Biology (B2 - Genes and health)
1/39
Earn XP
Description and Tags
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
40 Terms
Gas Exchange Surfaces

Cell respiration creates a constant demand to obtain oxygen for aerobic respiration and release carbon dioxide from aerobic respiration. Gas exchange is the transfer of gases (oxygen and carbon dioxide between the organism and the environment. Gas exchange takes place in the lungs across the cell membrane by simple diffusion. Animals need to maintain a concentration gradient across their exchange surfaces. Mammals achieve this through ventilation and blood circulation.
The amount of oxygen required for respiration is determined by:
•the amount of living cells in the organism (i.e. size) the rate at which the cells are respiring (metabolic rate and activity) so the amount of oxygen required depends (partly) on the volume of the organism.
The amount of oxygen that can enter the organism by diffusion is governed by the surface area of the organism. Thus the efficiency of gas exchange depends on the surface area: volume ratio.
Fick’s Law
rate of diffusion is proportional to:
Rate of diffusions proportional to: surface area * concentration gradient/diffusion distance (membrane thickness)
Cell membranes
Why do cells need membranes?
•Control entry and exit of substances
•Contain the cell contents and organelles; compartmentalize individual cell processes
•Communication with the external environment
•Site of chemical reactions
•Allows cell to change shape
•Transport and packaging within a cell
What are cell membranes made of?
Most of the cell membrane is impenetrable to water-soluble substances – it is composed of LIPIDS. Water-soluble substances and other molecules can pass into the cell through passageways composed of PROTEINS
Lipids
Fats and water do not mix. Water is a polar molecule (the oxygen end is slightly negative and the hydrogen end is slightly positive). Fats are non-polar and do not form hydrogen bonds with water. Fats are said to be hydrophobic and lie on the surface of the water to reduce the surface area in contact between the fat and the water.
Dietary lipids
•The fats that make up the cell membrane are different in structure from dietary fats. Dietary fats are composed of one glycerol backbone with three fatty acids attached to it. The fatty acids are non-polar and do not form hydrogen bonds with water so they are insoluble
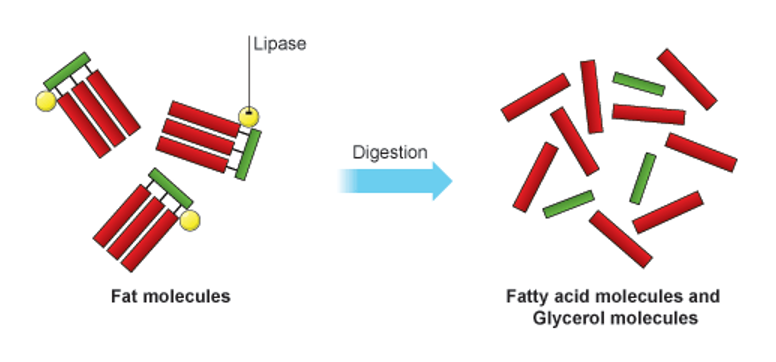
Lipids in the cell membrane
•These are phospholipids. They are composed of a glycerol backbone, two fatty acid chains, and a phosphate group. The fatty acid chains are non-polar. The phosphate group is polar.
Phospholipids
•The head (phosphate group) is hydrophilic. The tail (fatty acids) are hydrophobic
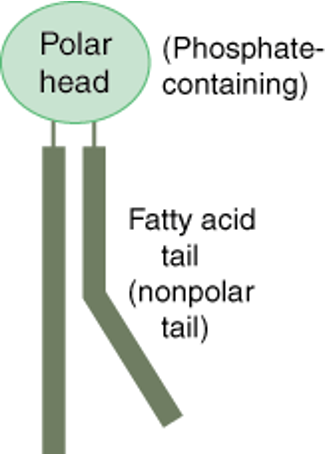
Fluid Mosaic Model
•The phospholipid bilayer is not a fixed structure
•Although the bilayer will always remain arranged in this configuration, the individual phospholipids are able to move, hence why it is also known as the fluid mosaic model
•The phospholipids rotate on their axis
•They can ‘swap' places with the phospholipid next to them
•They can occasionally ‘swap’ places with the phospholipid directly opposite them, though this is rare
Components of Cell membrane
•Glycolipids - maintain the stability of the cell membrane and facilitate cellular recognition, also aids cell to cell interaction/binding
•Glycoprotein –found on the cells surface, are protein molecules with a carbohydrate group attached These allow cells to communicate with one another. Individual groups of cells have their own glycoproteins, recognisable by the immune system.
•Cholesterol – regulates membrane fluidity
•Extrinsic / peripheral protein – cells recognition, cell communication, enzymes etc also act as receivers for incoming messages, such as hormones
•Intrinsic / integral protein – transport of substances

Transport across the cell membrane
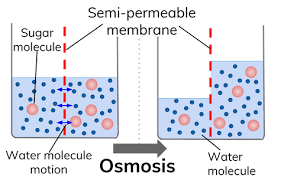
Osmosis:
Osmosis is the overall net movement of free water molecules from an area of high water potential to an area of low water potential across a partially permeable membrane. It is a passive process.
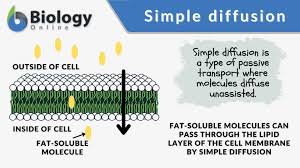
Simple diffusion:
The movement of molecules from an area of high concentration to an area of low concentration. The process is passive
Facilitated diffusion:
•The movement of molecules from an area of high concentration to an area of low concentration. Using carrier proteins that change shape to move larger molecules across. Channel proteins produce a polar hydrophilic channel for small charged molecules. The process is passive. EG Larger molecules = glucose and amino acids – carrier. Charged/polar molecules = ions K+ Na+ Cl- - channel
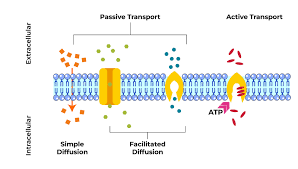
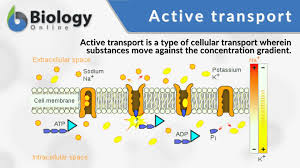
Active transport:
•The movement of molecules from an area of low concentration to an area of high concentration. Using carrier proteins. The active process requires ATP. Anything against the concentration gradient (except water). EG nitrates in the soil (plants), the last bit of glucose in digestion, Na+ and K+ in nerve impulses
Cytosis:
Endocytosis – into the cell, the cell membrane engulfs contents and ‘buds off’ to form a vesicle
→Phagocytosis, solids,
→Pinocytosis, liquids,
Exocytosis – out of the cell, the vesicle moves to the surface of the cell, The vesicle membrane fuses with the cell surface membrane, and Contents are released out of the cell
Uses ATP
Osmosis in RBCs
Animal cells/RBCs:
When placed in an isotonic solution, there is no net loss or gain of water and the cell’s water content remains steady.

When placed in a hypertonic solution, the cells lose water by osmosis and shrink. They are described as being CRENATED.

When placed in a hypotonic solution, the cells gain water by osmosis and burst. The bursting of red cells is called HAEMOLYSIS.
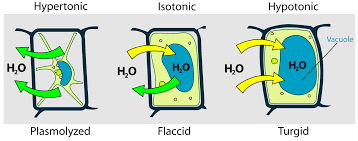
Osmosis (plant cells)
This fully turgid plant cell has been placed in a hypotonic solution. The net movement of water by osmosis is into the cell and the contents swell and presses against the cell wall.

This plant flaccid cell has become plasmolyzed as it has been placed in a hypertonic solution. The net movement of water by osmosis is out of the cell and the contents shrink.
Cystic fibrosis (treatments, symptoms and issues)
Symptoms:
Produce thicker mucus
Block the air passage- bronchi
Block tubes in the digestive system (pancreatic duct) & reproductive system (sperm duct)
Pancreatic cysts
Less energy
Salty sweat
Symptom | Explanation |
Lung infections Infertility Poor growth and malnutrition Emphysema Salty sweat Inability to carry out strenuous exercise | Pathogens are trapped in the thick mucus and not cleared to the back of the throat for swallowing to the stomach acid Mucus blocks the oviduct and sperm duct Pancreatic duct is blocked, meaning digestive enzymes are not released into the small intestine (may also damage pancreas) Coughing damages the alveoli Chloride ions are not reabsorbed from the sweat by the CFTR protein Gas exchange is reduced in the lungs, so inefficient uptake of oxygen into the blood, less aerobic respiration, less energy released |
Treatments:
Antibiotics & enzymes
Inhalants
Physiotherapy
CF (explanation)
CFTR is a gated chloride channel. The CFTR protein allows chloride ions to move into the mucus. This lowers the water potential of the mucus. So water moves into the mucus by osmosis down a water potential gradient. This keeps the mucus nice and watery. Cystic Fibrosis is a recessive genetic disease resulting in a non-functioning CFTR protein. A non-functioning CFTR protein means chloride ions are not transported into the mucus. So water does not move into the mucus. This causes thick sticky mucus which blocks airways, pancreatic duct, oviduct & sperm duct.
USING FICK’S LAW:
Diffusion is proportional to surface area.
Surface area reduces due to blocked alveoli/burst alveoli so less gas exchange.
Diffusion is proportional to concentration gradient.
Concentration gradient is less steep due to less ventilation occurring because of blocked airways/bronchi- diffusion happens more slowly.
Diffusion is inversely proportional to membrane thickness.
Membrane thickness increases due to thicker mucus covering the alveoli so has a greater distance to diffuse across.
Polymers. monomers and proteins
Proteins act as enzymes(transporting substances to cells), hormones, antibodies (hair and outer layer of skin), and structural proteins ((responsible for the formation of bones/teeth).
Definitions:
Polymer:- large molecules consisting of large numbers of repeating units connected by covalent bonds
Monomer:- a small molecule that may become chemically bonded to other monomers to form a polymer
→ Amino acids are monomers, and polypeptides are polymers.
Peptides
A protein may consist of one or more polypeptide chains.
Dipeptide: 2 amino acids
Tripeptides: 3 amino acids
Oligopeptide: A short chain of a few amino acids
Polypeptide: A chain of many amino acids
Amino acids
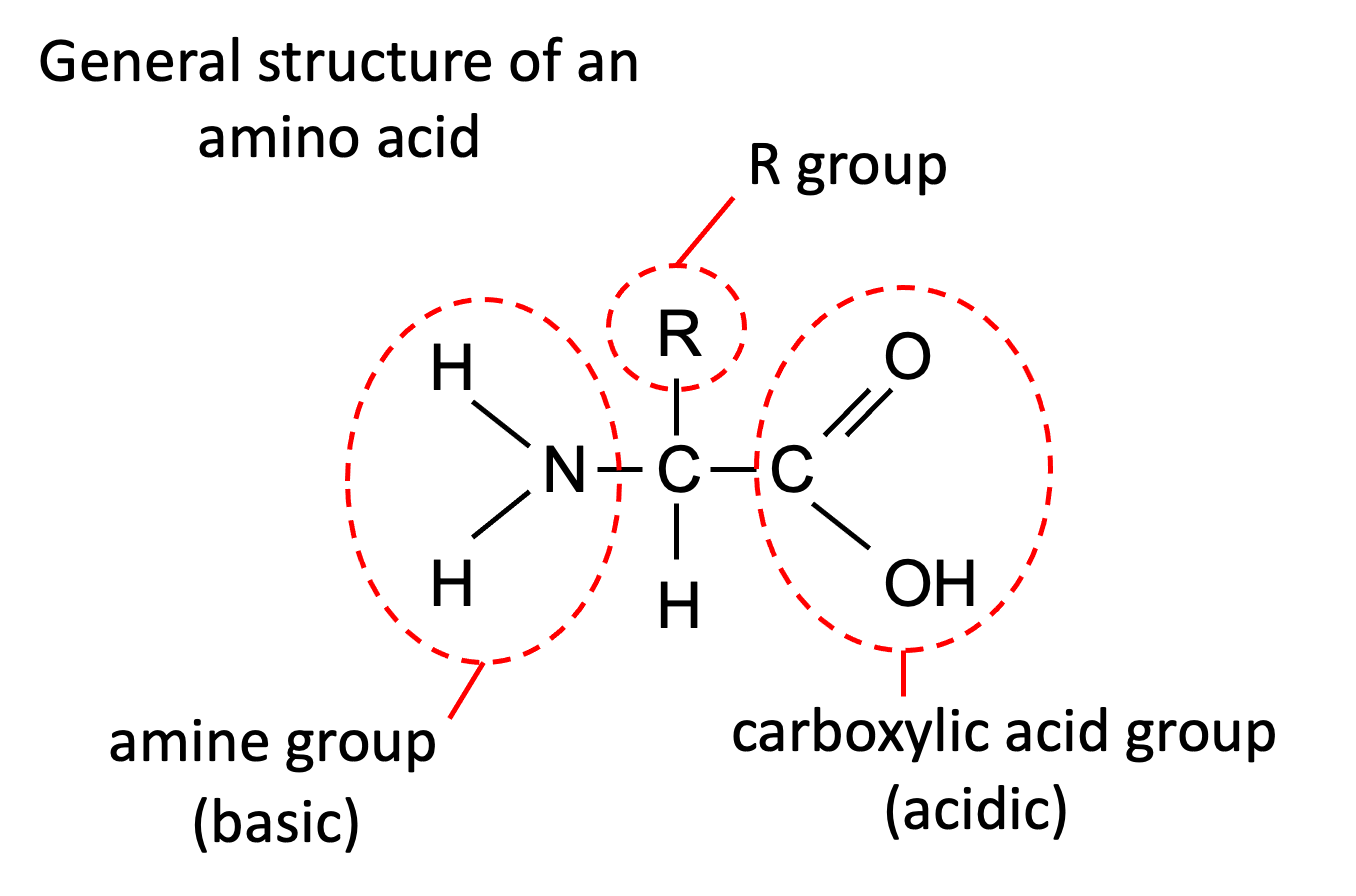
Amino acids consist of
Amine group: NH2
Carboxylic acid: COOH
Hydrogen atom: H
R group
The Amine group needs to be opposite the carboxylic group.
The carboxylic group is acidic - can donate a hydrogen
Amine group is basic - can accept hydrogen
Zwitterion: ions that have both permanent positive and negative charge, though the compound is overall neutral.
Reactions
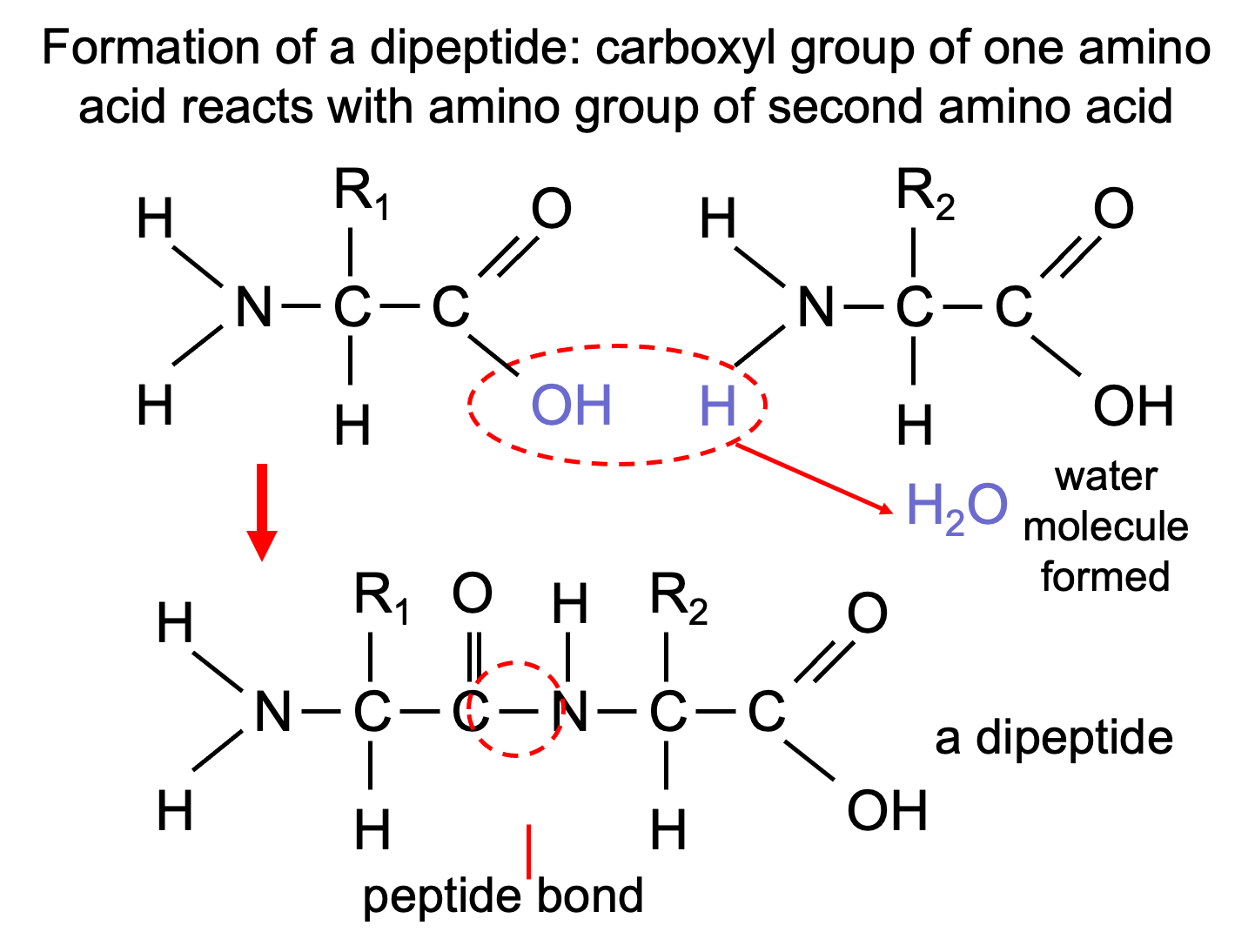
This is a condensation reaction.
2 amino acids → dipeptide + water
Hydrolysis is the reverse reaction.
dipeptide + water → 2 amino acids
Protein structure
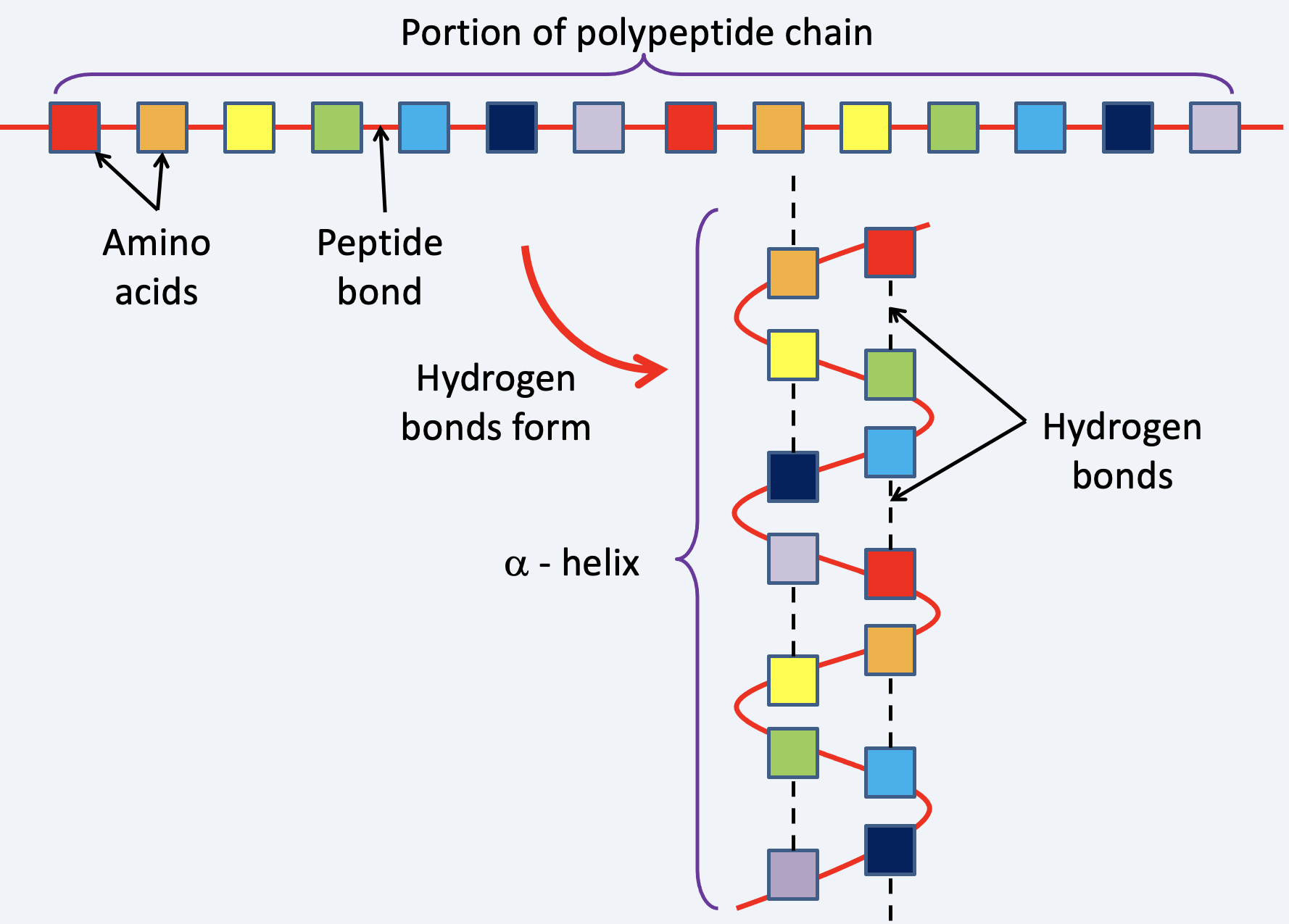
Primary structure: the sequence of amino acids in a polypeptide chain joined by peptide bonds.
Secondary structure: Primarily α-helices and β-pleated sheets. Secondary structures form as a result of hydrogen bonding between different amino acids in the chain.
- hydrogen bonds can form: the –CO (carboxyl group) of one amino acid and the –NH (amine group) of another amino acid. The –CO of one amino acid and the –OH (hydroxyl group) of another amino acid.
Tertiary structure:the secondary structures fold up to form a very precise three-dimensional structure.
Quarternary structure: The interaction of R groups withing multiple polypeptide chains
Globular vs Fibrous proteins
Globular:
Compact
Hydrophilic groups on the outside, hydrophobic on the inside
Soluble
Compact, highly folded with complex
tertiary/quaternary structures.
Soluble, forms colloids in water. They are useful for hormones, antibodies, etc.
Eg. Haemoglobin
Fibrous:
Long, structural proteins/Long polypeptide chains, folded in parallel
Insoluble in water,
providing support and strength
Very little tertiary/quaternary structure aside from cross-linkages for strength.
This makes them insoluble, and useful for providing structure.
Eg collagen
Haemoglobin vs Collagen
Haemoglobin. Water-soluble, with a complex quaternary structure. Contains four haem groups that oxygen can bind to. It is therefore used to carry oxygen in the blood to respiring tissues.
Collagen. Hydrogen and covalent bonds make it very strong. Polypeptide chains form a triple helix which creates fibres. This makes it useful in bones, cartilage and other connective tissue.
Enzymes
Biological catalysts – speed up reactions in living things, without getting used up themselves
Enzymes are proteins with a specific three-dimensional shape
Active site is complementary to the substrate
They are highly specific to their substrate due to their active site's shape.
This is the lock & key hypothesis
It means every enzyme works on a specific substrate
Enzyme models
• Lock-and-key hypothesis: enzyme active site fits exactly with the substrate.
• Induced fit model: enzyme active site molds itself around the substrate.
lexibility:
Lock and Key: Active site is rigid.
Induced Fit: Active site is flexible and molds to the substrate.
Specificity:
Lock and Key: Highly specific, substrate must perfectly match the active site shape.
Induced Fit: Less rigid specificity, as the enzyme adjusts to fit the substrate.
Enzyme Dynamics:
Lock and Key: No change in enzyme shape.
Induced Fit: Enzyme changes shape upon substrate binding for a better interaction.
The induced fit model is now more widely accepted due to its explanation of greater flexibility and efficiency in enzyme-substrate interactions.
The conformational change puts strain on the bonds within the substrate(s) molecule, lowering the activation energy, i.e. it does not take much more energy to break the bonds when they are under strain. The lowering of the activation energy exponentially increases the rate of reaction or even enables reactions to take place that otherwise would not.
Enzyme graphs
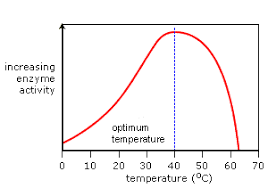
Temperature:
Increase temperature increases kinetic energy so more enzyme substrate collisions & complexes form increasing reaction rate
Above the optimum temperature, enzymes denature, so rate of reaction falls
At low temperatures there is low kinetic energy, so very few enzyme substrate collisions . . . reaction rate is low
Increased temperature increases kinetic energy and enzyme substrate collisions, so reaction rate increases
Optimum temperature when reaction rate is highest
Temperature is too high, atoms within the enzyme vibrate, breaking the bonds.
The active site changes shape, so the substrate no longer fits.
The enzyme is denatured
Optimum temperature is different for every enzyme
e.g. Arctic cod’s enzymes work best at 5°C
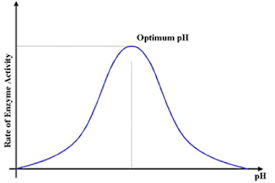
pH:
Above and below the optimum pH the enzymes denature and the reaction rate falls
Enzymes have an optimum pH.
If the pH changes too much from the optimum, the bonds within the enzyme break
This is due to changes in H+ and OH- molecules which interfere with the bonds holding the tertiary structure in place.
The active site changes shape and the enzyme will be denatured
The substrate no longer fits in the active site
Optimum pH is different for every enzyme
e.g. enzymes in the stomach work best at acid pH
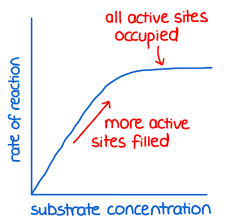
Substrate concentration:
At low substrate concentrations increasing the substrate concentration increases reaction rate due to more enzyme substrate collisions and more complexes forming
The graph levels off because the enzyme concentration becomes the limiting factor and all the active sites are full
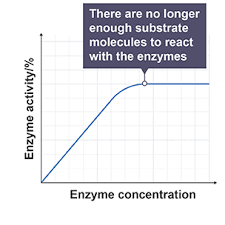
Enzyme concentration:
At low enzyme concentrations increasing the enzyme concentration increases reaction rate as more enzyme substrate collisions occur, so more complexes form
The graph levels out because the substrate level becomes the limiting factor not all active sites are full
Intracellular vs extracellular enzymes
Intracellular enzymes:
Operate within cells to catalyze reactions in the cytoplasm or organelles.
Examples:
Catalase: Breaks down hydrogen peroxide into water and oxygen within peroxisomes.
DNA polymerase: Replicates DNA inside the nucleus.
Extracellular enzymes:
Secreted by cells to catalyze reactions outside the cell.
Examples
Amylase: Secreted by salivary glands to break down starch into maltose in the mouth.
Trypsin: Produced by the pancreas, breaks down proteins in the small intestine.
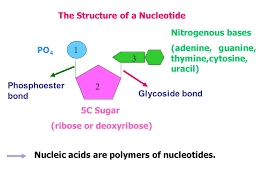
Nucleotides
Nucleic acids:
Polymer
Two types- DNA and RNA
Made up of repeating units unit called nucleotides (monomer).
NUCLEOTIDES:
Pentose sugar
Phosphoric acid
Nitrogenous base
SUGAR:
5 carbon sugar:
DNA- deoxyribose
RNA- ribose
PHOSPHATE GROUP:
As part of the nucleotide it joins to 5th carbon of the pentose sugar.
NITROGENOUS BASE:
Purines
Double ringed structure
Pyrimidines
Single ringed structure
Complimentary base pairs:
Adenine & Thymine (& Uracil)
Cytosine & Guanine
Structure of DNA
The double-stranded structure of DNA was determined in 1953 by the Watson and Crick.
X-ray diffraction studies by Rosalind Franklin strongly suggested that DNA was a helical structure.
The Austrian chemist Erwin Chargraff had earlier showed that DNA contained a 1:1 ratio of pyrimidine:purine bases.
There is around 2 m of DNA in a cell, so to fit it needs to be tightly coiled and folded.
Eukaryotic DNA is associated with proteins called histones. Together, these form chromatin – the substance from which chromosomes are made.
In prokaryotic cells, DNA is loose in the cytoplasm – there are no histones or chromosomes.
Structure of eukaryotic chromosomes:
DNA replication
Unzipping
DNA helicase unwinds the double helix.
H bonds break.
Bases become exposed
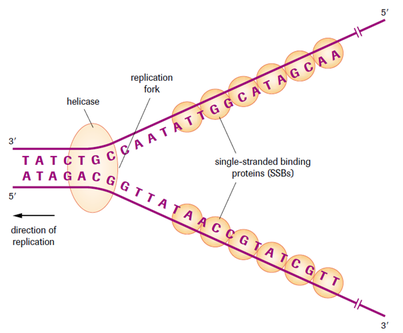
Free nucleotides bind
DNA strands act as templates.
Free nucleotides H bond to their complementary base pair.
DNA polymerase causes the sugar-phosphate backbone to form between the nucleotides by a condensation reaction (Phosphodiester bonds).
Rewinding
DNA ligase bonds the segments together.
DNA polymerase proof-reads the DNA.
The two strands (half new and half original DNA) rewind: Semi-conservative replication.
Steps of replication
Replication starts at a specific sequence on the DNA molecule.
DNA helicase unwinds and unzips DNA, breaking the hydrogen bonds that join the base pairs, and forming two separate strands.
The new DNA is built up from the four nucleotides (A, C, G and T) that are abundant in the nucleoplasm.
These nucleotides attach themselves to the bases on the old strands by complementary base pairing. Where there is a T base, only an A nucleotide will bind, and so on.
DNA polymerase joins the new nucleotides to each other by strong covalent bonds, forming the phosphate-sugar backbone.
A winding enzyme winds the new strands up to form double helices.
The two new molecules are identical to the old molecule.
Meselsohn and Stahl
Firstly…E.coli bacteria were grown on ‘known’ forms of nitrogen
These were the control tests
DNA was then extracted from the bacteria of each dish to be examined
The DNA was extracted
Then spun in a high speed centrifuge
Meselsohn & Stahl then examined the contents of the control tubes
Parent generation N15
A culture of E.coli bacteria was grown where the available nitrogen contained ONLY the ‘heavy’ 15N
All the bacteria became entirely “heavy”
A sample of DNA was extracted & spun
First generation N14
The bacteria were then transferred back to a medium ONLY providing ‘light nitrogen’ (14N)
Grown for one generation, then a DNA sample was extracted and spun
Second generation in N14
The bacteria were grown for one more generation in N14
A DNA sample was extracted and spun
RNA vs DNA
RNA and DNA
RNA:
Like DNA, RNA is assembled as a chain of nucleotides, but unlike DNA it is single-stranded rather than double-stranded.
Uracil (U) replaces Thymine (T)
Cellular organisms use messenger RNA(mRNA) to convey genetic information (using the letters G, A, U, and C to denote the bases guanine, adenine, uracil and cytosine) that directs synthesis of specific proteins.
Many viruses encode their genetic information using an RNA genome
Feature | DNA | mRNA | tRNA |
Double/Single | Double | Single | Single |
Size | Largest | Variable | Smaller |
Shape | Double helix | Single helix | Clover shape |
Sugar | Deoxyribose | Ribose | Ribose |
Bases | A, G, T, C | A, G, U, C | A, G, U, C |
Quantity in cells | Constant | Varies | Varies |
Stability | Very stable | Unstable | More stable than mRNA, less than DNA |
Transcription
Transcription takes place in the nucleus where the DNA double helix unwinds by DNA helicase and the hydrogen bonds break.
Once the strands are separated, the sequence on the template strand is used to produce a mRNA molecule.
mRNA is produced by free nucleotides in the nucleoplasm.
RNA polymerase is involved in the synthesising of the mRNA with the same base sequence as the coding strand (sense strand).
Complementary base pairing ensures that the order of bases on the DNA exactly determines the order on RNA.
The completed mRNA exits the nucleus through a pore in the nuclear envelope into the cytoplasm.
This is where the second stage takes place called translation
How is a molecule of mRNA made during transcription?
Translation
Translation is the process in which the mRNA is ‘read’ and translated into a protein.
Translation occurs on the ribosomes
The process involves another type of RNA molecule known as Transfer RNA or tRNA
tRNA serves as a link between the nucleotide sequence in mRNA and the free nucleotides that carry the amino acids which make up the polypeptides
It attaches and transports an amino acid to the ribosome as directed by the codon.
A codon is a three nucleotide sequence
Its anticodon is complementary to the codon
Question: Starting with mRNA, describe how the process of translation leads to the production of a polypeptide.
- mRNA attaches to ribosome;
- codon on mRNA;
- binds to an anti-codon on tRNA;
- each tRNA brings a specific amino acid;
- sequence of codons/bases on mRNA determines order of amino acids;
- formation of peptide bonds/amino acids - Joined by condensation reactions;
The triplet code
DEGENERATE:
More than one codon for most amino acids.
e.g. AUA, AUC or AUU = Isoleucine
NON-OVERLAPPING:
Each set of 3 bases (codons) are read only once.
i.e. AUA|GCU|AAU|CCG|UGG
UNIVERSAL
All organisms use the same genetic codes, DNA and RNA
Rough E.R and Golgi Body
Flattened sacs and tubules encrusted with ribosomes.
Proteins that are to be secreted by the cell are synthesised on the rough E.R.
They are then passed on to the Golgi body.
Golgi body: packages and modifies proteins
A group of flattened fluid sacs.
Vesicles containing newly synthesised proteins fuse with the golgi.
The golgi processes the protein e.g. adds a carbohydrate chain = glycoprotein.
Vesicles containing the finished product pinch off at the end of the golgi body.
The contents of the vesicle are then discharged outside = Exocytosis
Mutations
If there is a change in the sequence of nucleotides in the DNA, this is called a mutation.
A mutation is a random change in the genetic make-up of a cell.
Changes are more likely during DNA replication than any other time.
Mutations can either be base mutations eg. cystic fibrosis or full chromosome mutations eg. Downs Syndrome
Gene mutations:
A change in the sequence of bases in DNA
Substitution- A base is changed
Usually only changes one amino acid if any
Addition- A base is added (frame shift)
Every amino acid is changed from the point of mutation
Deletion- A base is removed (frame shift)
Every amino acid is changed from the point of mutationA change in the sequence of bases in DNA
Mutagens
Radiation
Radiation can break chemical bonds. This produces ions and particles that can combine with any other molecules. Radiation acts indirectly on DNA
Chemicals
Hundreds of chemicals acting in hundreds of ways. Some can interfere with base sequences and some can change the bases from one to another.
Viruses
Some viruses insert their DNA into that of the host cell e.g. Hepatitis
Application to real world - mutations
Most, not all, are harmful. Alleles have already been “tested” by natural selection. That means any random change is likely to be harmful.
EXCEPT in changing conditions.
Most mutations are recessive so will only be expressed in homozygous individuals.
They are rare and can occur in any cell at anytime
They are essential for evolution. Recombination can only explain variation within a species. For new species to arise, new alleles must be produced.
Cancer
Cancer is a special case of mutation. The cells multiply in an unstrained fashion and spread to other parts of the body.
A key event in cancer is the mutation of a gene into an oncogene.
Over 20 oncogenes have been discovered and derive from proto-oncogenes
All proto-oncogenes have a key role in cell division
Mark scheme qs and answers
Q1 - Explain how a gene mutation would cause an enzyme that doesn’t work:
The change in the base sequence of DNA
Changes the sequence of amino acids in a protein
Change in primary structure
Which changes its shape – tertiary structure (denatures the enzyme)
Active site doesn’t fit the substrate
So the enzyme doesn’t work & the reaction rate falls
Q2 - Explain why a substitution mutation is less serious than a deletion mutation (6 marks)
Degenerate code means a substitution can still code for the same amino acid, so has no effect
Particularly if it is the 3rd base of a codon
Even if the amino acid changes, only one amino acid is different in the sequence, may just cause a less functional protein
Especially if the new amino acid has a similar R group, or is not involved in important structural R group interactions
Deletion causes a shift in the reading frame
Which alters every amino acid after the point of mutation, so changes the primary structure & the specific 3D shape
Much more likely to result in a non functional protein
Inheritance (definitions)
Dominant – only one allele needed to show the phenotype
Recessive – two recessive alleles needed to show the phenotype
Homozygous – two alleles the same
Heterozygous – the two alleles are different
Phenotype – the set of observable characteristics determined by the genotype and the environment
Genotype – the alleles an individual has
Allele – a different version of the same gene, found at the same location (locus) on a chromosome