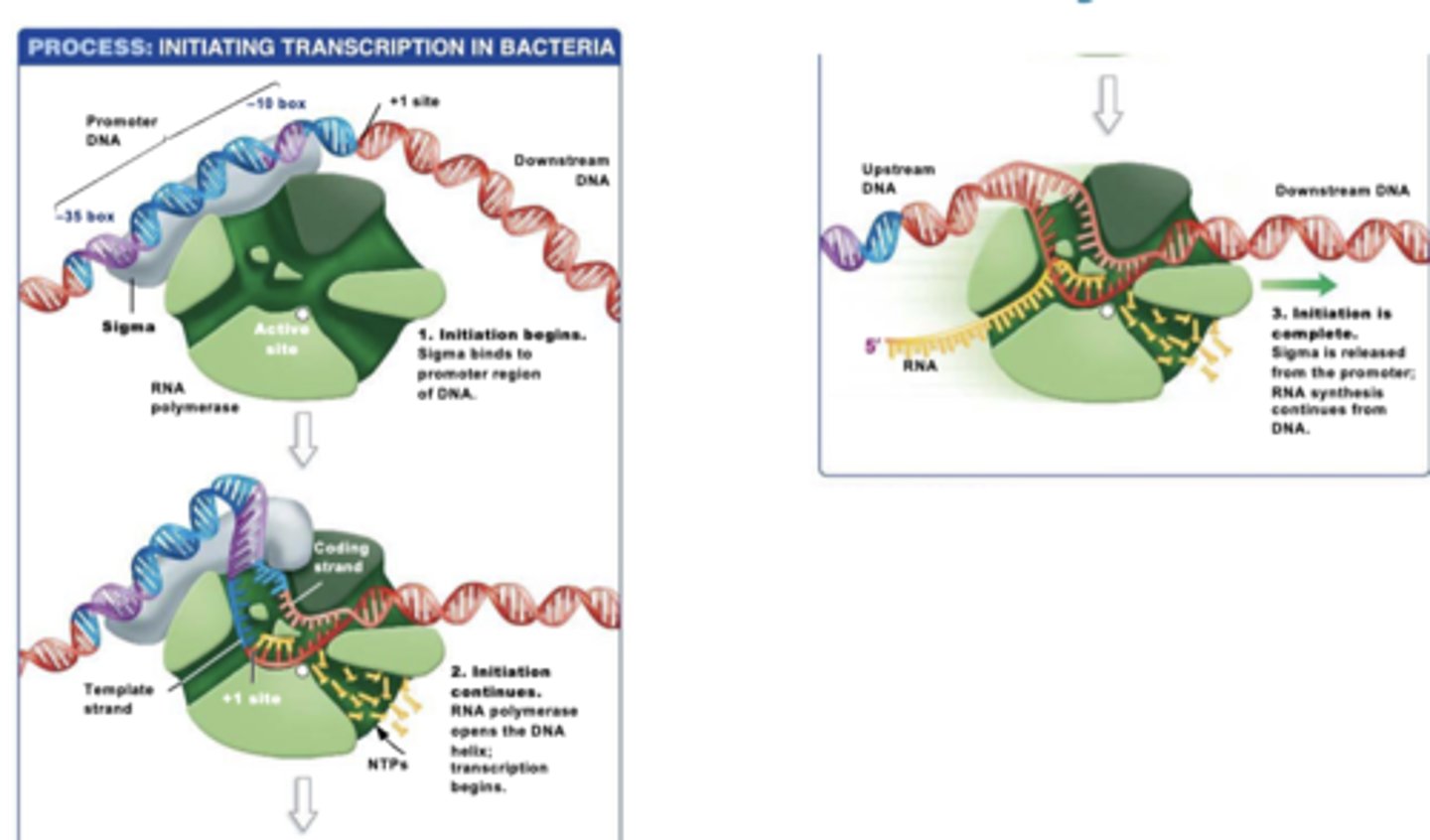
initiation steps in PROKARYOTES
1.)sigma factor binds to core enzyme to form the holoenzyme
2.) sigma directs holoenzyme to bind to promoters to promote the start of transcription
3.)diff sigma proteins bind to promoters with diff DNA base sequences (allows organism to activate certain genes in response to environment)
4.) RNA polymerase opens DNA helix and transcription begins
5.) initiation complete and sigma is released from promoter and RNA synthesis continues down RNA
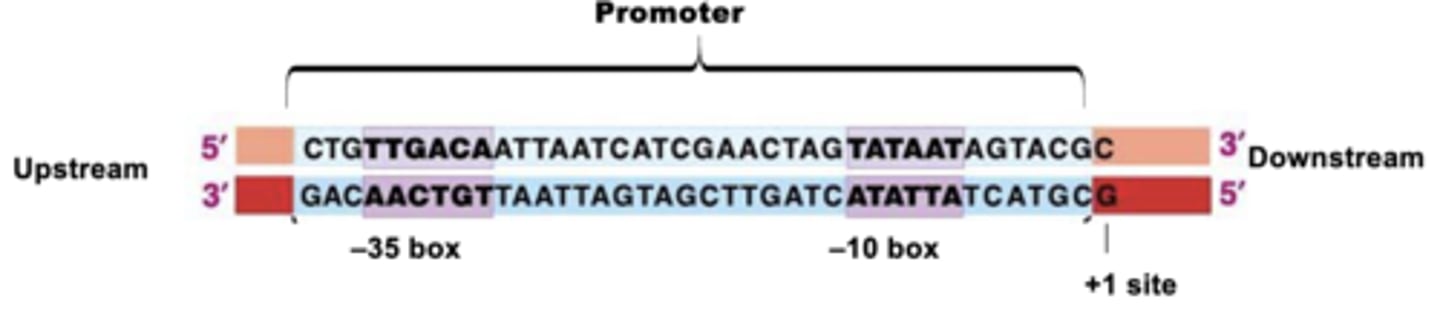
promoters in PROKARYOTES
stronger the promoter is the more product there will be
ribonucleoside triphosphate (NTP)
pairs with a complementary base on the DNA template strand, and RNA polymerization begins
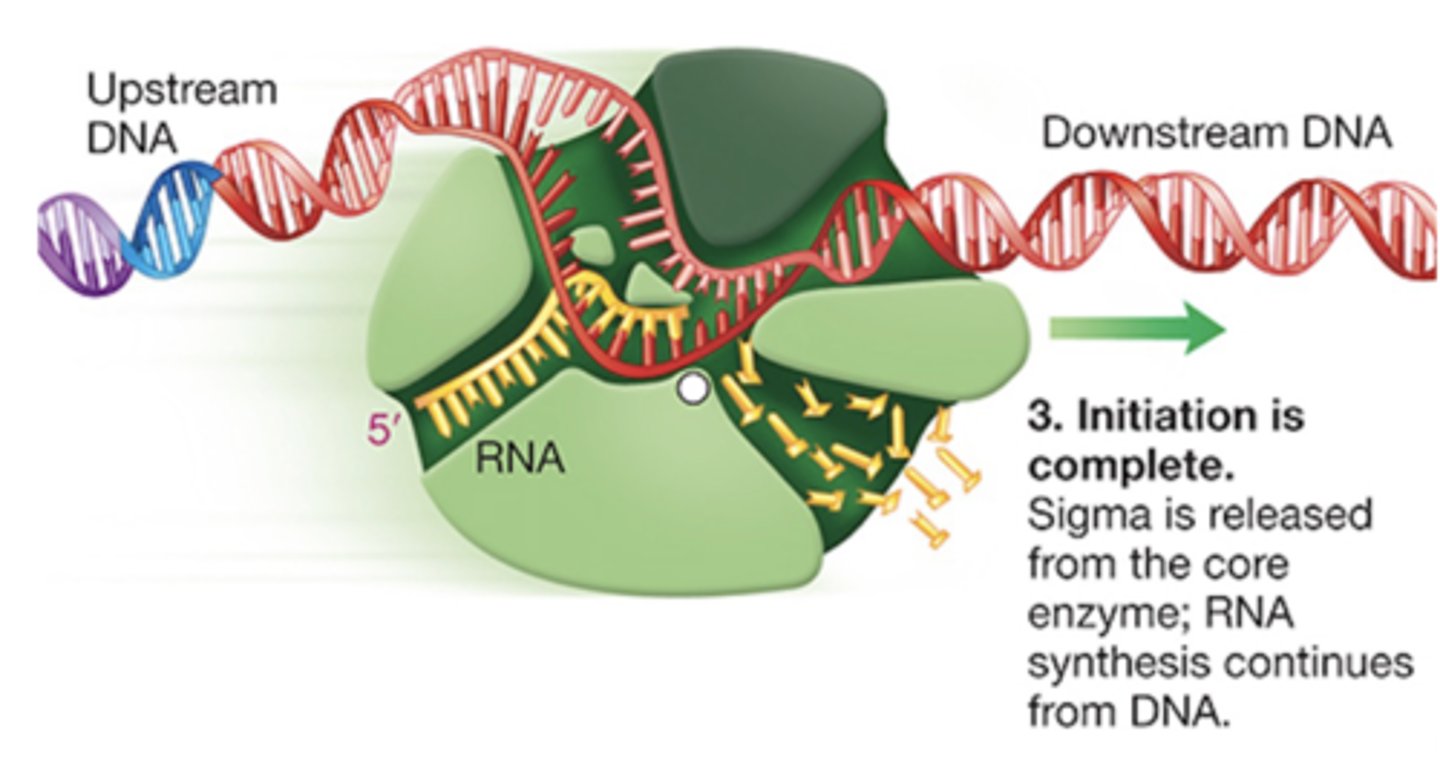
elongation in PROKARYOTES
1.) RNA polymerase moves along DNA template
2.) synthesies RNA in 5' -> 3' direction
3.) RNA polymerase has proofreading activity
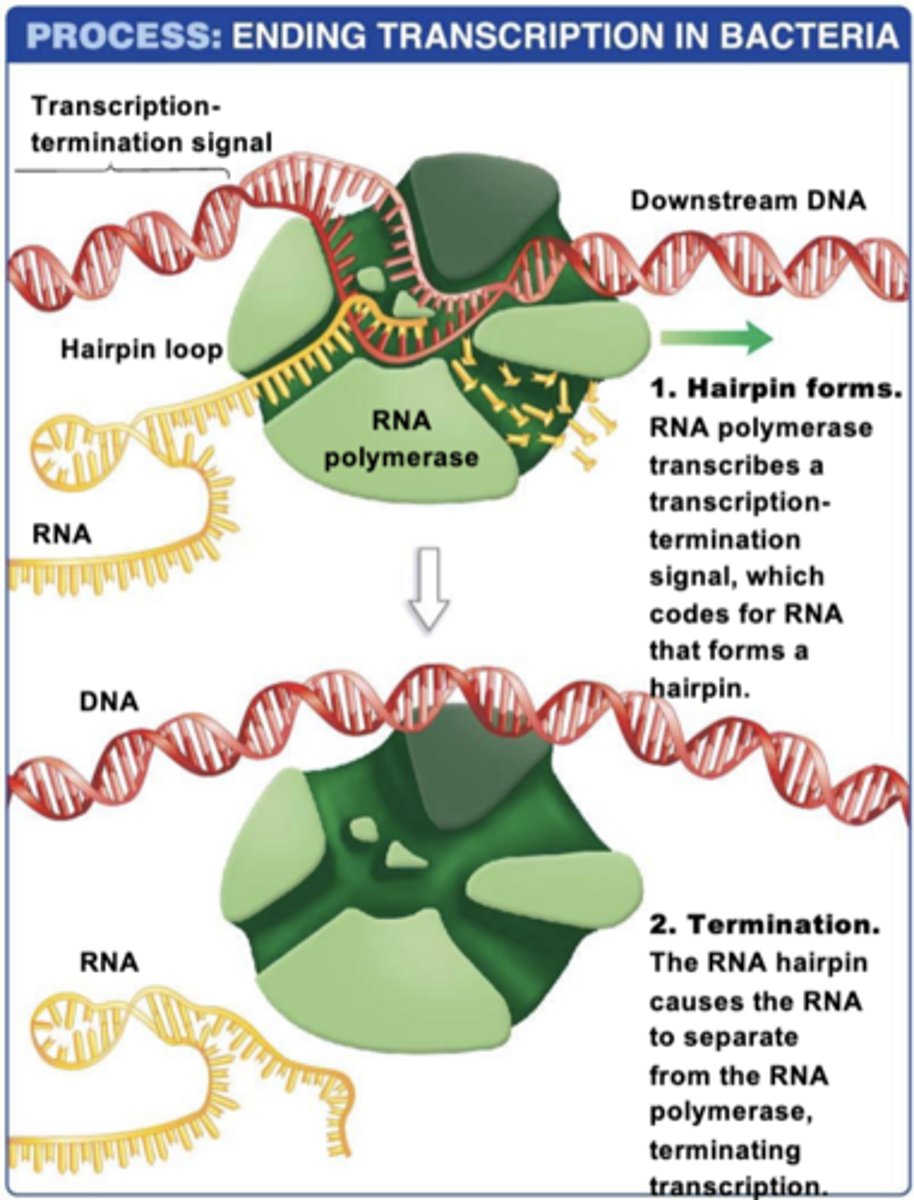
termination in PROKARYOTES
1.)transcribes a transcription-termination signal that forms a hairpin structure
2.)RNA hairpin causes the RNA to separate from the RNA polymerase, terminating transcription
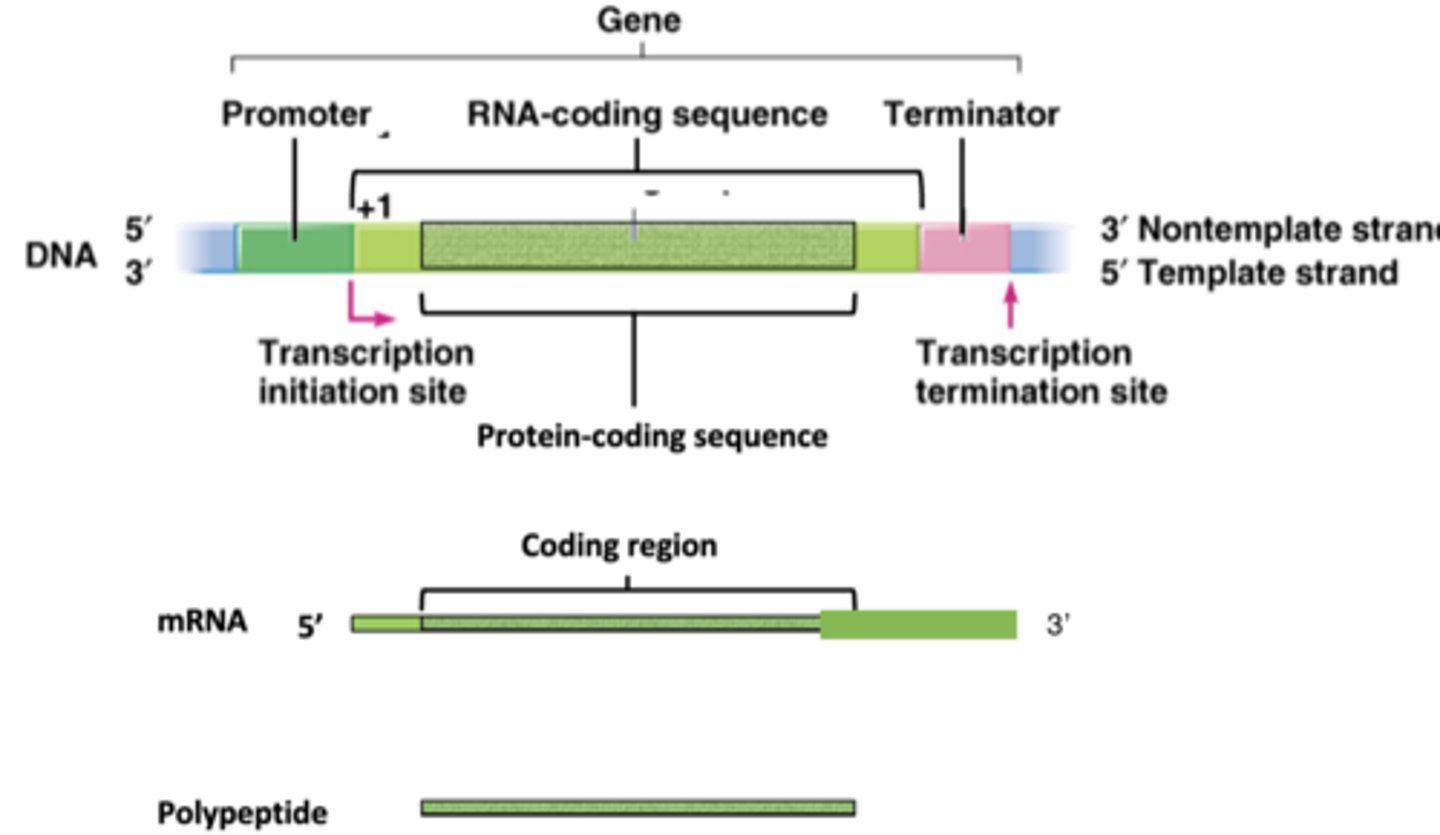
structure of prokaryotic gene
-promoter is not transcribed
- 1+ is first base to be incorporated in RNA chain
-terminator DOES get transcribed
transcription in eukaryotes
-more complex than in prokaryotes
-3 different RNA polymerases
RNA polymerases in EUKARYOTIC transcription
RNA polymerase I, RNA polymerase II, RNA polymerase III
RNA polymerase I
rRNAs and transcribes 3 largest RNAs
RNA polymerase II
transcribes protein-coding genes to pre-mRNAs and some snRNS
RNA polymerase III
transcribes tRNAs and everything not transcribed by RNA polymerase I and II

transcription of protein coding genes in EUKARYOTES
-transcribed by RNA polymerase II
-controlled by promoter and other regulatory elements
-promoters more diverse than in prokaryotes
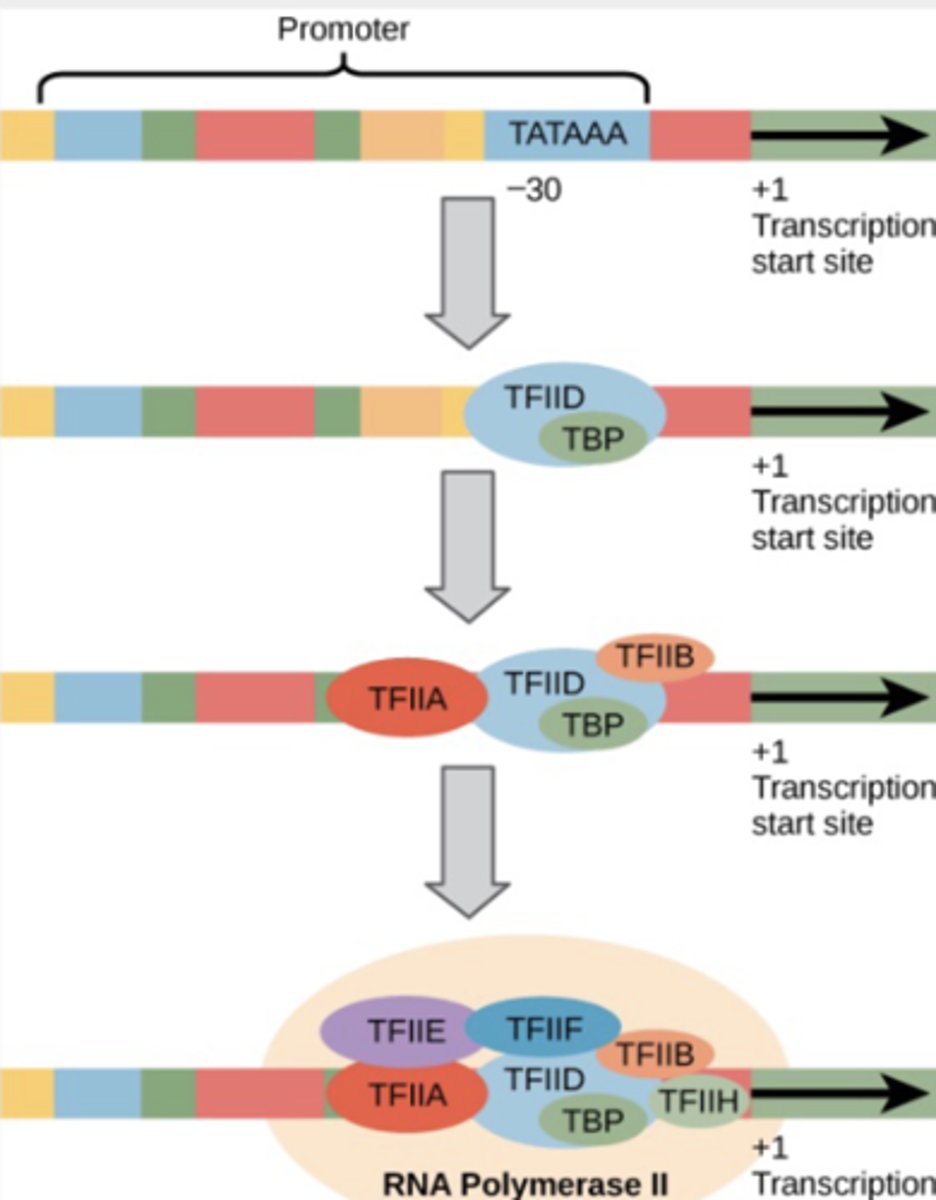
Initiation in EUKARYOTES
-requires core or basal promoter
-basal transcription factors (TF, TATA-binding proteins) bind to the core promoter
-RNA polymerase binds and starts transcribing
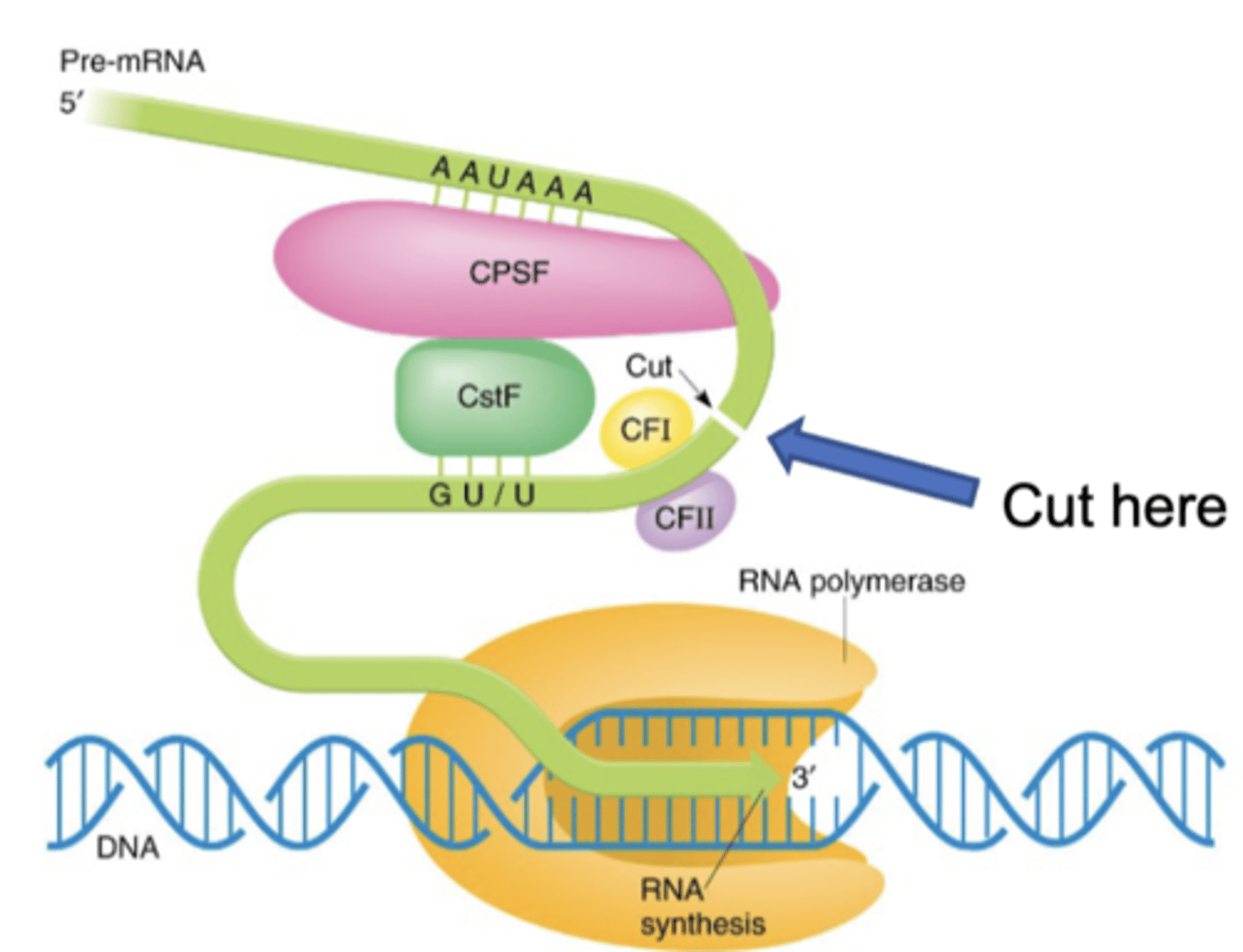
termination in EUKARYOTES
-special sequence in transcribed, the Poly-A signal
-enzymes cut the mRNA at the poly(A) signal
-eventually, RNA polymerase falls off the DNA
exons
expressed regions of eukaryotic genes that will be part of the final mRNA product
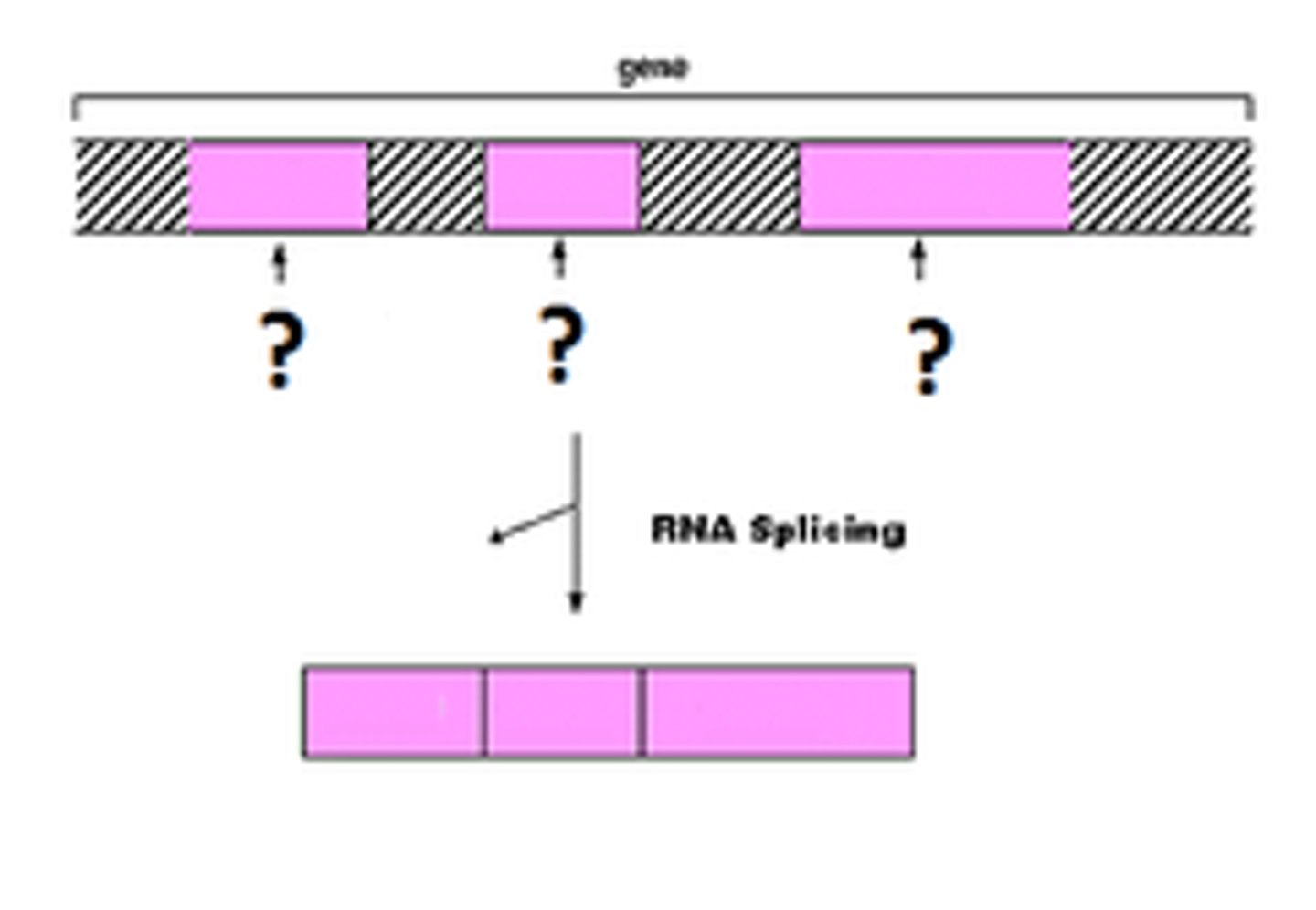
introns
intervening noncoding sequences that will not be in the final mRNA product
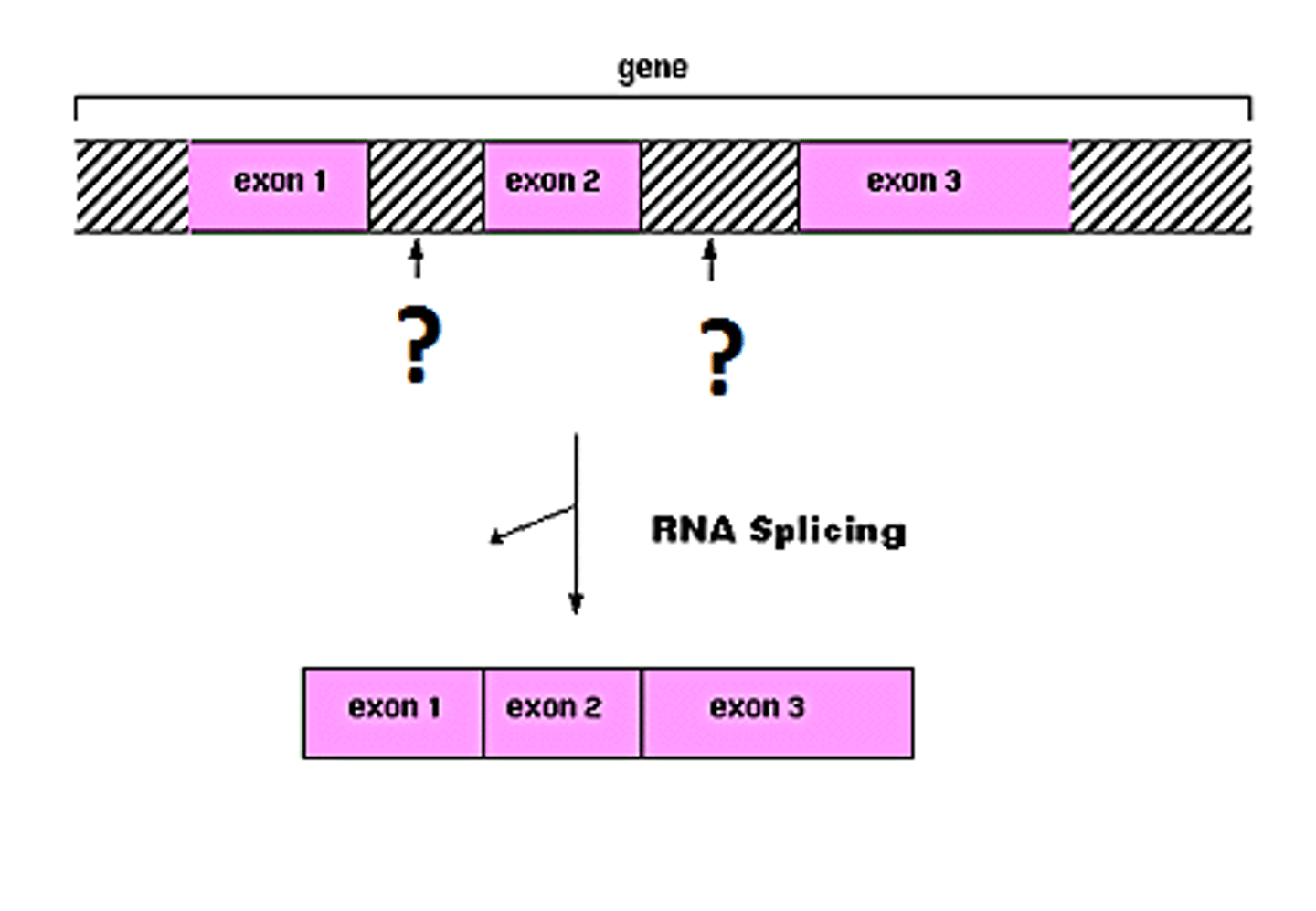
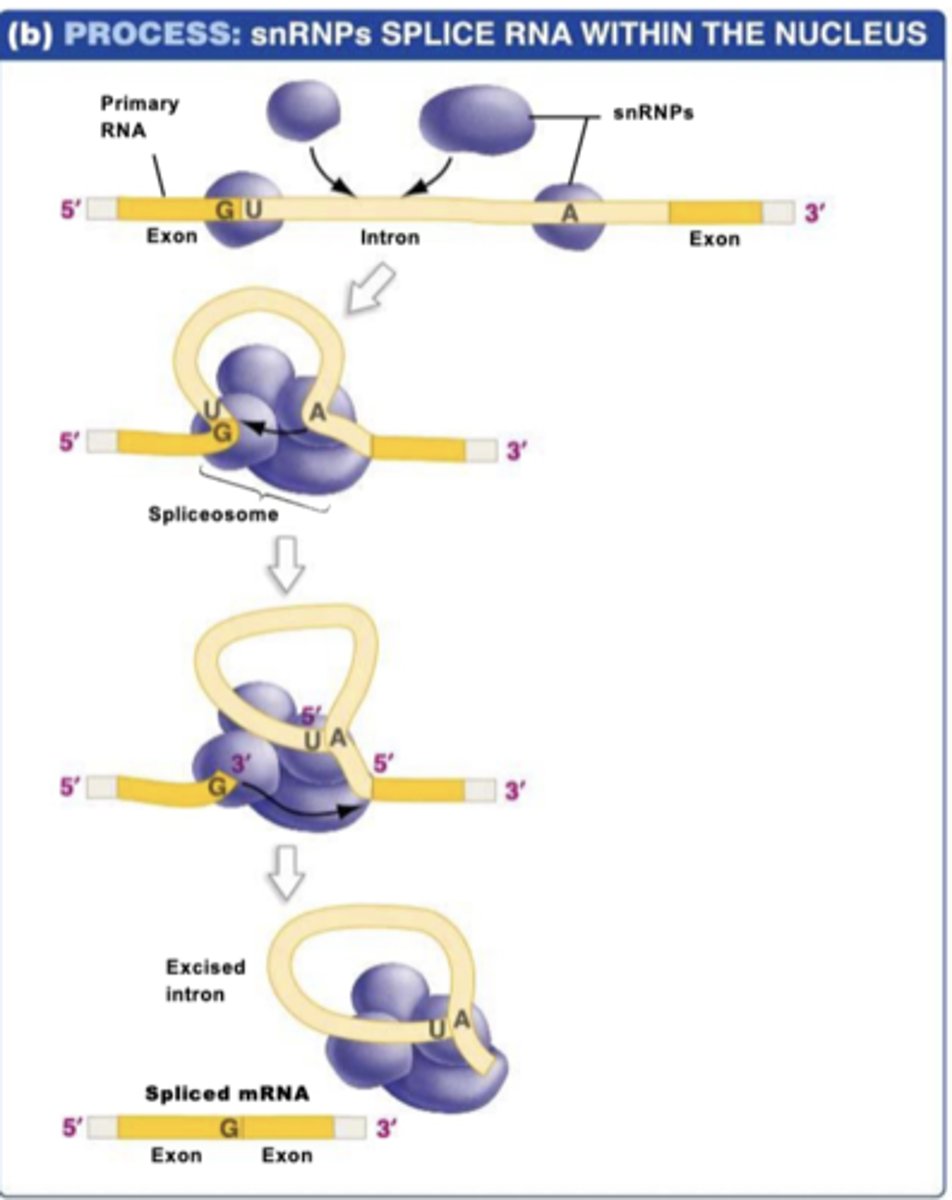
splicing
how introns are removed
1.) snRNPS bind to start and end of intron
2.) snRNPs assemble to form the spliceosome
3.) intron is cut, loop forms
4.) intron is released and exons are joined together
mRNA processing in prokaryotes
mRNAs are translated immediately, sometimes before transcription is even complete
mRNA processing in eukaryotes
-initial product of transcription is an immature primary transcript (pre-mRNA)
-primary transcripts must undergo RNA processing before they can be translated (3' Poly(A) tail and 5' GTP cap must be added post-transcription)
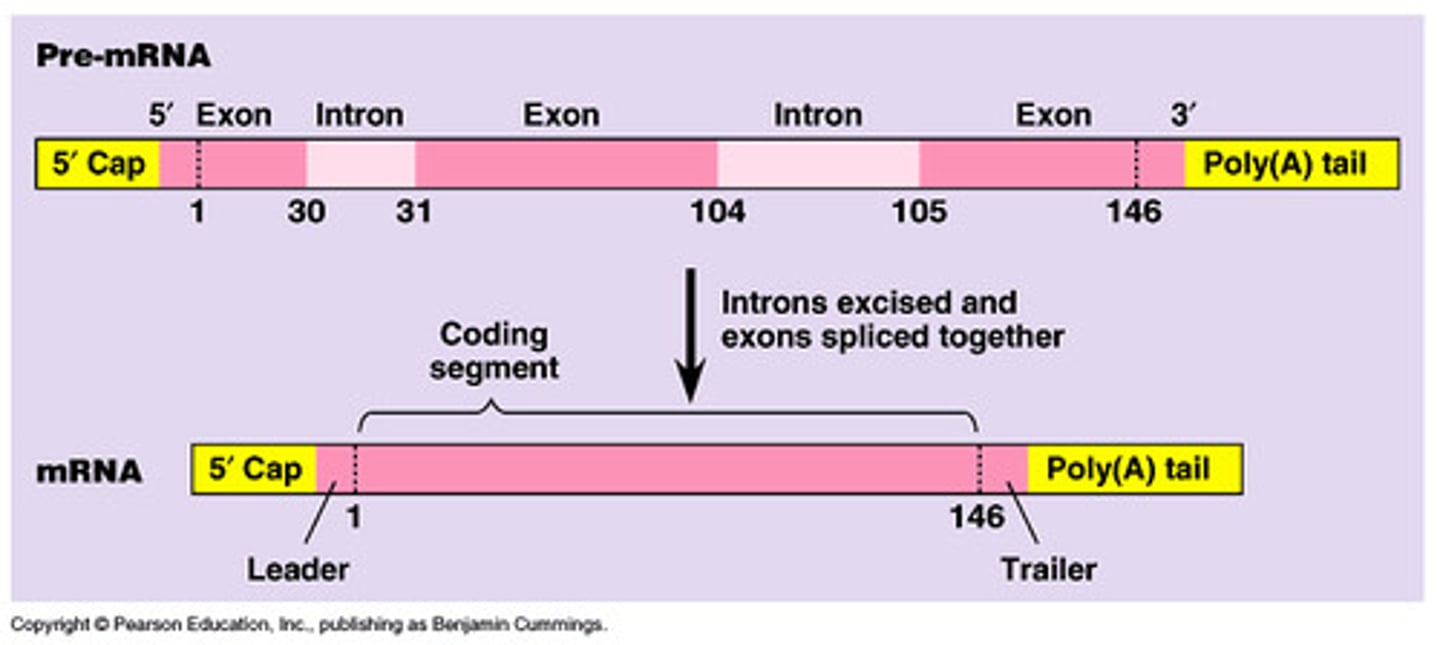
mRNA production in eukaryotes
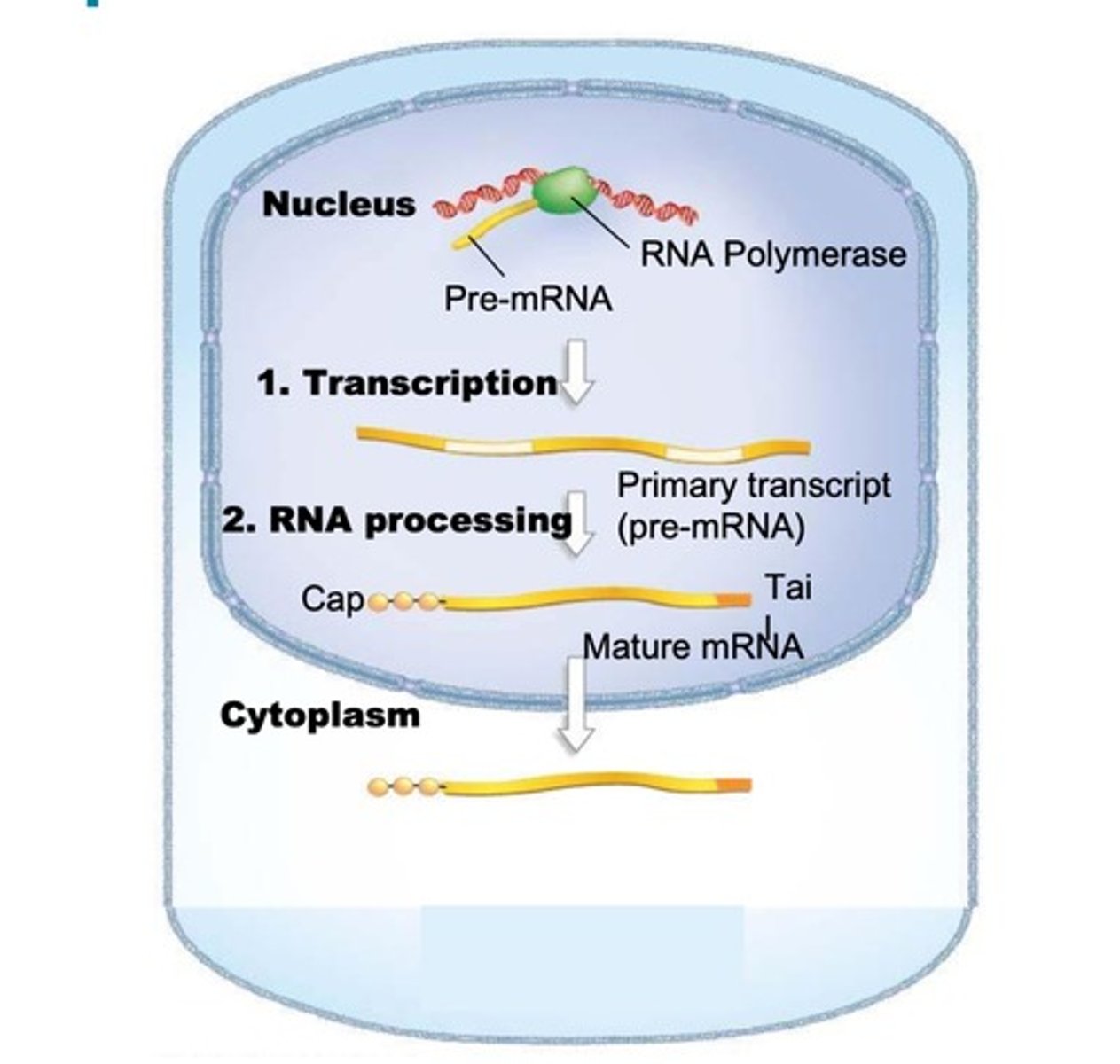
processing of tRNA
tertiary structure is an upside folded L with 5' CCA